Abstract
Morphogenesis in the fungal pathogen Candida albicans is an important virulence-determining factor, as a dimorphic switch between yeast and hyphal growth forms can increase pathogenesis. We identified CaCDC5, a cell cycle regulatory polo-like kinase (PLK) in C. albicans and demonstrate that shutting off its expression induced cell cycle defects and dramatic changes in morphology. Cells lacking CaCdc5p were blocked early in nuclear division with very short spindles and unseparated chromatin. GFP-tagged CaCdc5p localized to unseparated spindle pole bodies, the spindle, and chromatin, consistent with a role in spindle elongation at an earlier point in the cell cycle than that described for the homologue Cdc5p in yeast. Strikingly, the cell cycle defects were accompanied by the formation of hyphal-like filaments under yeast growth conditions. Filament growth was determinate, as the filaments started to die after 24 h. The filaments resembled serum-induced hyphae with respect to morphology, organization of cytoplasmic microtubules, localization of nuclei, and expression of hyphal-specific components. Filament formation required CaCDC35, but not EFG1 or CPH1. Similar defects in spindle elongation and a corresponding induction of filaments occurred when yeast cells were exposed to hydroxyurea. Because CaCdc5p does not appear to act as a direct repressor of hyphal growth, the data suggest that a target of CaCdc5p function is associated with hyphal-like development. Thus, an internal, cell cycle–related cue can activate hyphal regulatory networks in Candida.
INTRODUCTION
Morphogenesis of fungal pathogens is intimately linked with virulence, supporting the need for a comprehensive understanding of the regulation of fungal cell morphology. Candida albicans, a common fungal pathogen of humans, exists in yeast, pseudohyphal and hyphal growth forms, and can enhance virulence in part by exploiting the hyphal growth mode (Lo et al., 1997). The environmentally stimulated yeast to hyphal transition in Candida is mediated by several signaling pathways (Whiteway, 2000), the most characterized of which involve MAP kinase and cAMP-based signaling. However, several factors influence hyphal formation independent of the known pathways in Candida (Whiteway, 2000; Liu, 2001), and hyphal growth involves regulation beyond the level of transcription (Torralba and Heath, 2001), supporting the existence of additional regulatory networks and reinforcing the complexity of the response systems.
In the model fungus Saccharomyces cerevisiae, morphogenesis is tightly coordinated with cell cycle progression. Bud growth and the corresponding polarization of actin, synthesis of DNA, and duplication of spindle pole bodies occurs at the G1/S transition, whereas elongated or pseudohyphal growth is associated with a block in G2 mediated by Swe1p-dependent negative regulation of Cdc28p (Kron and Gow, 1995; Rua et al., 2001). Polar or isometric growth of buds can be maintained by overexpression of G1 or mitotic cyclins, respectively (Lew and Reed, 1995). In filamentous and dimorphic fungi, the connection between cell cycle factors and true hyphal growth is less clear. Hyphae continue to grow whether their apical-most nuclei are in interphase or mitosis (Kron and Gow, 1995), and it was recently demonstrated that the duration of cell cycle stages was similar in yeast, germlings, and apical hyphal cells of C. albicans (Hazan et al., 2002). However, the coordination between nuclear localization, division, and septation with the initiation and maintenance of hyphal growth in Candida requires a relationship between aspects of the cell cycle and hyphal growth. For example, during the yeast to hyphal transition, the nucleus and septins move to a hyphal specific position out in the developing germ tube before the first mitosis, after which one daughter nucleus migrates with the growing tip, while the other moves back into the mother yeast cell (Gale et al., 2001; Sudbery, 2001). Septation follows each nuclear division in established hyphae, creating uni-nucleate subapical compartments of similar length that remain arrested in G0 or G1 until branching takes place (Kron and Gow, 1995). A few cell cycle factors have been characterized in Candida; these include the G1 cyclin CLN1, which is required for maintaining hyphal growth (Loeb et al., 1999) and the CDC2-related kinase CRK1, which can promote hyphal formation (Chen et al., 2000). In addition, a forkhead transcription factor that regulates B-cyclin gene expression is required for hyphal growth in Candida (Bensen et al., 2002).
The polo-like kinases (PLKs) comprise a family of cell cycle regulators with the potential to influence hyphal morphogenesis in Candida, because they function at various stages during the initiation and progression through mitosis and are required for septation/cytokinesis (reviewed in Nigg, 1998; Alexandru et al., 2001; Toyoshima-Morimoto et al., 2001). Furthermore, the PLK homologue Cdc5p in S. cerevisiae physically interacts with septins and Swe1p (Bartholomew et al., 2001; Song and Lee, 2001) and can alter cell morphology by generating elongated buds upon overexpression (Song et al., 2000).
To explore the relationship between cell cycle factors and hyphal morphogenesis in C. albicans, we investigated the role of a PLK homologue, CaCDC5. We demonstrate that CaCdc5p is required for spindle elongation and that gene repression under yeast growth conditions leads to dramatic hyphal-like growth. Similar inhibition of spindle elongation and a corresponding induction of filaments with hydroxyurea suggest there is an endogenous mechanism in place to connect aspects of the cell cycle and the hyphal signaling networks in C. albicans.
MATERIALS AND METHODS
Strains, Oligos, and Media
Strains and oligos used in the investigation are listed in Table 1. Cells were grown at 30°C in 0.67% yeast nitrogen base containing 2% sodium succinate (SS medium) or 2% glucose (SD medium) for induction and repression of the Candida PCK1 promoter, respectively (Leuker et al., 1997). Control strains including RM1000 and CB102 were grown in the same media supplemented with uridine and histidine as required. To repress CaCDC5 expression from the PCK1 promotor, yeast cells were grown for 24 h in SS medium, washed with dH2O, and then diluted into SD medium to an OD600 of 0.25. SD medium with or without methionine and cysteine was used to repress and induce expression, respectively, from the Candida MET promotor (Care et al., 1999). Hyphal-inducing conditions involved the addition of 10% heat-inactivated fetal calf serum (GIBCO/Invitrogen, Burlington, Ontario) to growth medium and incubation at 37°C. For hydroxyurea (HU) treatment, cells of strain SC5314, CB110, CR216, JKC19, HLC52, and HLC54 were grown overnight in yeast extract/peptone/dextrose medium (YPD) or SD medium and then diluted to an OD600 of 0.4 in fresh medium containing 200 mM HU (Sigma Chemical Co., St. Louis, MO). Overexpression of CaCdc5p-GFP was performed in S medium containing 2% casaminoacids (Leuker et al., 1997).
Table 1.
Candida albicans strains and oligos used in this study
| Genotype | Source | |
|---|---|---|
| Strain | ||
| SC5314 | CaCDC5/CaCDC5 URA3/URA3 HIS1/HIS1 | Fonzi and Irwin (1993) |
| CA14 | ura3Δ::imm434/ura3Δ::1 imm434 | Fonzi and Irwin, 1993 |
| RM1000 | ura3Δ::imm434/ura3Δ::1 imm434 his1Δ::hisG/his1Δ::hisG | Negredo et al. (1997) |
| JCK19 | cph1Δ::hisG/cph1Δ::hisG ura3Δ::imm434/ura3Δ::1 imm424 | Lo et al., (1997) |
| HLC52 | efg1Δ::hisG/efg1Δ::hisG URA3-hisGΔ::imm434/ura3Δ::1 imm434 | Lo et al., 1997 |
| HLC54 | HLC52 cph1Δ::hisG/cph1Δ::hisG | Lo et al., 1997 |
| HLC69 | cph1Δ::hisG/cph1Δ::hisG efg1Δ::hisG/efg1Δ::hisG ura3Δ:: imm434/ura3Δ::1 imm434 | Lo et al., 1997 |
| CR216 | CAI4 cdc35Δ::hisG-URA3-hisG/cdc35Δ::hisG | Rocha et al., 2001 |
| CR276 | cdc35Δ::hisG/cdc35Δ::hisG | Rocha et al., 2001 |
| CB102 | cacdc5Δ::hisG/CaCDC5 PCK1::CaCDC5-URA3 | This study |
| CB104 | cacdc5Δ::hisG/cacdc5Δ::HIS1 PCK1::CaCDC5-URA3 | This study |
| CB105 | cacdc5Δ::hisG/cacdc5Δ::HIS1 PCK1::CaCDC5-hisG | This study |
| CB108 | CAI4 cacdc5Δ::hisG/MET::CaCDC5-URA3 | This study |
| CB109 | cacdc5Δ::hisG/CaCDC5 MET::(URA3) | This study |
| CB110 | RM1000 (TUB1-GFP-URA3) | This study |
| CB112 | CB105 (TUB1-GFP-URA3) | This study |
| CB113 | CB105 (CDC12-GFP-URA3) | This study |
| CB115 | RM1000 (PCK1::CaCDC5-GFP-URA3) | This study |
| CB116 | RM1000 (CaCDC5-GFP-URA3) | This study |
| CB303 | CB108 cdc35Δ::hisG/cdc35Δ::hisG | This study |
| CB305 | CB108 cph1Δ::hisG/cph1Δ::hisG efg1Δ::hisG/efg1Δ::hisG | This study |
| CB400 | RM1000 (pRM100 URA3+, HIS1+) | This study |
| CB401 | CB102 (pCB102 HIS1+) | This study |
| Oligos | ||
| CB1F (KpnI) | TCGAGCAGGACCAATTGC | |
| CB1R (PstI) | CTGTGGTGGGTGAAGCGA | |
| CB3F (BamHI) | CGAAGCGCCGACATATCA | |
| CB3R (BamHI) | CATGAAAATGTTCCGG | |
| CB4F (Not1) | CGAAGCGCCGACATATCA | |
| CB4R (BglII) | CATGAAAATGTTCCGG | |
| CB5F (KpnI) | TGATATGTCGGCGCTTCG | |
| CB5R (KpnI) | GCTTTGCAAAATGCATGTTCC | |
| CB6F (PstI) | TGATATGTCGGCGCTTCG | |
| CB6R (BamHI) | GGAAATTCACTGGCATCAAC | |
| CB50F (BglII) | CCAAAGGGAGGAGAAGAA | |
| CB50R (SacI) | ACAAATAAATAAATCGCTCGG | |
| CB51F (SpeI) | GTAGTATGGTTGATCGTGTTC | |
| CB51R (XhoI) | ATATTCTTCTTCTTCTTCAGGGAAAGAATCAGT | |
| CB43F (SpeI) | AGACAACGCTACTTGATATTT | |
| CB43R (XhoI) | AGCTTCTTTAAATGCTTTTTTC | |
| CB44F (XhoI) | ATGAGTAAAGGAGAAGAACTTTTC | |
| CB44R (SpeI) | TTATTTGTATAGTTCATCCATGCC |
Transformation and Southern and Northern Analyses
Cells were transformed using lithium acetate (Chen et al., 1992), and DNA and RNA were extracted according to Rose et al. (1990) and Köhrer and Domdey (1991), respectively. Southern analysis was performed using the DIG Hybridization System (Roche Diagnostics, Mannheim, Germany). To determine the level of PCK1::CaCDC5 expression, cells from strains RM1000, CB102, and CB104 were grown to an OD600 of 0.8 in SS media at 30°C or to an OD600 of 0.2 followed by washing and transferring to SD media. Strain CB104 was collected after 4, 7, or 24 h in SD medium, whereas strains RM1000 and CB102 were collected at 4 h, because these dividing yeast cells entered stationary phase with longer incubation periods. Total RNA, 20 μg, was analyzed using a 32P-labeled (T7 Quick Prime Kit, Amersham Pharmacia Biotech, Piscataway, NJ) PCR product of CaCDC5 and hybridization conditions described by Srikantha and Soll (1993). 32P-labeled PCR fragments from HWP1, DDR48, ECE1, and ALS1 were used to probe total RNA from strains CB102, CB104, SC5314 grown in SD medium, and SC5314 grown in SD medium plus 200 mM HU. An ACT1 probe (Rocha et al., 2001) was used as a loading control. Northern blots were visualized with a phosphoimager.
Cloning and Plasmid and Strain Production
To regulate expression of CaCDC5, a strain containing a single copy of CaCDC5 under control of the PCK1 promotor was created. Sequence data for C. albicans was obtained from the Stanford Genome Technology Center website at http://www-sequence.stanford.edu/group/candida. Sequencing of C. albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund. The open reading frame plus 800 and 1500 base pairs of 5′ and 3′ flanking sequence, respectively, of CaCDC5 was PCR amplified from genomic DNA derived from strain SC5314 using primers CB1F and CB1R and cloned into the KpnI/PstI sites of vector pTZ18R, creating plasmid pCB100. Primers CB3F and CB3R reverse-amplified the flanking and vector sequences from pCB100, into which the BamHI/BglII URA3 blaster cassette (Fonzi and Irwin, 1993) was cloned, creating plasmid pCB101. Primers CB4F and CB4R were also used to amplify the flanking DNA and vector sequence from pCB100, into which a NotI/BamHI PCR-amplified CaHIS1 ORF was cloned, creating plasmid pCB102. The URA3-containing deletion construct was liberated from pCB101 with KpnI and PstI and transformed into strain RM1000. Transformants were screened by PCR and Southern analyses to confirm the complete elimination of one copy of CaCDC5 and creation of strain CB100. CB100 was plated on 5-fluoorotic acid (5-FOA) to select for the URA3-strain CB101. The ORF of CaCDC5 plus 300 base pairs of terminator sequence was PCR amplified from genomic DNA using primers CB5F and CB5R and cloned into the KpnI site following the PCK1 promotor in the URA3 blaster-containing plasmid pJA24 (a kind gift from J. Ash, derived from plasmid p5921 from Fonzi and Irwin, 1993), creating plasmid pCB103. pCB103 was cut at a unique XhoI site within the PCK1 promotor sequence to direct integration at the PCK1 promotor locus in strain CB101, creating strain CB102. The empty plasmid was transformed as a control, creating control strain CB103. The HIS1-containing deletion construct was liberated from pCB102 with SphI and SacI and transformed into strain CB102 to replace the second endogenous copy of CaCDC5 while in the presence of succinate, creating strain CB104. Removal of the URA3 marker by plating onto 5-FOA resulted in strain CB105. All strains were analyzed by PCR and Southern analyses to confirm correct integration of transforming DNA and replacement of both endogenous copies of CaCDC5 (our unpublished results), and all PCR-derived clones were sequenced. For additional controls, strain RM1000 was transformed with plasmid pRM100, which contains Candida URA3 and HIS1, producing strain CB400. Strain CB401 consisted of one of several transformants from strain CB102 which did not correctly integrate the HIS1 knockout cassette, remaining heterozygous for CaCDC5 but HIS1+. These transformants behaved identically to strains RM1000 and CB102 (supplemented with uridine and histidine) when switching between glucose and succinate-containing medium (our unpublished results).
To regulate expression of CaCDC5 in another strain using a different promotor, strain CAI4 was transformed with the URA3-based deletion construct from plasmid pCB101, to create strain CB106. CB107 was created by plating CB106 on 5-FOA. A 3′-truncated copy of CaCDC5, lacking 1000 base pairs upstream from the stop codon, was PCR amplified with primers CB6F and CB6R and cloned into the PstI/BamHI sites following the MET promotor in plasmid pCa-DIS (Care et al., 1999), creating plasmid pCB106. The plasmid was cut with ClaI to direct integration at the remaining endogenous copy of CaCDC5 in strain CB107. The resulting strain, CB108, contained a full-length copy of CaCDC5 under control of the MET promotor and a 3′ truncated copy with no terminal processing sequence. As a control, strain CB107 was transformed with empty pCaDIS plasmid, creating strain CB109.
To regulate expression of CaCDC5 in cacdc35Δ/cacdc35Δ and efg1Δ/efg1Δ, cph1Δ/cph1Δ mutant backgrounds, strains CR276 and HLC69, respectively, were transformed with the URA3-containing knockout cassette liberated from plasmid pCB101. Colonies heterozygous for CaCDC5 were grown on 5-FOA and then transformed with ClaI-cut plasmid pCB106 to allow integration at the remaining endogenous copy of CaCDC5, resulting in strains CB303 and CB305. Analysis by Southern and Northern analyses confirmed the correct integration of constructs and proper regulation of the remaining copy of CaCDC5 under control of the MET promotor (our unpublished results).
A GFP-tagged α-tubulin strain was created by PCR-amplifying a 5′-truncated copy of CaTUB1, containing 1000 base pairs upstream from the STOP codon and 300 base pairs of terminator sequence, from genomic DNA isolated from strain SC5314 with primers CB50F and CB50R. The PCR fragment was cloned into the BglII/SacI sites of plasmid p5921 (Fonzi and Irwin, 1993), creating plasmid pCB104. Primers CB51F and CB51R were designed to bind immediately up and downstream of the STOP codon, respectively, and to reverse-amplify the plasmid sequence, into which a XhoI/SpeI PCR fragment (produced with primers CB44F and CB44R) of GFP (Morschhauser et al., 1998) was cloned, creating plasmid pCB105. pCB105 was digested with BstEII for directed integration at the endogenous TUB1 locus, creating full-length 3′-tagged TUB1-GFP, under control of its own promotor, and a 5′-truncated copy. pCB105 was integrated into strains RM1000, CB101, and CB105 to create strains CB110, CB111, and CB112, respectively.
CaCdc5p was tagged with GFP according to the same protocol. A 5′-truncated BamHI/PstI fragment of CaCDC5, containing 1000 base pairs upstream of the STOP codon and 300 base pairs of terminator sequence, was cut from plasmid pCB100 and cloned into plasmid p5921, creating plasmid pCB107. Primers CB43F and CB43R annealed immediately upstream and downstream of the STOP codon of 5′-truncated CaCDC5 in plasmid pCB107 and reverse amplified the plasmid, into which the XhoI/SpeI-containing GFP PCR fragment was cloned, creating plasmid pCB108. pCB108 was cut at BstEII for site-directed integration at the endogenous CaCDC5 locus in strain RM1000, creating a 3′-tagged copy of CaCDC5-GFP and a 5′-truncated copy in strain CB114. PCK1::CaCDC5-GFP was constructed in a similar way, using primers CB43F and CB43R to reverse amplify the gene plus vector sequence from plasmid pCB103, into which the XhoI/SpeI-containing GFP PCR fragment was cloned, creating plasmid pCB109. pCB109 was integrated at the PCK1 promotor in strain RM1000, creating strain CB115. CDC12-GFP in plasmid pVEC, a kind gift from Dr. Ursula Oberholzer, was transformed into strain CB105 to create strain CB113.
Cell Staining and Microscopy
Nuclei and septa were visualized by fixing cells in 70% ethanol for 1 h, followed by incubation in 1 μg/ml 4′,6′diamidino-2-phenylindole dihydrochloride (DAPI, Sigma) for 20 min and 1 μg/ml calcofluor white (Sigma) for 10 min. Immunofluorescence was performed by fixing cells in an equal volume of double-strength fixative solution, containing 8% paraformaldehyde (Sigma) in 80 mM PIPES buffer, pH 6.8, 5% DMSO (Sigma), 10 μg/ml leupeptin (Sigma), 4 mM AEBSF (Roche Diagnostics), and 1 μM aprotinin (Roche Diagnostics), for 30 min, followed by washing with 1× PIPES buffer, pH 6.8 (40 mM). For immunolocalization of α-tubulin, cell walls were then digested for 10 min at 37°C with 10 μg/ml zymolase (ICN Biomedicals, Aurora, OH) in 40 mM PIPES buffer, pH 6.8, 5% BSA (Sigma), and protease inhibitors described above. Membranes were permeabilized with 0.1% Nonidet P-40 (BDH, Poole, England) in 40 mM PIPES, pH 6.8, for 5 min. Immunolocalization of 16B1-F10 (Marot-Leblond et al., 2000) omitted these two steps. Cells were incubated in 1/100 dilution of monoclonal anti–α tubulin clone B-5–1-2 (Sigma), or 1/500 dilution of Mab 16B-F10, in 1× PIPES buffer, 0.05% sodium azide (Sigma), and 5% BSA overnight. After washing in PIPES buffer, cells were incubated in 1/100 dilution of donkey anti-mouse FITC-coupled secondary antibody (Sigma) for 2 h, washed, and stained with DAPI and calcofluor white as described above. Nonspecific binding of the secondary antibody was investigated by preparing a paired sample without primary antibody. A signal was not detected (our unpublished results). To determine cell viability, unfixed cells were stained with 10 μg/ml propidium iodide (Sigma) and immediately visualized.
Cells were examined on a Leitz Aristoplan microscope using 10×, 40×, or 100× (1.32 NA) objectives with Nomarski differential interference contrast (DIC) or epifluorescence optics and on a Leica DMIRE2 inverted microscope using a 100× (1.32 NA) objective with phase contrast and fluorescence optics, using the appropriate filter sets.
Flow Cytometry
Cells were prepared for FACS analysis according to Lew et al. (1992), with some modifications. Cells (5 × 106–1 × 107 cells/ml) were fixed overnight in 70% ethanol, washed with 0.2 M Tris buffer, pH 7.5, treated with 0.8 mg/ml RNaseA (Pharmacia, Piscataway, NJ) for 2 h at 37°C, washed, and incubated in 50 μg/ml propidium iodide (Sigma) overnight. Cells were analyzed with a Becton-Dickinson FAC-Scan. Results are based on 10,000–20,000 nongated events.
RESULTS
C. albicans Contains a Polo-like Kinase, CaCDC5
A single gene resembling a polo-like kinase was identified in the Stanford C. albicans sequencing data base (http://sequence-www.stanford.edu/group/candida), and cloned. CaCDC5 is 50% identical to CDC5 from S. cerevisiae and contains the conserved domains of PLKs, including the carboxy-terminal polo box, and the amino terminal catalytic domain. CaCDC5 contains the GeGGFArC motif in subdomain I of the catalytic domain, comparable to other PLKs. However, in place of the conserved ExxT motif between subdomain VII and VIII of the catalytic region in other PLKs, CaCDC5 contains an SxxT sequence, resembling kinases like MEK. CaCDC5 contains potential destruction box sequences in the amino-terminus (RSQPLQPLN and KEKLSALCK), similar to those in CDC5 (RSKLVHTPI, REKLSALCK) of S. cerevisiae (Shirayama et al., 1998).
Repression of CaCDC5 Induces Yeast Cells to Grow into Filaments
A strain containing a single copy of CaCDC5 under control of the regulatable PCK1 promotor (Leuker et al., 1997) was created to manipulate CaCDC5 expression (see MATERIALS AND METHODS). Northern analysis demonstrated that CaCDC5 was overexpressed in SS-inducing medium and repressed in SD repressing medium (Figure 1). To confirm that the effects of repressing CaCDC5 were not due to changes in carbon source, the heterozygote strain CB102 and parental strain RM1000 were subjected to identical changes in medium. In addition, a second strain containing a single copy of CaCDC5 under control of the CaMET promotor (Care et al., 1999) in strain CAI4 was created for comparison of phenotype.
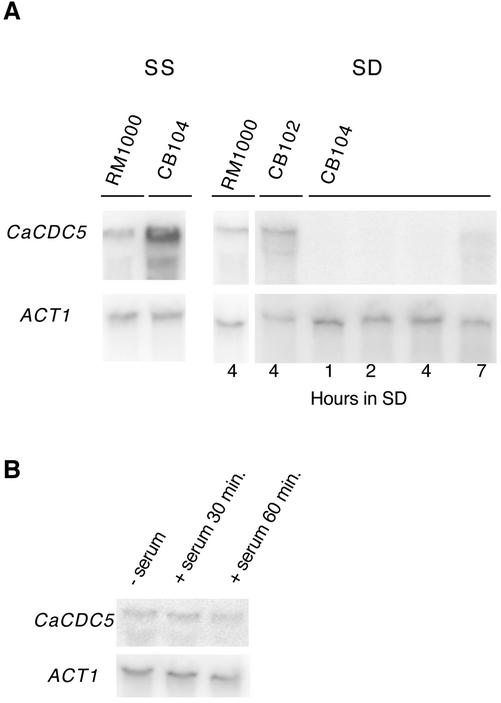
Figure 1.
(A) Northern analysis of CaCDC5 expression under control of the Candida PCK1 promotor. Strains CB104 (cacdc5Δ::hisG/cacdc5Δ:: HIS1/PCK1::CaCDC5), CB102 (cacdc5Δ::hisG/CaCDC5 PCK1::CaCDC5) and RM1000 (CaCDC5/CaCDC5) were grown in SD medium for 1, 2, 4, or 7 h, or in SS medium. Total RNA, 20 μg, was analyzed with a CaCDC5-specific probe, followed by an ACT1 probe to compare RNA loading. CaCDC5 is overexpressed in SS medium and repressed in SD medium at 1–7 h of incubation, although some leakiness is present at 7 h. (B) Northern analysis of CaCDC5 expression in cells grown in YPD at 30°C without serum or at 37°C with serum for 30 and 60 min.
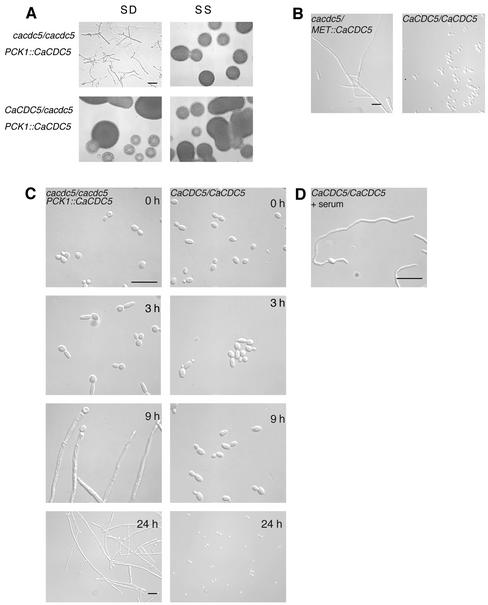
Under yeast growth conditions at 30°C, overexpressing CaCDC5 in the PCK1-regulated strain (CB104) did not result in any gross changes in phenotype, because the morphologies of yeast cells and colonies were normal (Figure 2A). In contrast, shutting off CaCDC5 expression by streaking onto solid SD medium induced dramatic changes in morphology and proliferation. Yeast cells changed shape into filaments, which became highly elongate but did not branch out to form mycelial colonies. Repressing CaCDC5 expression in liquid medium at 30°C also induced the formation of filaments. By 3 h, the majority of cells contained an elongated extension, which continued to grow in a polarized manner, creating hyphal-like filaments (Figure 2B; Table 2). The filaments were initially wider in diameter (Figure 2, B and D) and grew at a mean rate of one third of that of serum-induced hyphae incubated at 37°C (0.13 ± 0.01 μm/min, n = 45, vs. 0.41 ± 0.01 μm/min, n = 32), although the presence of some longer filaments indicated an ability to grow at rates approaching that of serum-induced hyphae. Different environmental conditions and activation mechanisms could account for such differences in growth characteristics. In contrast to strain CB104, the heterozygote strain (our unpublished results) and parental strain (Figure 2B) grew as yeast under repressing conditions. The cells demonstrated a transient pseudohyphal growth stage upon switching from SS to SD medium but resumed normal yeast growth and morphology by 7 h (Figure 2B). The heterozygote strain appeared more pseudohyphal than the parental strain and contained some elongated cells, suggesting a gene-dosage effect. Repressing CaCDC5 with the MET promotor also produced filaments (Figure 2D), supporting that the phenotype is due to manipulation of CaCDC5.
Figure 2.
Repressing CaCDC5 expression results in the formation of filaments under conditions favoring yeast growth. (A) Strains CB104 (cacdc5Δ::hisG/cacdc5Δ:: HIS1/PCK1::CaCDC5) and CB102 (cacdc5Δ::hisG/CaCDC5 PCK1:: CaCDC5) were incubated for 24 h on solid SD or SS medium. Bar, 30 μm. (B) Time course analysis of the formation of filaments in liquid SD medium. Strains CB104 and RM1000 were grown in SS medium (0 h), washed, diluted into SD medium, and fixed in 70% ethanol after 3, 9, and 24 h of incubation at 30°C. Bars, 10 μm. (C) Filament formation upon repressing CaCDC5 with the MET promotor. Strains CB108 (cacdc5Δ::hisG/MET:: CaCDC5) and CB109 (cacdc5Δ::hisG/MET::) were grown in SD medium lacking methionine for promotor induction, then diluted to an OD600 of 0.25 in SD medium containing methionine and cysteine to repress the promotor for 24 h. Bar, 10 μm. (D) Hyphal growth in strain RM1000 incubated in the presence of 10% serum at 37°C for 4 h. Bar, 10 μm.
Table 2.
Proportion of cells (%) in a single, budded, or elongated morphology upon incubation in SD medium
| Single
|
Buddeda
|
Elongatedb
|
||||
|---|---|---|---|---|---|---|
| Hours | CB104 | RM1000 | CB104 | RM1000 | CB104 | RM1000 |
| 0 | 88.4 | 99.5 | 11.6 | 0.5 | 0 | 0 |
| 3 | 2.0 | 4.4 | 26.0 | 83.2 | 72.0 | 10.6 |
| 5 | 0 | 7.9 | 0 | 91.1 | 100 | 0 |
| 8 | 0 | 47.0 | 2.5 | 53.0 | 97.5 | 0 |
Cells from strains CB104 (cacdc5Δ::hisG/cacdc5Δ::HIS1/PCK1p::CaCDC5) and strain RM1000 (CaCDC5/CaCDC5) were grown at 30°C in SD medium and fixed after 3, 5, and 8 h. More than 200 cells were counted in each time point.
Budded cells include those cells containing a bud equal to or smaller than the mother yeast cell in size and budded pseudohyphal cells.
Elongated cells contain an elongated secondary extension with a length greater than the diameter of the mother yeast cell or equal to the mother cell but more narrow.
The relative absence of branching and mycelial colony formation suggested that filament growth was determinate. In support of this, transferring filaments from repressing medium back to inducing medium after 24 h allowed some reversion to yeast growth, but many cells remained trapped in a filamentous state. Several CaCDC5-repressed filaments became highly vacuolated and approximately half of the cells stained with propidium iodide after 24 h of repression (our unpublished results), indicating that the cells were dying. Therefore yeast cells switch to a new active growth mode upon repression of CaCDC5, but the cells loose viability at a later time, suggesting that either CaCDC5 is essential, or maintenance of the growth mode and the signals that generate it eventually become toxic to the cells.
CaCdc5p Is Not a Direct Negative Regulator of Hyphal Growth like Tup1p or Nrg1p
Because the depletion of CaCdc5p results in the production of hyphal-like filaments, the possibility that CaCdc5p is a direct negative regulator of hyphal growth comparable to Nrg1p or Tup1p (Braun and Johnson, 1997; Braun et al., 2001; Munir et al., 2001) was investigated by analyzing CaCDC5 mRNA levels in yeast and serum-induced hyphae and determining whether overexpression of CaCdc5p could inhibit serum-induced hyphal growth. In contrast to that observed with Nrg1p or Tup1p, CaCDC5 mRNA expression did not change between yeast and hyphal cells (Figure 1B), and serum-induced hyphal growth was not inhibited by CaCdc5p overexpression in strain CB115 (our unpublished results). Therefore CaCdc5p does not appear to act like general repressors of serum-induced hyphal growth, suggesting that an aspect of CaCdc5p function is involved in generating and/or transmitting a signal to activate the hyphal-like growth mode.
Filament Initiation upon Repression of CaCDC5 Is Associated with an Early Block in Nuclear Division
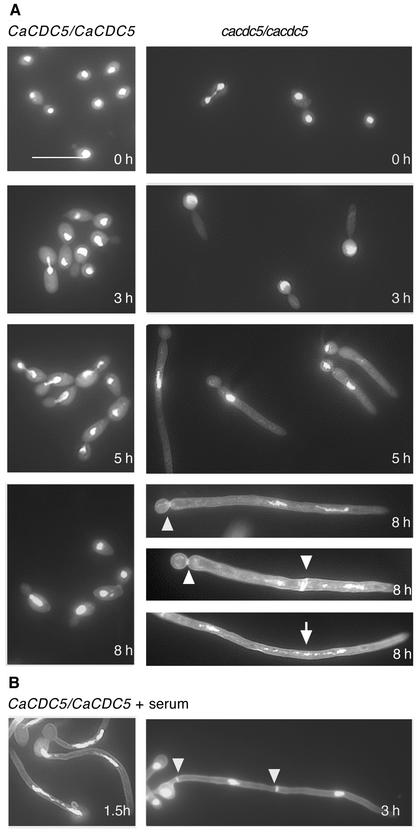
Because CaCdc5p did not appear to be a direct negative regulator of hyphal growth, the filaments could be forming in response to a cell cycle defect induced by repression of CaCDC5. PLKs are essential for nuclear division, as compromised PLK function results in a block at G2/M in most organisms (Llamazares et al., 1991; Okhura et al., 1995; Lane and Nigg, 1996), or at anaphase B in S. cerevisiae (Kitada et al., 1993). To determine whether defects in nuclear division accompanied filament formation upon depletion of CaCdc5p, cells incubated in SD medium were fixed at several time points and stained with DAPI. At 3 h in repressing medium, control yeast cells were budding, dividing, and undergoing mitosis (Figures 2 and 3). In contrast, nuclei in the majority of CaCDC5-repressed cells did not divide during the formation of filaments up to 5 h, after which nuclei in ∼45% of the cell population escaped the division block (Table 3; Figure 3). The ability of nuclei to divide could reflect leakiness in the regulated expression of CaCDC5. At 8 h, when the control yeast strains CB102 (our unpublished results) and RM1000 had undergone four or five rounds of nuclear division, 65% of CaCDC5 repressed cells contained two nuclei, whereas 28% of the cells contained fragmented DNA (Table 3; Figure 3). Only 3.5% of the cells contained three nuclei. Fragmentation of DNA progressed up to 24 h of repression, preventing an accurate quantification of nuclei at the later time point. The filaments were 28.9 ± 1.5 μm (SEM; n = 45) in length at 5 h of repression, when approximately half of the cells had escaped the block in nuclear division, and 62.1 ± 2.6 μm (SEM; n = 45) after 8 h, when the majority of cells contained two nuclei. In contrast, the nucleus in serum-induced hyphae at 37°C divided approximately every 1.5 h, corresponding to hyphal lengths of 37.3 ± 1.1 μm (SEM; n = 32) and 92.8 ± 2.4 μm (SEM; n = 32) for the first and second rounds of division, respectively (Figure 2B; Table 3). The lack of similar coordination between filament length and nuclear division in CaCDC5-repressed cells suggests that the block in nuclear division is due to repression of CaCDC5 and not the initiation of a hyphal-like growth mode. Although the nucleus eventually escapes the block in division, normal rates of mitosis are not resumed and DNA fragmentation occurs over time while filamentous growth continues. These results suggest CaCDC5 is required for the early stages of nuclear division and chromatin separation, similar to its counterparts in Schizosaccharomyces pombe and higher organisms, but in contrast to that in S. cerevisiae.
Figure 3.
(A) Nuclear division and septation are impaired upon repressing CaCDC5. Strains CB104 (cacdc5Δ::hisG/cacdc5Δ::HIS1/PCK1:: CaCDC5) and RM1000 (CaCDC5/CaCDC5) were grown in SS medium, washed, transferred to SD medium, and then fixed and stained with DAPI and calcofluor at various time points. Septa (small arrowheads) and fragmented DNA (arrow) were observed after 5 h. Note the absence of nuclei in the mother yeast cell after 5 h. (B) Nuclei and septa in serum-induced hyphae grown at 37°C from strain RM1000 after 1.5 and 3 h. Bar, 10 μm.
Table 3.
Proportion of cells demonstrating nuclear division and positioning upon incubation in SD medium or serum
| No. of nuclei
|
Position of nuclei
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Strain | 1 | 2 | 3 | >3 | Fraga | Mother | Mother and tube | Tube |
| Hours in SD | ||||||||
| CB104 | ||||||||
| 0 (n = 242) | 100 | 0 | 0 | 0 | 0 | 100 | 0 | 0 |
| 3 (n = 212) | 92.0 | 8.0 | 0 | 0 | 0 | 68.0 | 8.0 | 24.0 |
| 5 (n = 319) | 55.0 | 45.0 | 0 | 0 | 0 | 1.0 | 36.7 | 62.4 |
| 8 (n = 197) | 2.5 | 65.5 | 3.6 | 0 | 28.4 | 0 | 1.5 | 70.0 |
| RM1000b | ||||||||
| 0 (n = 200) | 100 | 0 | 0 | 0 | 0 | — | — | — |
| 3 (n = 236) | 88.6 | 11.4 | 0 | 0 | 0 | — | — | — |
| 5 (n = 238) | 89.5 | 10.5 | 0 | 0 | 0 | — | — | — |
| 8 (n = 225) | 86.2 | 13.8 | 0 | 0 | 0 | — | — | — |
| Hours in serum at 37°C | ||||||||
| RM1000 | ||||||||
| 0 (n = 100) | 100 | 0 | 0 | 0 | 0 | 100 | 0 | 0 |
| 1.5 (n = 106) | 52.0 | 48.0 | 0 | 0 | 0 | 33.3 | 34.5 | 32.2 |
| 3.0 (n = 59) | 0 | 20 | 58 | 22.0 | 0 | 0 | 100 | 0 |
Values are expressed as percentage. Cells from strain CB104 or RM1000 were incubated in SD medium at 30°C for the indicated times, fixed, and stained with DAPI. Cells from strain RM1000 were incubated in SD medium plus 10% serum at 37°C for the indicated time points and fixed and stained with DAPI.
Fragmentation of nuclei prevented an accurate quantification of the total number of nuclei per cell after 8 h of repression.
Cells containing 2 nuclei were doublets.
Staining the CaCDC5-repressed cells with calcofluor white demonstrated that septa or chitin deposition did not occur until later time points, consistent with when nuclei started to escape the block in division (Figure 3; Table 4). Taken together with the inhibition of chromatin separation, this observation suggests that CaCDC5-repressed cells were blocked at an early stage in the cell cycle, preceding septation, and that filament formation correlated with this early block.
Table 4.
Proportion of CaCDC5-repressed cells and serum-induced hyphae containing septa
| No. of septa
|
Positioning of septaa
|
|||||
|---|---|---|---|---|---|---|
| Strain | 0 | 1 | 2 | Neck | Neck and tube | Tube |
| Hours in SD | ||||||
| CB104 | ||||||
| 0 (n = 242) | 100 | 0 | 0 | 0 | 0 | 0 |
| 3 (n = 212) | 98.1 | 1.9 | 0 | 100 | 0 | 0 |
| 5 (n = 227) | 86.3 | 10.6 | 3.1 | 77.4 | 22.6 | 0 |
| 8 (n = 217) | 40.3 | 50.0 | 9.7 | 83.8 | 16.2 | 0 |
| Hours in serum 37°C | ||||||
| RM1000 | ||||||
| 0 (n = 100) | 100 | 0 | 0 | 0 | 0 | 0 |
| 1.5 (n = 106) | 98.9 | 1.2 | 0 | 0 | 0 | 100 |
| 3.0 (n = 59) | 19.0 | 47.0 | 34.0 | 17.6 | 41.2 | 40.7 |
Values are expressed as percentage. Cells from strain CB104 and RM1000 were incubated in SD medium at 30°C and in SD medium plus 10% serum at 37°C, respectively, for the indicated times, fixed, and stained with calcofluor.
Represents % of total number of cells containing septa.
CaCdc5p Is Required for Spindle Elongation
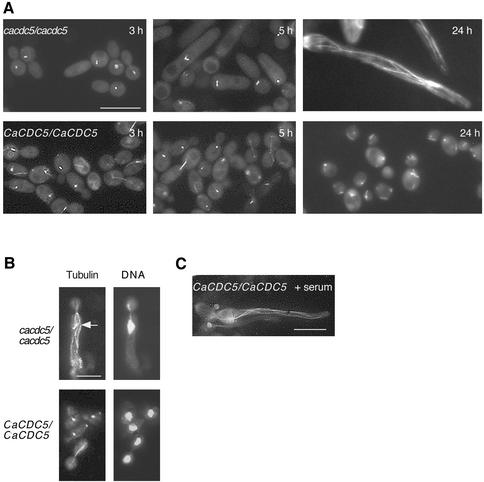
Tub1p-GFP spindle patterns were analyzed to determine the specific stage in nuclear division at which CaCdc5p is required. Tub1p was tagged with GFP in strains RM1000, CB102, and CB104, and the resulting strains (CB110, CB111, and CB112, respectively) responded to repressing medium in a manner similar to the nontagged strains. The GFP-tagged CaCDC5-regulated strain (CB112), however, was somewhat more sensitive to the absence of CaCdc5p than strain CB104, as the filaments were shorter in length at the various time points. The majority of cells from strain CB112 depleted of CaCdc5p for 3 h were elongated or large doublets (Figure 4A; Table 5) and contained spindles in the form of distinct spots or two spots side by side, corresponding to unseparated spindle pole bodies or very short spindles in S-G2 phases of the cell cycle (Barton and Gull, 1988; Hazan et al., 2002). Despite the elongated and large doublet morphology of cells at 3 h, only 2.2% of the population contained an extended mitotic spindle. After 4–7 h, the majority of filamentous cells still contained spot or short bar-like spindles, but an increasing proportion of cells contained a slightly longer bar-like spindle, probably representative of G2/M, and 6% of the filaments contained an extended mitotic spindle (Figure 4A; Table 5). Spindle orientation was also disturbed in several cells, regardless of whether the nucleus was in the mother yeast cell or in the filament. Filaments that survived after 24 h of CaCDC5 repression contained extensive cytoplasmic microtubule arrays resembling those in serum-induced hyphae of Candida (Figure 4C) and hyphae of other filamentous fungi (Han et al., 2001; Hazan et al., 2002), although the intensity of the Tub1p-GFP signal was greater in CaCdc5p-depleted cells at 30°C compared with serum-induced hyphae at 37°C. The remaining dead cells either did not stain or contained a diffuse signal. Similar patterns of spindles and cytoplasmic microtubules were observed using immunofluorescence with anti–α tubulin antibody in the nontagged strains incubated in repressing medium for 3 h (Figure 4B) or 24 h (our unpublished results), respectively, indicating that the microtubule patterns were not artifacts of GFP-tagged tubulin. Immunofluorescence demonstrated that cytoplasmic microtubules similar to those in established hyphae were present in the CaCdc5p-depleted filaments as early as 3 h of repression (Figure 4B). Their relative absence in the GFP-tagged strain at early time points is likely a reflection of the amplified signal with immunofluorescence. In contrast, control yeast cells of strain CB110 grown at 30°C were cycling at the different time points and demonstrated the expected spindle patterns (Figure 4, A and B; Table 5). For example, of the proportion of cells demonstrating a large budded morphology, the majority contained a mitotic spindle (Table 5). Slightly elongated cells were present at early time points but were pseudohyphal intermediates since they eventually budded. When the control strain CB110 was grown in serum at 37°C for 2–3 h to induce hyphal growth, nuclei in the apical regions of hyphae were cycling, with 8% containing mitotic spindles (Table 5). The decrease in the proportion of CaCdc5p-depleted cells containing spot-like spindles and the corresponding increase in cells containing slightly longer bar spindles at a later time point (28% at 7 h compared with 13% in serum-induced hyphae) supports the notion that the majority of CaCDC5-repressed cells were inhibited at an early stage in spindle elongation but eventually began to leak through the block.
Figure 4.
Spindle elongation is blocked in CaCDC5-repressed filaments. (A) Tub1p-GFP was visualized in strains CB110 (CaCDC5/CaCDC5, TUB1-GFP) and CB112 (cacdc5::hisGΔ/cacdc5Δ:: HIS1/PCK1::CaCDC5, TUB1-GFP) grown in SD medium for 3, 5, and 24 h. Bar, 10 μm. (B) Paired images of immunolocalized α-tubulin and DAPI-stained DNA in strains CB104 (cacdc5Δ::hisG/cacdc5Δ:: HIS1/PCK1::CaCDC5) and RM1000 (CaCDC5/CaCDC5) grown in SD medium for 3 h. Note the cytoplasmic microtubules in the filaments. The short spindle in strain CB104 is indicated by an arrow. Bar, 5 μm. (C) Microtubule organization in serum-induced hyphae. Cells of strain CB110 (CaCDC5/CaCDC5, TUB1-GFP) were incubated in SD medium containing 10% fetal calf serum at 37°C for 2 h to induce hyphal formation and visualize microtubules. Bar, 10 μm.
Table 5.
Spindle elongation and cell morphology in cells depleted of CaCdc5p, exposed to HU or treated with serum
| Spindle elongation | Cell Morphology | ||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| Strain | Hours in SD | S/early G2a | G2/Mb | Mc | Single or small bud | Large Bud | Elongated |
| CB112 | 3 (n=226) | 92.0 | 5.8 | 2.2 | 31.0 | 11.0 | 58.0 |
| 4 (n=267) | 84.0 | 9.7 | 6.3 | 0 | 17.0 | 83.0 | |
| 7 (n=186) | 66.0 | 28.0 | 5.9 | 0 | 0 | 100 | |
| CB110 | |||||||
| 3 (n=236) | 87.0 | 1.8 | 11.2 | 63.0 | 24.0 | 13 | |
| 4 (n=180) | 85.0 | 4.6 | 10.4 | 76.0 | 18.9 | 5.6 | |
| 7 (n=190) | 89.5 | 1.0 | 9.5 | 87.8 | 12.2 | 0 | |
| Hours in HU | |||||||
| CB110 | 3 (n=274) | 97.7 | 2.3 | 0 | 0 | 5.4 | 94.6 |
| 7 (n=397) | 81.0 | 18 | 1.0 | 0 | 0 | 100 | |
| Hours in serum at 37°C | |||||||
| CB110 | 3 (n= 75) | 81.3 | 10.7 | 8.0 | 0 | 0 | 100 |
Values are expressed as percentages.
Unseparated spindle pole bodies and early spindles represented by a spot or two spots side by side.
Short bar spindles equivalent to 3 to 4 spots in length.
Extended mitotic spindles, greater than 4 spots in length.
CaCdc5p Localizes to the Spindle Pole Bodies, Spindle, DNA, and Bud Neck in Yeast
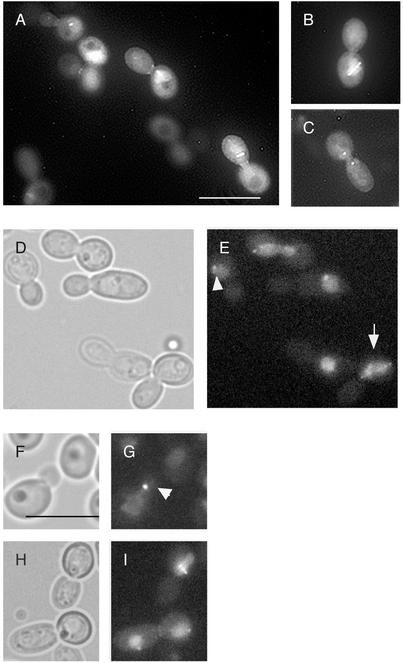
PLKs localize to spindle pole bodies/centrosomes, the spindle, chromosomes, and sites of cytokinesis in diverse organisms (Glover et al., 1998; Song et al., 2000). The localization of CaCdc5p-GFP was analyzed by integrating PCK1::CaCDC5-GFP into strain RM1000 and overexpressing the protein with synthetic medium containing 2% casaminoacids. The spindle, spindle pole bodies, DNA, and bud neck clearly demonstrated a signal (Figure 5, A–C), consistent with known PLK localizations. Similarly, a strain containing CaCdc5p-GFP under control of its endogenous promotor demonstrated identical localizations but with a weaker signal (Figure 5, D–I). Intriguingly, CaCdc5p-GFP under control of its own promotor localized to unseparated spindle pole bodies in cells with very small buds (Figure 5, E and G), suggesting a function for CaCdc5p in the early stages of spindle elongation.
Figure 5.
CaCdc5p localizes to the spindle pole bodies, spindle, DNA, and bud neck. Strain CB115 (A–C) containing PCK1:: CaCDC5-GFP was grown in S medium containing 2% casaminoacids for overexpression. Strain CB116 (CaCDC5-GFP-URA3) (D–I) demonstrates similar localization patterns. Note the staining of a spindle-like structure overlaying chromatin (arrow in E) and spindle pole bodies and chromatin in small budded cells (arrow heads in E and G). Bars, 10 μm
The DNA Synthesis Inhibitor Hydroxyurea Induces Similar Filament Formation and Impairs Spindle Elongation
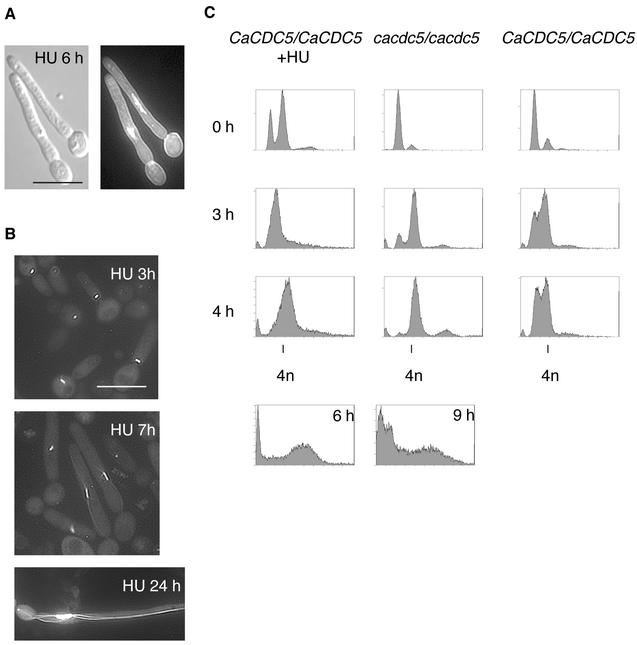
We and other groups (Bai et al., 2002; Hazan et al., 2002) found that exposing yeast cells of C. albicans to HU produced filaments under yeast growth conditions. Because the filaments closely resembled those of CaCDC5-repressed cells, we analyzed the filaments further to identify common perturbed features and therefore possible mechanisms involved in triggering filament initiation. After 2–3 h in 200 mM HU at 30°C, cells from strain SC5314 were elongated and developed into filaments resembling those of CaCDC5-repressed cells (Figure 6A). By 24 h, the filaments were shorter in length than the CaCdc5p-depleted filaments and more uniformly vacuolated. The response was cell density dependent, as yeast cells with an OD600 greater than 0.6 did not form filaments upon incubation in HU. DAPI staining (Figure 6A) and Tub1p-GFP (Figure 6B) patterns in cells from strains SC5314 and CB110, respectively, exposed to HU demonstrated that nuclear division and spindle elongation were blocked in a way similar to that of CaCDC5-repressed cells, although the HU-induced filaments retained the short spindle for a longer period (Table 5). FACS analysis demonstrated that HU-exposed cells were blocked in S phase but progressed to a G2 (4n) content of DNA, whereas CaCDC5-repressed cells progressed through S phase but were subsequently blocked in G2 (Figure 6C). Some fragmentation of chromatin occurred in both conditions at later time points (Figure 6C). These results demonstrate that impaired spindle elongation is a common, early defect in HU-treated and CaCDC5-repressed cells, and thus an aspect of spindle function may be linked to filament formation.
Figure 6.
Hydroxyurea induces filament formation under yeast growth conditions and impairs spindle elongation. (A) Strain SC5314 was grown in YPD medium overnight and then diluted to an OD600 of 0.4 in fresh YPD containing 200 mM HU. Cells were fixed at 6 h and stained with DAPI and calcofluor. (B) Strain CB110 (CaCDC5/CaCDC5, TUB1-GFP) was incubated in 200 mM HU for 3, 7, and 24 h. Note the similarity to filaments of CaCDC5-repressed cells (Figures 2B, 3A, and 4A). Bar, 10 μm. (C) FACS analysis demonstrating S phase and G2 phase blocks in HU-treated and CaCDC5-repressed cells, respectively. Cells of strains CB104 (cacdc5::hisGΔ/cacdc5Δ::HIS1/PCK1::CaCDC5) and RM1000 (CaCDC5/CaCDC5) were transferred from SS to SD medium, collected at the indicated time points, and processed for FACS analysis. Strain SC5314 was grown in YPD containing 200 mM HU and processed for FACS analysis. Note the fragmentation of DNA at later time points of 9 and 6 h.
CaCDC5-repressed and HU-exposed Filaments Demonstrate Hyphal Characteristics and Express Factors Normally Regulated by Hyphal Transcription Pathways
The filaments described here demonstrated several hyphal-like characteristics. A distinguishing feature of hyphal development in C. albicans is the migration of the nucleus into the germ tube before mitosis (Sudbery, 2001). Nuclei in both CaCDC5-repressed and HU-exposed cells demonstrated this behavior (Figures 3 and 6; Table 3), where the nucleus moved out of the mother cell and into the filament. The filaments also contained extensive cytoplasmic microtubules, comparable to serum-induced hyphae.
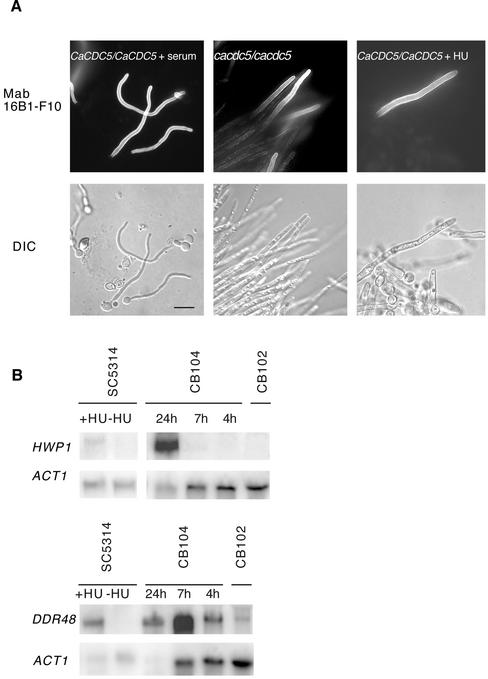
Further evidence for similarity between the filaments and true hyphae was obtained by investigating the expression of a protein that is specifically induced by the hyphal-generating condition of serum and high temperature, using the MAb 16B1-F10 (Marot-Leblond et al., 2000). Immunolocalization with 16B1-F10 demonstrated that the antigen was produced by several filaments depleted of CaCdc5p for 7 h or exposed to HU for 24 h (Figure 7A). The filaments did not stain along their entire length, as seen with serum-induced hyphae, indicating the antigen was not expressed immediately upon filament initiation.
Figure 7.
Filaments produced by repression of CaCDC5 or exposure to HU are similar to hyphae in expression of a serum-induced antigen and RNA. (A) Serum-induced hyphae from strain SC5314, filaments from strain CB104 (cacdc5Δ::hisG/cacdc5Δ:: HIS1/PCK1::CaCDC5) depleted of CaCdc5p for 7 h, and filaments from strain SC5314 incubated in HU for 24 h were fixed and processed for immunolocalization of MAb16B1-F10. Bar, 10 μm. (B) Northern analysis demonstrating the filament-induced expression of RNA normally induced by the hyphal-generating conditions of serum and high temperature. Total RNA, 20 μg, from strain SC5314 incubated in YPD with 200 mM HU for 6 h, SC5314 without HU, CB104 (cacdc5::hisGΔ/cacdc5Δ:: HIS1/PCK1::CaCDC5) grown in SD medium for 24, 7, and 4 h, and CB102 (cacdc5Δ::hisG/CaCDC5 PCK1:: CaCDC5) grown in SD medium for 4 h was hybridized with probes specific for HWP1, DDR48, and ACT1 as a loading control.
Northern analysis of cells depleted of CaCdc5p for 4, 7, or 24 h or exposed to HU for 6 h revealed the expression of factors normally induced by the hyphal signaling pathways in response to serum, including HWP1 (Sharkey et al., 1999) and DDR48 (Lane et al., 2001). The factors were not all induced at similar stages of filament development (Figure 7B), and certain other hyphal-specific factors, including ECE1 and ALS1, were not expressed (our unpublished results). Therefore, repression of CaCDC5 and exposure to HU leads to the formation of filaments and the activation of aspects of the hyphal transcription program, suggesting a link between spindle function and hyphal development.
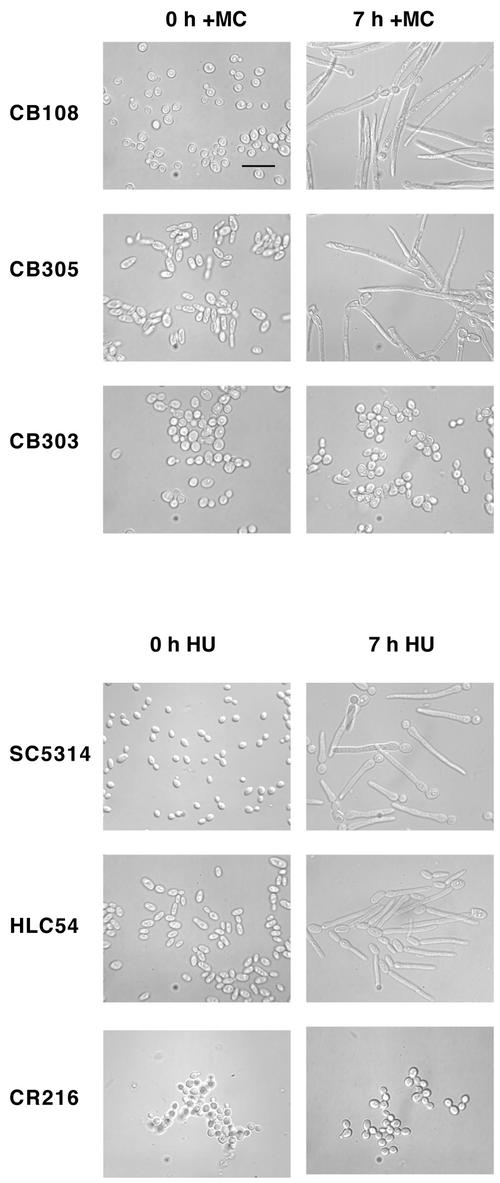
Filament Formation Requires CaCDC35, but not EFG1 or CPH1
Several hyphal signaling pathways in C. albicans converge on the transcription factors Efg1p and Cph1p (Ernst, 2000). The absence of both these factors prevents hyphal formation under most conditions (Lo et al., 1997), whereas CaCdc35p is required for hyphal formation under all conditions tested (Rocha et al., 2001). To determine whether these factors are required for the formation of filaments described here, strains lacking CPH1, EFG1, and CaCDC35 were depleted of CaCdc5p or exposed to HU. After 7 h of CaCDC5 repression or exposure to HU, filaments were able to form normally in the double mutant lacking both Efg1p and Cph1p (Figure 8).Efg1p and Cph1p single mutants (strains JCK19 and HLC52) also formed filaments in response to HU (our unpublished results). However, the absence of CaCdc35p severely compromised filamentous growth in both conditions. The cells treated with HU or depleted of CaCdc5p looked identical and resembled large budded yeast with some isotropically enlarged daughter cells containing a distal polarized evagination. After 24 h, filamentous growth was still severely compromised, but more cells contained abnormal shapes and a short polarized extension in the CaCDC5-repression vs. HU-treated condition (our unpublished results). This difference likely reflects the more toxic effects of hydroxyurea and inhibition of DNA synthesis, causing cell death more quickly than when CaCDC5 is repressed. The results suggest that absence of CaCdc5p and exposure to HU act through a similar pathway to produce hyphal-like filaments and communicate with the hyphal signaling networks at the level of CaCdc35p.
Figure 8.
Filamentous growth in cells depleted of CaCdc5p or exposed to HU require CaCdc35p, but not Efg1p and Cph1p. Cells from strains CB108 (cacdc5Δ::hisG/MET:: CaCDC5-URA3), CB305 (cacdc5Δ::hisG/MET:: CaCDC5-URA3 cph1Δ::hisG/cph1Δ::hisG efg1Δ::hisG/efg1Δ::hisG) and CB303 (cacdc5Δ::hisG/MET:: CaCDC5-URA3 cdc35Δ::hisG/cdc35Δ::hisG) were incubated in SD medium–methionine for overexpression of MET:: CaCDC5, or SD medium + methionine and cysteine to repress MET:: CaCDC5 expression for 7 h. Cells from strains SC5314 (+/+), HLC54 (cph1Δ::hisG/cph1Δ::hisG efg1Δ::hisG/efg1Δ::hisG -URA3-hisG) and CR216 (cdc35Δ::hisG/cdc35Δ::hisG-URA3-hisG) were grown in SD medium, diluted to an OD600 of 0.4 in fresh medium containing 200 mM HU, and incubated for 7 h. Cells were fixed in 70% EtOH before collection of images. Bar, 10 μm.
DISCUSSION
Regulation of hyphal formation in C. albicans involves signaling pathways that ultimately converge to control both the expression of hyphal-specific genes and the activation/recruitment of mechanical factors required for hyphal development. We show that the polo-like kinase homologue CaCDC5 is required for spindle elongation and that perturbing spindle function through repression of CaCDC5 or exposure to HU is associated with activation of the transcription program and machinery required for hyphal-like formation. Thus a mechanism is in place in C. albicans to allow communication between an internal, cell cycle–related cue and hyphal development.
CaCDC5 Is Required for Spindle Formation during S Phase
CaCDC5, along with PLKA from Aspergillus nidulans (Bachewich and Osmani, unpublished results) are the first characterized polo-like kinase homologues in hyphal-producing fungi. The short spindles and unseparated chromatin that result from repression of CaCDC5 and the localization of CaCdc5p to unseparated spindle pole bodies in cells with small buds support a role for CaCdc5p in the early stages of spindle elongation. Such a role is consistent with known PLK functions in most other organisms, but the demonstration that a PLK is required as early as S phase has not been previously reported. In contrast to higher organisms and S. pombe, spindle initiation occurs during S phase in both C. albicans (Barton and Gull, 1988; Hazan et al., 2002) and S. cerevisiae (Winey and O'Toole, 2001). Several lines of evidence suggest a requirement for Cdc5p in spindle formation and DNA replication during S phase in S. cerevisiae (Hardy and Pautz, 1996; Cheng et al., 1997; Bartholemew et al., 2001), but the cdc5–1 mutant arrests with elongated spindles in mitosis with partially separated chromatin (Kitada et al., 1993) and early spindle defects associated with compromised Cdc5p function have yet to be reported. Therefore CaCdc5p acts earlier in the cell cycle and/or has some different functions than Cdc5p.
Depletion of CaCdc5p Induces a Dramatic Switch in Morphology from Yeast Cells to Actively Growing Filaments with Hyphal Characteristics in the Absence of Serum or High Temperature
The development of filaments upon repression of CaCDC5 is consistent with a switch to a hyphal-like growth mode, as opposed to the terminal phenotype of slowly expiring elongated buds, because the filaments were actively extending at one third of the rate of serum-induced hyphae incubated at the higher temperature of 37°C, and demonstrated several hyphal characteristics, including movement of the nucleus out of the yeast cell into the filament, development of an extensive organization of cytoplasmic microtubules, and expression of factors lying downstream of the hyphal signaling pathways. In filamentous fungi, mutations in a variety of genes result in wide diameter hyphae (Harris et al., 1997; Kaminskyj and Hamer, 1998; Momany et al., 1999), supporting that the wide filaments described here are closely related to true hyphae. The filaments are also not analogous to elongated buds of S. cerevisiae that form in response to an inability to deposit a septum (Jimenez et al., 1998), because the early block in nuclear division and spindle elongation, and ability to produce filaments with HU, support the notion that filaments emerge during an early stage in the cell cycle, before the timing of septation. Septin rings, visualized with Cdc12p-GFP, appeared normal in CaCdc5p-depleted cells at 7 h and were localized at the yeast/filament junction (our unpublished results), supporting that septin-related defects did not generate the filamentous growth described here. The localization of septin rings at the neck of the filaments is consistent with filament emergence during late S/G2 of the cell cycle and is comparable to the localization of the first septa in serum-induced hyphae, which initiated growth later than G1/S (Hazan et al., 2002).
The eventual death of the filaments suggests that CaCDC5 is essential. This possibility exists despite the fact that active hyphal-like growth occurs upon gene repression, because hyphae of filamentous fungi can grow for a determinate period of time in the absence of nuclear division. However, at a later point the nucleus must divide in order for growth to continue. For example, mutations in some essential cell cycle genes such as nimXcdc2p34 in A. nidulans do not prevent spore germination and determinate hyphal growth (d'Enfert, 1997), and blocking nuclear division in established hyphae of C. albicans also does not prevent hyphal growth (Yokoyama et al., 1990). Therefore, depletion of CaCdc5p and the associated block in nuclear division trigger a change in growth mode to a hyphal-like state, which continues for some time, but eventually the cells loose viability in the absence of proper nuclear division.
Intriguingly, several hyphal characteristics appeared at different stages in the development of CaCDC5-repressed filaments. Increased expression of DDR48, movement of the nucleus into the filament, and initiation of a cytoplasmic microtubule network were observed as early as 3–4 h after repressing CaCDC5. HWP1 expression was delayed but normally does not increase in serum-induced hyphae until later in development, after 60 min (Nantel et al., 2002). Microarray analysis of the filaments indicates the expression of additional factors that are normally induced by serum and regulated by the hyphal signaling pathways (our unpublished results). The differences in timing of gene expression could be due to the fact that an internal cue, as opposed to serum and high temperature, initiated the hyphal-like growth process, and different forks in the signaling networks were utilized. Indeed, there are some differences in the expression of transcripts (Nantel et al., 2002) and utilization of known components of the hyphal signaling pathways in hyphae produced under different environmental conditions (Giusani et al., 2002). Delayed expression of certain genes, especially surface or secreted factors that are not required for hyphal development but turn on as a consequence of hyphal growth, could also reflect differences in growth rate, feedback regulation from the developing hypha to the transcriptional pathways, and/or the involvement of factors required for initiation vs. maintenance of hyphal growth, examples of which have been identified (Nantel et al., 2002). These results suggest that internal signaling, as opposed to external environmental cues such as serum, may activate some transcriptional and other regulatory aspects governing hyphal initiation.
Defects in Spindle Elongation and the Corresponding Generation of Filaments in CaCDC5-repressed and HU-exposed Cells Suggest a Link between Spindle Function and Activation of Hyphal Growth
CaCdc5p did not act as a direct negative regulator of hyphal formation like Tup1p (Braun and Johnson, 1997) or Nrg1p (Braun et al., 2001; Munir et al., 2001), suggesting the induction of hyphal-like growth upon CaCdc5p depletion could be due to a CaCdc5p-dependent cell cycle–related function. The high degree of similarity between HU-induced and CaCDC5-repressed filaments suggests that filamentous growth was triggered by the same cue. A common feature of CaCDC5 repression and HU treatment is impaired spindle elongation, suggesting a link between spindle function and the hyphal regulatory program. Consistent with this, repression of DpbIIp, a subunit of DNA polymerase, results in determinate filament formation in C. albicans (Backen et al., 2000), and defects in DNA synthesis can inhibit spindle elongation. In addition, nocodazole induces similar filaments in C. albicans that can be partially suppressed by deletion of the spindle checkpoint factor MAD2 (Bai et al., 2002), suggesting filamentous growth is partially, but not fully, dependent on the kinetochore attachment/spindle assembly branch of the spindle checkpoint. Activation of this branch of the spindle checkpoint alone, however, is not sufficient to induce filament formation because deletion of a homologue of the centromere protein CENP-A in C. albicans did not result in filamentous growth (Sanyal and Carbon, 2002). Although the data support the idea that spindle function is involved in the cue leading to activation of hyphal-like growth, we cannot rule out other pathways or some additional role for CaCdc5p itself, because polo-like kinases can be negatively regulated by spindle and DNA damage checkpoints (Sanchez et al., 1999; Smits et al., 2000; Hu et al., 2001) and nocodazole and HU activate spindle checkpoints (Hu et al., 2001; Garber and Rine, 2002). The nature of the internal signal and mechanism of transmission to the hyphal regulatory pathways is currently under investigation.
The morphogenic effects of inhibiting different stages in the cell cycle of Candida are not known, but repression of another essential cell cycle factor in C. albicans, CaCdc42p, resulted in isometric growth under yeast growth conditions (Ushinsky et al., 2002). In addition, applying different stresses to yeast cells of Candida at 30°C did not elicit the filamentous response described here (Martchenko and Whiteway, unpublished results; Enjalbert et al., 2003), indicating the phenotype is not a reaction to general cell stress.
Activation of Filament Formation Is Dependent on CaCdc35p but not Efg1p or Cph1p
The formation of both CaCDC5-repressed and HU-exposed filaments is dependent on CaCdc35p but not Efg1p/Cph1p, suggesting that depletion of CaCdc5p and exposure to HU may act through similar pathways leading to hyphal-like formation, and communication with the hyphal signaling networks occurs at the level of CaCdc35p. The facts that CaCdc35p is predicted to act upstream of Efg1p (Rocha et al., 2001), Efg1p in turn was not required for filament formation, and the filaments expressed HWP1, a factor lying down-stream of Efg1p (Sharkey et al., 1999), suggest that additional pathways feed into and out of CaCdc35p for filament formation. Transcript profiling of the cacdc35/cacdc35 strain (Harcus, Nantel, and Whiteway, unpublished results) also supports a role for CaCdc35p outside of Efg1p regulation.
Function of a Link between Spindle Elongation and Hyphal Development
The recent demonstration that initiation of hyphal growth in C. albicans is not limited to one cell cycle phase suggests that the cell cycle is not a direct regulator of hyphal growth (Hazan et al., 2002). The ability to induce hyphal-like formation upon depletion of CaCdc5p and perturbation of spindle function in yeast cells, however, suggests that there is a connection between aspects of the cell cycle and the hyphal signaling machinery. This regulatory relationship could exist as a type of checkpoint, perhaps with the duration and/or extent of spindle formation being monitored. Interestingly, blocks in S phase can prevent hyphal growth in A. nidulans, whereas mutations in some other essential cell cycle factors do not prevent short-term hyphal growth (d'Enfert, 1997). Regardless of variations in different organisms, the demonstration that lack of CaCdc5p and perturbation of spindle function influences hyphal-like growth and transcription in Candida indicates that hyphal-like growth can be activated by internal, cell cycle–related cues, as opposed to external signals like serum, and introduces a new level within the hyphal signaling networks of C. albicans.
Acknowledgments
We thank all members of the Whiteway lab for discussions and assistance, Dr. A. Marot-Leblond for Mab16B1-F10, Lucie Bourget for assistance with the FACS analysis, and Ursula Oberholzer, James MaGee, Josee Ash, Peter Sudbery, Joachim Morschhauser, and William Fonzi for plasmids. This work was supported in part by a Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship and Visiting Fellowship to C.B. and by the National Research Council Genomics Health Initiative to M.W. Sequence data for C. albicans was obtained from the Stanford Genome Technology Center website at http://www-sequence.stanford.edu/group/candida. Sequencing of C. albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.02-05-0076. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.02-05-0076.
References
- Alexandru, G., Uhlmann, F., Mechtler, K., Poupart, M., and Nasmyth, K. (2001). Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell 105, 459–472. [DOI] [PubMed] [Google Scholar]
- Backen, A.C., Broadbent, I.D., Fetherston, R.W., Rosamond, J.D.C., Schnell, N.F., and Stark, M.J.R. (2000). Evaluation of the CaMAL promotor for regulated expression of genes in Candida albicans. Yeast 16, 1121–1129. [DOI] [PubMed] [Google Scholar]
- Bai, C., Ramanan, N., Wang, Y.M., and Wang, Y. (2002). Spindle assembly checkpoint component CaMad2p is indispensable for Candida albicans survival and virulence in mice. Mol. Microbiol. 45, 31–44. [DOI] [PubMed] [Google Scholar]
- Bartholomew, C.R., Woo, S.H., Chung, Y.S., Jones, C., and Hardy, C. (2001). Cdc5 interacts with the Wee1 kinase in budding yeast. Mol. Cell. Biol. 21, 4949–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, R., and Gull. K. (1988). Variation in cytoplasmic microtubule organization and spindle length between two forms of the dimorphic fungus Candida albicans. J. Cell Sci. 91, 211–220. [DOI] [PubMed] [Google Scholar]
- Bensen, E.S., Filler, S.G., and Berman, J. (2002). A forkhead transcription factor is important for true hyphal growth as well as yeast morphogenesis in Candida albicans. Eukaryotic Cell 1, 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, B.R., and Johnson, A.D. (1997). Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277, 105–109. [DOI] [PubMed] [Google Scholar]
- Braun, B.R., Kadosh, D., and Johnson, A.D. (2001). NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 20, 4753–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Care, R.S., Trevethick, J., Binley, K.M., and Sudbery, P.E. (1999). The MET3 promotor: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34, 792–798. [DOI] [PubMed] [Google Scholar]
- Chen, J., Zhou, S., Wang, Q., Chen, X., Pan, T., and Liu, H. (2000). Crk1, a novel Cdc2-related protein kinase, is required for hyphal development and virulence in Candida albicans. Mol. Cell. Biol. 20, 8596–8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D.C., Yang, B.C., and Kuo, T.T. (1992). One-step transformation of yeast. Curr. Genet. 21, 83–84. [DOI] [PubMed] [Google Scholar]
- Cheng, L., Collyer, T., and Hardy, C.F. (1997). Cell cycle regulation of DNA replication initiator factor Dbf4p. Mol. Cell. Biol. 19, 4270–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Enfert, C. (1997). Fungal spore germination: insights from the molecular genetics of Aspergillus nidulans and Neurospora crassa. Fungal Genet. Biol. 21, 163–172. [Google Scholar]
- Ernst, J.F. (2000). Transcription factors in Candida albicans-environmental control of morphogenesis. Microbiology 146, 1763–1764. [DOI] [PubMed] [Google Scholar]
- Enjalbert, B., Nantel, A., and Whiteway, M. (2003). Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell 14, 1460–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi, W.A., and Irwin, M.Y. (1993). Isogenic strain construction and gene mapping in Candida albicans. Genetics 134, 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, C., Gerami-Nejad, M., McClellan, M., Vandoninck, S., Longtine, M.S., and Berman, J. (2001). Candida albicans Int1p interacts with the septin ring in yeast and hyphal cells. Mol. Biol. Cell 12, 3538–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber, P.M., and Rine, J. (2002). Overlapping roles of the spindle assembly and DNA damage checkpoints in the cell-cycle response to altered chromosomes in Saccharomyces cerevisiae. Genetics 161, 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusani, A.D., Vinces, M., and Kumamoto, C.A. (2002). Invasive filamentous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics 160, 1749–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover, D.M., Hagan. I.M., and Tavares, A.A.M. (1998). Polo-like kinases: a team that plays throughout mitosis. Genes Dev. 12, 3777–3787. [DOI] [PubMed] [Google Scholar]
- Han, G., Liu, B., Zhang, J., Zuo, W., Morris, N.R., and Xiang, X. (2001). The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr. Biol. 11, 719–724. [DOI] [PubMed] [Google Scholar]
- Hardy, C.F., and Pautz, A. (1996). A novel role for Cdc5 in DNA replication. Mol. Cell. Biol. 16, 6775–6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S.D., Hamer. L., Sharpless, K.E., and Hamer, J.E. (1997). The Aspergillus nidulans sepA gene encodes an FH1/2 protein involved in cytokinesis and the maintenance of cellular polarity. EMBO J. 16, 3474–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan, I., Sepulveda-Becerra, M., and Liu, H. (2002). Hyphal elongation is regulated independently of cell cycle in Candida albicans. Mol. Biol. Cell 13, 134–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, F., Wang, Y., Liu, D., Li, Y., Qin, J., and Elledge, S.J. (2001). Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell 107, 655–665. [DOI] [PubMed] [Google Scholar]
- Jimenez, J., Cid, V.J., Cenamor, R., Yuste, M., Molero, G., Nombela, C., and Sanchez, M. (1998). Morphogenesis beyond cytokinetic arrest in Saccharomyces cerevisiae. J. Cell Biol. 143, 1617–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminskyj, S.G.W., and Hamer. J.E. (1998). hyp loci control cell pattern formation in vegetative mycelium of Aspergillus nidulans. Genetics 148, 669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada, K., Johnson, A.L., Johnston, L.H., and Sugino, A. (1993). A multicopy suppressor gene of the Saccharomyces cerevisiae G1 cell cycle mutant gene dbf4 encodes a protein kinase and is defined as CDC5. Mol. Cell. Biol. 13, 4445–4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhrer, K., and Domdey, H. (1991). Preparation of high molecular weight RNA. Methods Enzymol. 194, 398–405. [DOI] [PubMed] [Google Scholar]
- Kron, S.J., and Gow, N.A.R. (1995). Budding yeast morphogenesis: signaling, cytoskeleton, and cell cycle. Curr. Opin. Cell Biol. 7, 845–855. [DOI] [PubMed] [Google Scholar]
- Lane, H.A., and Nigg, E.A. (1996). Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in functional maturation of mitotic centrosomes. J. Cell Biol. 135, 1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, S., Birse, C., Zhou, S., Matson, R., and Liu, H. (2001). DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida. J. Biol. Chem. 276, 48988–48996. [DOI] [PubMed] [Google Scholar]
- Leuker, C.E., Sonneborn, A., Delbrück, S., and Ernst, F. (1997). Sequence and promotor regulation of the PCK1 gene encoding phosphoenolpyruvate carboxykinase of the fungal pathogen Candida albicans. Gene 192, 235–240. [DOI] [PubMed] [Google Scholar]
- Lew, D.J., Marini, N.J., Reed, S.I. (1992). Different G1 cyclins control the timing of cell cycle commitment in mother and daughter cells of the budding yeast S. cerevisiae. Cell 69, 317–327. [DOI] [PubMed] [Google Scholar]
- Lew, D.J., and Reed, S.I. (1995). A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J. Cell Biol. 129, 739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. (2001). Transcriptional control of dimorphism in Candida albicans. Curr. Opin. Microbiol. 4, 728–735. [DOI] [PubMed] [Google Scholar]
- Llamazares, S., Moreira, A., Tavares, A., Girdham, C., Spruce, B.A., Gonzales, C., Karess, R.E., Glover, D.M., and Sunkel, C.E. (1991). polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev. 5, 2153–2165. [DOI] [PubMed] [Google Scholar]
- Lo, H., Köhler, J.R., DiDomenico, B., Loebnberg, D., Cacciapuoti, A., and Fink, G.R. (1997). Nonfilamentous C. albicans mutants are avirulent. Cell 90, 939–949. [DOI] [PubMed] [Google Scholar]
- Loeb, J.D.J., Sepulveda-Becerra, M., Hazan, I., and Liu, H. (1999). A G1 cyclin is necessary for maintenance of filamentous growth in Candida albicans. Mol. Cell. Biol. 19, 4019–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marot-Leblond, A., Grimaud, L., Nail, S., Bouterige, S., Apaire-Marchais, V., Sullivan, D.J., and Robert, R. (2000). New monoclonal antibody specific for Candida albicans germ tube. J. Clin. Microbiol. 38, 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momany, M., Westfall. P., and Abramowsky, G. (1999). Aspergillus nidulans swo mutants show defects in polarity establishment, polarity maintenance, and hyphal morphogenesis. Genetics 151, 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morschhauser, J., Michel, S., and Hacker, J. (1998). Expression of a chromasomally integrated single copy GFP gene in Candida albicans and its use as a reporter of gene regulation. Mol. Gen. Genet. 257, 412–420. [DOI] [PubMed] [Google Scholar]
- Munir, A. et al. (2001). NRG1 represses yeast-hypha morphogenesis and hypha specific gene expression in Candida albicans. EMBO J. 20, 4742–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantel A. et al. (2002). Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell. 13, 3452–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg, E.A. (1998). Polo-like kinases: positive regulators of cell division from start to finish. Curr. Opin. Cell Biol. 10, 776–783. [DOI] [PubMed] [Google Scholar]
- Negrado, A., Gil. C., Pla, J., and Nombela, C. (1997). Cloning analysis and one step disruption of the ARG5,6 gene of Candida albicans. Microbiology 143, 297–302. [DOI] [PubMed] [Google Scholar]
- Okhura, H., Hagan, I.M., and Glover, D.M. (1995). The conserved Schizosaccharomyces pombe kinase plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes Dev. 9, 1059–1073. [DOI] [PubMed] [Google Scholar]
- Rocha, C.R., Schroppel, K., Harcus, D., Marcil, A., Dignard, D., Taylor, B.N., Thomas, D.Y., Whiteway, M., and Leberer, E. (2001). Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12, 3631–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M.D., Winston, F., and Hieter, P. (1990). Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Rua, D., Tobe, B., and Kron, S.J. (2001). Cell cycle control of yeast filamentous growth. Curr. Opin. Microbiol. 4, 720–727. [DOI] [PubMed] [Google Scholar]
- Sanchez, Y., Bachant, J., Wang, H., Hu, F., Liu, D., Tetzlaff, M., and Elledge, S.J. (1999). Control of the DNA damage checkpoint by Chk1 and Rad53 protein kinases through distinct mechanisms. Science 286, 1166–1171. [DOI] [PubMed] [Google Scholar]
- Sanyal, A., and Carbon, J. (2002). The CENP-A homolog CaCse4p in the pathogenic yeast Candida albicans is a centromere protein essential for chromosome transmission. Proc. Natl. Acad. Sci. USA 99, 12969–12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey, L.L., McNemar, M.D., Saporito-Irwin, S.M., Sypherd, P.S., and Fonzi, W.A. (1999). HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J. Bacteriol. 181, 5273–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama, M., Zachariae, W., Ciosk, R. Nasmyth, K. 1998. The polo-like kinase cdc5p and the WD repeat protein Cdc20/fizzy are regulators and substrates of the anaphase-promoting complex in Saccharomyces cerevisiae. EMBO J. 17, 1336–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits, V.A.J., Klommaker, R., Arnaud, L., Rijksen, G., Nigg, E.A., and Medema, R.H. (2000). Polo-like kinase is a target of the DNA damage checkpoint. Nat. Cell Biol. 2, 672–676. [DOI] [PubMed] [Google Scholar]
- Song, S., Grenfall, T.Z., Garfield, S., Erikson, R.L., and Lee, K.S. (2000). Essential function of the polo box of Cdc5 in subcellular localization and induction of cytokinetic structures. Mol. Cell. Biol. 20, 286–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, S., and Lee, K.S. (2001). A novel function of Saccharomyces cerevisiae CDC5 in cytokinesis. J. Cell Biol. 152, 451–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantha, T., and Soll, D.R. (1993). A white-specific gene in the white-opaque switching system of Candida albicans. Gene 131, 53–60. [DOI] [PubMed] [Google Scholar]
- Sudbery, P.E. (2001). The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol. Microbiol. 41, 19–31. [DOI] [PubMed] [Google Scholar]
- Torralba, S., and Heath, IB. (2001). Cytoskeletal and Ca2+ regulation of hyphal tip growth and initiation. Curr. Top. Dev. Biol. 51, 135–187. [DOI] [PubMed] [Google Scholar]
- Toyoshima-Morimoto, F., Taniguchi, E., Shinya, N., Iwanatsu, A., and Nishida, E. (2001). Polo-like kinase 1 phosphorylates cyclin B and targets it to the nucleus during prophase. Nature 410, 215–220. [DOI] [PubMed] [Google Scholar]
- Ushinsky, S.C., Harcus, D., Ash, J., Dignard, D., Marcil. A., Morchhauser, J., Thomas, D.Y., Whiteway, M., and Leberer, E. (2002). CDC42 is required for polarized growth in human pathogen Candida albicans. Eukaryotic Cell 1, 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway, M. (2000). Transcriptional control of cell type and morphogenesis in Candida albicans. Curr. Opin. Microbiol. 3, 582–588. [DOI] [PubMed] [Google Scholar]
- Winey, M., and O'Toole, E.T. (2001). The spindle cycle in budding yeast. Nat. Cell Biol. 3, E23–E27. [DOI] [PubMed] [Google Scholar]
- Yokoyama, K., Kaji, H., Nishimura, K., and Miyaji, M. (1990). The role of microfilaments and microtubules in apical growth and dimorphism of Candida albicans. J. Gen. Microbiol. 136, 1067–1075. [DOI] [PubMed] [Google Scholar]