Abstract
NFATc (a member of the family of nuclear factors of activated T cells) is a transcriptional factor responsible for the Ca2+-inducible activation of cytokine genes during the immune response. In resting T cells, NFATc is retained in the cytoplasm by a mechanism that depends on multiple phosphorylations in an N-terminal regulatory domain. Physical interaction with and dephosphorylation by Ca2+-activated calcineurin (Cn) allows the protein to enter the nucleus, where it binds to specific sites in cytokine gene promoters. Previous studies had identified a peptide segment in NFATc that binds Cn stably. Here we report the identification of a second Cn-binding element in NFATc, which synergizes with the previously identified element. Although these sequences are conserved in all isoforms of NFAT, we find that the two sites contribute differentially to the overall affinity for Cn in an isoform-dependent manner. The regulatory domain of NFAT also was found to be entirely devoid of structure, both in the phosphorylated and unphosphorylated state. This finding suggests that the NFAT regulatory domain does not undergo phosphorylation-induced conformational switching, but instead requires partner proteins to control accessibility of the NFAT nuclear localization and nuclear export signals.
The nuclear factor of activated T cells (NFAT) defines a family of Ca2+-inducible transcription factors discovered for their critical role in regulating the transcription of cytokine genes (1–3). Four individually encoded members of the NFAT family are known: NFATc (also known as NF-ATc1 or NFAT2) (4), NFATp (NF-ATc2/NFAT1) (5), NFAT3 (NF-ATc4), and NFAT4 (NFATx/NF-ATc3) (6, 7). [There is disagreement as to whether a fifth NFAT-related protein, NFAT5 (8), should be included within the family.] Each family member serves a distinct biological role, as revealed by their unique expression patterns, together with the markedly different phenotypes of mice carrying targeted disruptions of the NFATc, NFATp, and NFAT4 genes (9–12).
NFAT proteins share a conserved domain located toward the C terminus (13) that binds DNA and also participates in cooperative protein–protein interactions with other transcription factors (14–16). Immediately N terminal to this domain lies a second conserved module of ≈300 residues known as the NFAT homology region (NFAT-h). The N terminus of NFAT, including the NFAT-h, regulates nuclear/cytoplasmic trafficking in response to changes in intracellular Ca2+ concentrations. This function is apparently self-contained, as Ca2+-dependent trafficking can be transferred to unrelated proteins through fusion to the NFAT regulatory domain (17–20). In resting T cells, the protein is retained in the cytoplasm and its NFAT-h is heavily phosphorylated. Engagement of the T cell antigen receptor or treatment of cells with Ca2+-ionophores activates calcineurin (Cn), a Ca2+/calmodulin-dependent Ser/Thr phosphatase, which dephosphorylates the NFAT-h, resulting in rapid translocation of the protein to the nucleus (21).
The clinically important immunosuppressive drugs FK506 and cyclosporin A (CsA) block this translocation process by inhibiting Cn activity in T cells (refs. 22 and 23, reviewed in ref. 24). These drugs also inhibit Cn activity in nonhemopoietic cells, a likely cause of side effects that seriously limit immunosuppressive therapy in a significant fraction of organ transplant patients. Consequently, interest in new drugs that act by a more cell-specific mechanism runs high. Elucidating the detailed molecular mechanism of phosphorylation-dependent nuclear trafficking by the NFAT N-terminal domain would further this goal.
Previous studies have characterized signal sequences for both nuclear import (NLS) (17, 25, 26) and nuclear export (NES) (27, 28) in the NFAT-h and flanking regions. The prevailing model holds that the phosphorylation state of the NFAT-h controls switching between two alternative conformational states that are either import or export conducive. Direct structural evidence for or against this model has not been reported to date. As for the NFAT-h⋅Cn interaction, it has been shown that Cn stably associates with NFATc, NFATp, and NFAT4, even in the absence of substrate phosphate groups (18, 29). This interaction has been mapped to a conserved ≈13-aa segment (Cn-binding peptide A, CnBP-A) within the N-terminal domain of NFATp (30). Substitution of key residues within this segment by alanine results in loss of binding to Cn in vitro and substantially reduces dephosphorylation and nuclear translocation after ionomycin treatment in vivo. Similar results were obtained for NFAT4 (26), suggesting that this peptide motif may be a common Cn binding site for all isoforms of NFAT.
In this study we present evidence that NFATc has an additional Cn-binding site (CnBP-B), which synergizes with the previously identified docking site (CnBP-A) to increase the overall binding affinity for Cn. A peptide corresponding to CnBP-B from NFATp does not detectably bind Cn, whereas CnBP-B peptides from NFAT3 and NFAT4 bind somewhat more tightly than the NFATc counterpart. The existence of a second Cn binding site in only select isoforms of NFAT raises the possibility that a difference in the mode and strength of interaction with Cn may give rise to isoform-dependent differences in the activation of NFAT.
Materials and Methods
Preparation of Phosphorylated NFAT1–414/CD Spectroscopy.
The N-terminal fragment of human NFATc containing the first 414 residues (NFAT1–414) and tagged with the hexa-his sequence at the C terminus was overexpressed in Escherichia coli BL21(DE3) cells and purified by using Ni-nitrilotriacetic acid resin (Qiagen, Chatsworth, CA) according to the manufacturer's recommended procedure. Priming phosphorylation by protein kinase A (PKA, a gift from S. Taylor, University of California, San Diego) was carried out at 30°C for 2 h in a buffer containing 5 μM NFAT1–414, 0.1 μM PKA, 50 mM potassium phosphate (pH 7.3), 125 mM NaCl, 1 mM DTT, 1 mM MgCl2, and 0.25 mM ATP. For full phosphorylation, recombinant rabbit glycogen synthase kinase-3β (GSK-3β) (a gift from P. Roach, Indiana University) then was added to a concentration of 0.3 μM, and the reaction was allowed to proceed for 15 h. CD spectra were measured on a Jasco J-710 spectrometer at 25°C, with a quartz cuvette of 1-mm path length. Eight scans were signal-averaged to obtain the final spectra for each sample and corrected for the buffer and the kinases, which introduced a 3–5% correction in the spectra.
Glutathione S-Transferase (GST) Binding Assay/Western Blot.
Full-length NFAT1–414 and various fragments of it were fused to GST, and the fusion proteins were purified by affinity chromatography with 300 μl glutathione resin per 500 ml of LB culture. GST resin containing bound GST-NFAT fusion polypeptide was equilibrated with 200 nM recombinant human Cn (purified to 95% homogeneity and high specific activity as described in ref. 31) in binding buffer containing 20 mM Tris⋅HCl (pH 8.0), 100 mM NaCl, 1.5 mM CaCl2, 6 mM MgCl2, 1 mM DTT, and 0.2% Triton X-100, for 30 min at 4°C. The resin then was washed five times in fresh binding buffer, and the bound fraction was analyzed by electrophoresis on a 12% SDS/PAGE gel. Bound Cn was detected by standard Western blot using monoclonal Cn antibody (PharMingen).
Purification of CnBP-B Peptide and NMR Spectroscopy.
A 21-aa 15N-labeled peptide containing CnBP-B was biosynthesized by modification of a published protocol (32). The peptide was purified by reversed-phase chromatography on C18 column (Beckman) using a linear gradient (0.1% trifluoroacetic acid, 20–60% CH3CN over 20 min). A single major peak eluted at 8 min. An unlabeled sample prepared in parallel was found to contain a peptide of the expected molecular mass (2,491 Da), as determined by electrospray ionization MS (Harvard Chemistry Department Mass Spectrometry Facility).
All NMR spectra were acquired on a Bruker DMX500 spectrometer equipped with a 5-mm triple-resonance probe. The peptide was dissolved in NMR buffer (37.5 mM deuterated Tris⋅acetate, pH 6.5/50 mM KCl/37.5 mM MgSO4/5 mM per-deuterated DTT) at a concentration of 0.5 mM. A 1H-15N heteronuclear single quantum correlation experiment (HSQC) was performed with the sweep widths of 14 (F2) and 34 (F1) ppm, by using the enhanced sensitivity method with water suppression. Peptide assignments were derived from a nuclear Overhauser effect spectroscopy and a total correlation spectroscopy experiment. HSQC experiments were performed on two samples, one with the 15N-labeled peptide alone, and the other with the 15N-labeled peptide in the presence of 50 μM Cn. Data were analyzed with the felix95 software package (Molecular Simulations, San Diego).
Secreted Alkaline Phosphatase (SEAP) Assays.
Wild-type and mutant versions of NFAT1–414 were cloned into the mammalian expression vector pBJ5 (4) under the control of the SR-α promoter. Jurkat cells stably transfected with the adenovirus large-T antigen (Jurkat-TAg cells, ref. 33) were maintained in RPMI media containing 10% FBS. The cells (5 × 105) were transiently transfected by using 3 μl of cationic liposome DMRIE-C (GIBCO/BRL) with a reporter plasmid encoding the mammalian SEAP, together with varying amounts of the pBJ5 derivatives. The cells were allowed to recover for 24 h before stimulation with 1 μM ionomycin (Sigma) and 20 ng/ml phorbol myristate acetate (Sigma). The SEAP activity was measured a day later by using 600 μM methylumbellyferyl phosphate as the fluorescent substrate.
Results
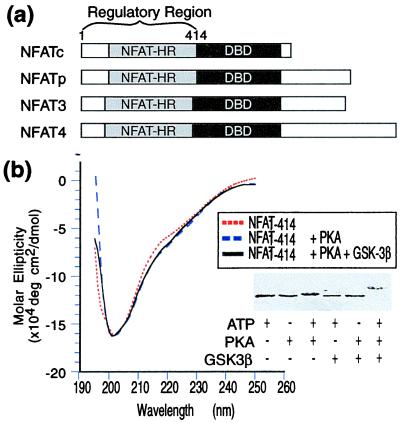
To characterize the NFATc regulatory domain biochemically, we overexpressed and purified a fragment of human NFATc spanning the NH2-terminal 414 amino acid residues (NFAT1–414, Fig. 1a). The CD spectrum of unphosphorylated NFAT1–414 (Fig. 1b) closely resembles that of an unfolded protein (34) and specifically lacks the spectral features characteristic of α-helical or β-sheet secondary structure (Fig. 1b). Consistent with the notion that unphosphorylated NFAT1–414 possesses little, if any, folded structure, the protein is degraded rapidly on exposure to 500 ng/ml of trypsin or chymotrypsin, whereas the folded core domain of Cn A remained intact under these conditions (data not shown).
Figure 1.
(a) Schematic diagram illustrating the domain organization of the four NFAT family members. The regulatory domain comprises the NFAT-h and the sequences N terminal to it. DBD, DNA binding domain. (b) CD spectra of NFAT in the unphosphorylated and phosphorylated forms. (Inset) SDS/PAGE gel showing the shift in electrophoretic mobility of NFAT1–414 caused by phosphorylation.
The kinases responsible for phosphorylating NFATc have been identified as PKA and GSK-3β (25). These enzymes act coordinately, with PKA catalyzing “priming” phosphorylations (35) that are necessary and sufficient for GSK-3β to further process the regulatory region and establish its fully phosphorylated state. In the case of NFAT4, MEK kinase 1 (MEKK1) and casein kinase Iα (CKIα) have been proposed as the intracellular kinases (26), but the phosphorylation sites for CKIα in vitro substantially overlap those for PKA plus GSK-3β. To test whether NFAT1–414 undergoes conformational reorganization after phosphorylation, we reacted the protein in vitro with PKA and GSK-3β. Although SDS/PAGE analysis indicated that the protein underwent phosphorylation after treatment with each of the two kinases, no change is evident in the CD spectra obtained after phosphorylation with either PKA alone or PKA plus GSK-3β (Fig. 1b). Thus, phosphorylation of the NFAT regulatory domain does not induce a detectable change in its secondary structure.
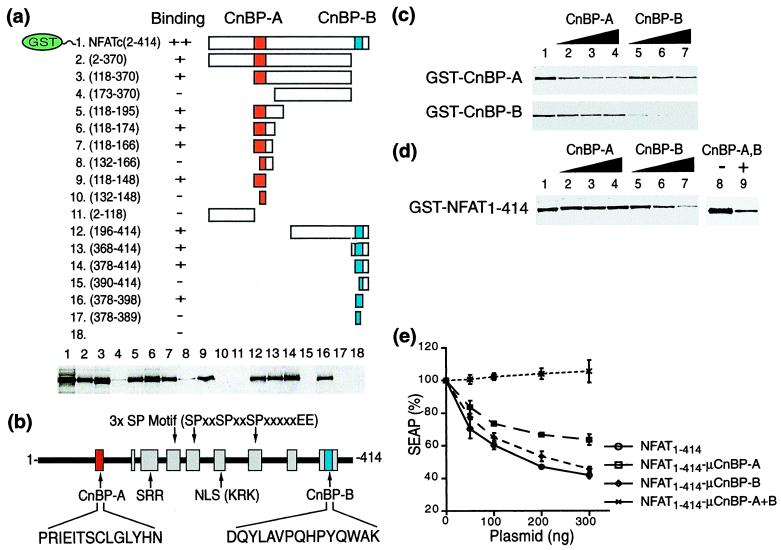
The observation that NFAT1–414 apparently lacks a well-defined three-dimensional fold, even in the phosphorylated form, raises the possibility that its interaction with Cn may be localized to one or more short contiguous polypeptide stretches in the regulatory region of NFATc. Such a short Cn-BP sequence has been identified in NFATp (30, 36) and NFAT4 (26). Furthermore, other proteins that bind Cn, including AKAP79 (37) and Bcl-2 (38), also have been shown to do so by using short peptide segments. To localize the Cn-BP sequences in NFATc, we created a series of GST fusion proteins bearing systematically truncated segments of NFAT1–414 (Fig. 2a). Their interaction with Cn was determined by pull-down assays using glutathione beads (Fig. 2a). Two polypeptide sequences, CnBP-A and another located near the CO2H-terminal end of the regulatory region, CnBP-B, were found to interact strongly with Cn (refer to Fig. 2b). Further truncations indicated the minimal CnBP-A and CnBP-B to comprise residues 118–131 and 384–398, respectively (data not shown). CnBP-A corresponds to the Cn-interacting sequence previously identified in NFATp (30) and NFAT4 (26). CnBP-B, on the other hand, represents a novel binding site. Both Cn-binding elements are conserved among the members of the NFAT family, corresponding to the outermost of the nine conserved motifs within the NFAT-h (39) (Fig. 2b). Using isothermal scanning calorimetry (40), we determined equilibrium dissociation constants (Kd) of 2.5 μM for the CnBP-A⋅Cn interaction and 1.3 μM for the CnBP-B⋅Cn interaction. A CnBP-A peptide from NFATp has been determined by surface plasmon resonance to bind Cn with an IC50 of ≈12 μM (30).
Figure 2.
(a) (Upper) Schematic representation of the GST fusion proteins used in the binding assay. The numbers in parentheses indicate the residues of NFATc fused to the C-terminal end of GST. The two Cn-binding sites, CnBP-A and CnBP-B, are highlighted. (Lower) Results of GST pull-down assays visualized by Western using an anti-Cn antibody. (b) CnBP-A and CnBP-B are located within the outermost motifs of the nine conserved in the NFAT homology region (39). SRR, serine-rich region; SP motif, serine-proline motif. Expansion brackets and parentheses show the amino acid sequences of certain key elements, in single-letter code. (c) Competition GST-pull-down assays. CnBP-A or CnBP-B fused to GST (GST-CnBP-A or CnBP-B) was incubated with Cn in the presence of either the CnBP-A or CnBP-B peptide in solution. (d) Same as in c, except the full-length NFAT regulatory domain fused to GST was used as bait. (Left) The results of competition by either peptide alone; (Right) competition by both. Concentrations in c and d are as follows: lanes 1 and 8, no peptide added; lanes 2–4, CnBP-A at 50, 100, and 300 μM, respectively; lanes 5–7, CnBP-B at 50, 100, and 300 μM, respectively; lane 9, CnBP-A and CnBP-B at 25 μM each; Cn at 100 nM in all lanes. (e) Inhibition of NFAT-dependent reporter gene activity by intracellular overexpression of the NFAT regulatory domain. A reporter construct having the SEAP cDNA linked to three tandem copies of the human IL-2 promoter was transfected into Jurkat-TAg cells, along with an expression vector encoding the N-terminal regulatory domain of human NFATc. Both wild-type regulatory region (NFAT1–414) and mutants having three alanine substitutions (R119A, E121A, and T123A) in CnBP-A (NFAT1–414–μCnBP-A) and/or a deletion (residues 378–414) of CnBP-B (NFAT1–414–μCnBP-B) were analyzed. SEAP activity was measured with 4-methylumbelliferyl phosphate as the substrate. All runs were normalized to 100% SEAP activity in the absence of NFAT plasmid.
The lack of any discernible sequence similarity between CnBP-A and CnBP-B (Fig. 2b) suggests that they might bind to different sites in Cn. To address this possibility, we assayed the ability of CnBP-A and CnBP-B peptides to compete for the binding of Cn to GST–CnBP-A and GST–CnBP-B fusion proteins. As expected, binding of Cn to each GST–peptide fusion protein to Cn was disrupted in a concentration-dependent manner by the identical peptide in solution (Fig. 2c). In a reciprocal experiment, however, neither peptide in solution significantly diminished the binding of Cn to the other GST–peptide fusion, thus indicating that CnBP-A and CnBP-B interact with distinct and nonoverlapping binding sites on Cn. CnBP-A alone failed to block the binding of full-length GST–NFAT1–414 to Cn, but CnBP-B showed some competition at high concentrations. However, the most efficient competition was observed when both peptides were present simultaneously (Fig. 2d).
To study the in vivo role of the two Cn binders, CnBP-A and CnBP-B, Jurkat-TAg cells were transiently transfected with a reporter construct in which NFAT drives expression of the SEAP gene. These cells were cotransfected with expression constructs encoding various competitors derived from the regulatory domain of NFATc (Fig. 2e). Overexpression of full-length NFAT1–414 in this assay system substantially decreases SEAP activity, presumably by blocking out the interaction of the endogenous Cn and NFAT. Similarly, overexpression of an NFAT1–414 mutant bearing alanine substitutions in CnBP-A or deletion of CnBP-B suppresses SEAP expression, suggesting that these mutant version of the regulatory domain are still able to bind Cn in vivo. On the other hand, mutations in both CnBP-A and CnBP-B abolish the ability to inhibit endogenous Cn activity, even at the highest concentrations of the overexpression plasmid. These in vivo results are fully in accord with the above in vitro observations that CnBP-A and CnBP-B interact with independent sites in Cn.
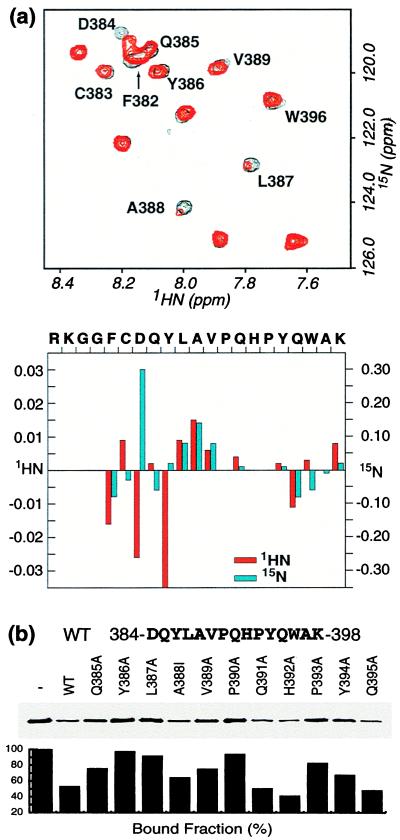
To identify the residues in CnBP-B that are critical for binding to Cn, we performed a 1H-15N HSQC NMR experiment. This experiment correlates the chemical shift of each backbone amide proton with that of the nitrogen atom to which it is attached, yielding a cross-peak for each amide H–N pair. The positions of these cross-peaks provide a sensitive and informative probe for the magnetic (and hence chemical) environment of the peptide backbone. The HSQC spectrum of the CnBP-B peptide alone shows a relatively narrow chemical shift dispersion (Fig. 3a, black), which indicates a lack of stable secondary structure, as is typical for peptides. Addition of substoichiometric amounts of Cn results in concentration-dependent positional shifts for some peaks (Fig. 3a, red), thus indicating that CnBP-B and Cn associate and dissociate rapidly on the NMR time scale. The observed chemical shift changes could be caused directly by intermolecular contacts of the peptide backbone to Cn or indirectly by folding of the peptide backbone induced on binding of side chains to Cn. Plotting the magnitude of the chemical shift perturbations as a function of the CnBP-B sequence reveals that the effects are largest around the center of the peptide (Fig. 3b). In addition to the positional shifts, the cross-peaks for L387 and A388 exhibit significant line broadening and loss of signal intensity in the presence of Cn, presumably because these residues are bound relatively tightly and therefore exchange slowly (41). Residues on the periphery of the peptide, for example G380, G381, A397, and K398 experience minimal line broadening, even at 0.3 mole equivalents of Cn. These NMR results are consistent with those of truncation assays, indicating that the segment of CnBP-B most important for binding to Cn comprises residues 384–398.
Figure 3.
(a) (Upper) 1H-15N HSQC of 500 μM CnBP-B in the absence and presence of 10 mol% Cn. The spectrum shows part of the amide proton/amide nitrogen region. (Lower) Quantification of chemical shift changes in the amide proton and amide nitrogen chemical shifts, measured in ppm. The following residues were not detected by HSQC: R378, K379, P390, H392, and P393. (b) (Upper) Sequences of synthetic peptides used to block against binding of GST–CnBP-B in a competition pull-down assay. (Lower) The amount of Cn pulled down in the presence of peptide competitors was quantified by SDS/PAGE and Western blotting using an anti-Cn antibody, followed by scanning and analysis using the macbas software package. WT refers to wild-type CnBP-B. All peptides were acetylated at the N terminus and amidated at the C terminus and were present at 200 μM concentration.
To study the interactions involving the side chains, we prepared wild-type and point-mutant synthetic peptides corresponding to the minimal interaction segment (NFATc residues 384–398, Fig. 3b). The relative interaction strength of these peptides for Cn was determined by a competition assay. The fraction of Cn remaining bound is plotted as a histogram in Fig. 3b (Lower). The point mutations that have the strongest effect correlate well with those suggested by NMR to be most important for the binding (e.g., Y386, L387, and V389). The two proline residues in the peptide, which are invisible in HSQC spectra, also seem to be important for the interaction with Cn, based on this mutational study.
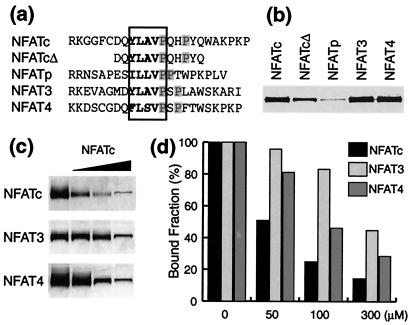
The sequence of CnBP-B is only moderately conserved among the known NFAT isoforms (Fig. 4a), thus raising the possibility of isoform-dependent differences in the interaction with Cn. To test this possibility, we fused the CnBP-B motifs from NFATp, NFAT3, and NFAT4 to GST and assayed their ability to pull down Cn from solution. As shown in Fig. 4b, the CnBP-B motifs from NFATc, NFAT3, and NFAT4 are effective at pulling down Cn, whereas that from NFATp is not. We also used a CnBP-B peptide to block the interaction of Cn with GST–CnBP-B fusions of the various NFAT isoforms (Fig. 4 c and d). The CnBP-B motif from NFATc was most effective at competing against itself and less effective at competing against the NFAT3 and NFAT4 CnBP-B. Taken together, these findings establish the following isoform-dependence of interaction strengths between Cn and the CnBP-B motif on NFAT: NFAT3 ≈ NFAT4 > NFATc >> NFATp.
Figure 4.
Isoform dependence of the interaction of CnBP-B with Cn. (a) Amino acid sequence of peptides corresponding to the CnBP-B region from the four known NFAT isoforms. NFATcΔ is a truncated version of the NFATc peptide. The “core” binding sequence is boxed, and the prolines that are critical for binding, as judged from Fig. 3b, are highlighted in gray. (b) NFAT peptides derived from the various isoforms were fused to the C terminus of GST and used in Cn pull-down assays. (c) The same NFAT baits as in b were used to pull down Cn in the presence of the NFATc CnBP-B peptide as competitor (50, 100, and 300 μM concentrations). (d) Quantification of the results in c analyzed by scanning and macbas.
Discussion
The intracellular partitioning of NFAT between the cytoplasm and nucleus is controlled by the presence or absence of phosphate groups on Ser/Thr residues in the N-terminal regulatory domain of the protein. The mechanism by which phosphorylation controls nuclear trafficking of NFAT remains unknown, but has been suggested to involve phosphorylation-dependent conformational switching in the regulatory domain, which leads to masking or unmasking of the NLS and NES (20, 26, 28). This mechanistic analysis is complicated by the fact that the regulatory domain of NFAT not only undergoes changes in its covalent structure as the result of phosphorylation/dephosphorylation, but also interacts with at least five known proteins: the phosphatase Cn (17, 30, 39), the kinases PKA and GSK-3β (25), the nuclear import receptor importin-α (42, 43), and the nuclear export receptor CRM-1 (28). Here we have focused on the interaction between Cn and the regulatory domain of human NFATc.
Structural Implications.
The prevailing model for regulation of NFAT nuclear trafficking (20, 26, 28) predicts that the phospho and dephospho forms of the regulatory domain should possess detectable differences in conformation, especially in the region of the phosphorylation sites and the NLS. However, our data failed to reveal any such phosphorylation-dependent conformational switch, as the CD spectra of the phosphorylated and unphosphorylated regulatory domains of NFAT are virtually superimposable. Perhaps most significantly, the CD spectra of the regulatory domain clearly indicate it to be unfolded, irrespective of phosphorylation state. This lack of a stable fold explains the exceptionally high proteolytic susceptibility of the regulatory domain over its entire ≈400-aa length, a property that is again unaffected by phosphorylation (data not shown).
The absence of a defined three-dimensional structure for the NFAT regulatory domain has important implications for understanding the mechanism of regulated nuclear trafficking. First, it challenges the notion that the regulatory domain itself exhibits phosphorylation-dependent conformational switching, to which potential interacting partners respond. Proteins that lack defined tertiary structure are inherently dynamical and therefore are incapable of being frozen in discrete states. Second, the absence of folding makes intramolecular masking of the NLS by the nonadjacent Ser/Thr-phosphates unlikely. In a folded protein, elements widely separated in sequence space may end up close together in three-dimensional space, thus favoring their interaction entropically. However, because the NFAT regulatory domain does not fold on phosphorylation, intramolecular contacts between the NLS and Ser/Thr phosphates are strongly disfavored by a large entropic penalty.
The present analysis applies equally well to intramolecular masking of the NES and indeed provides a rationale for the recent observation that Cn is required to prevent nuclear export of NFAT4 mediated by CRM-1 (28). In the regulatory domain of NFAT4, the CnBP-A Cn docking site lies close to the reported NES, such that Cn and CRM-1 bind in a mutually competitive manner. Cn thus functions as the requisite masking protein to control nuclear export of NFAT4 and presumably for the other NFAT isoforms as well. Just as Cn is required to mask the NES from access to CRM-1, we reason it likely that a cytoplasmic partner protein is required to mask the NLS from access to importin-α. Cn would seem to be an obvious candidate, although the weakness of its binding to NFAT in the absence of Ca2+ (44) renders this scenario less likely. CRM1 also must be considered a candidate, because it is an established interaction partner and because it translocates to the cytoplasm along with NFAT (45). Other candidates include the reported NFAT kinases PKA, GSK-3β (25), casein kinase Iα, and MEKK1 (26). Of these, only casein kinase Iα has been shown thus far to associate stably with the regulatory domain of NFAT4 (26).
Interactions Between Cn and the NFAT Signaling Domain.
It is now well established that the hypophosphorylated form of NFAT remains stably associated with Cn in the nucleus of Ca2+-stimulated cells. This association serves not only to provide a high local concentration of phosphatase activity in the sustained presence of the Ca2+ stimulus, but is also essential for the physical masking of the NES (28) (see above for discussion). Intriguingly, evidence also suggests the interaction with Cn is required for transcriptional activation by the nuclear form of NFAT (39). Previous work has mapped the Cn interaction region in NFAT to a short peptide stretch (CnBP-A) in the N-terminal end of the regulatory domain (see also Fig. 2b). In the present work, we have identified a second Cn-interacting peptide stretch (CnBP-B) in the C-terminal end of the NFAT regulatory region. CnBP-B exhibits the properties expected of a Cn-interacting ligand: (i) the sequence of CnBP-B is found in all four NFAT isoforms; (ii) CnBP-B peptides derived from NFATc, NFAT3, and NFAT4 interact strongly and specifically with Cn in vitro; and (iii) deletion of CnBP-B was required, along with mutations in CnBP-A, to abolish the interaction of the NFATc regulatory domain with Cn in vivo. Evidence for a second Cn binding site in NFAT is also apparent in coimmunoprecipitation studies on NFAT4, which localized the interaction to residues 310–407, a region containing CnBP-B (residues 390–403) (39); however, a second study on NFAT4 failed to identify CnBP-B, because the analysis did not extend to the C-terminal end of the regulatory region (26). In the case of NFATp, CnBP-B has not been identified as a Cn interaction site despite extensive analysis, perhaps because its CnBP-B motif binds Cn weakly, unlike the corresponding peptide from the other NFAT isoforms (Fig. 4a).
Our findings that NFAT proteins contain two independent Cn-BP motifs (Fig. 4e) raise the question of the relative roles of CnBP-A and CnBP-B. In this regard, it is noteworthy that the wild-type CnBP-A peptide alone from NFATp effectively blocks the interaction of Cn with NFATp both in vitro and in vivo (30). On the other hand, this same peptide gives only partial competition against the Cn interaction with NFATc or NFAT4 in vitro, and this cross-isoform competition has not been analyzed in vivo (30, 44). We find that neither the CnBP-A nor CnBP-B peptide alone from NFATc blocks the Cn⋅NFATc interaction, but both peptides together block effectively (Fig. 2c). These data are consistent with a model wherein Cn binds cooperatively to both CnBP-A and CnBP-B, but the strength of the interaction at each site varies among the individual isoforms. For instance, the CnBP-A sequence in NFATp is atypically strong and the CnBP-B sequence in this isoform atypically weak, as compared with the other NFAT isoforms, in which the binding energetics are more equally distributed between the two motifs. The variations in binding strength at the individual CnBP sites are integrated on the surface of Cn, resulting in unique thermodynamic and kinetic behavior for each isoform. These fundamental physical differences create a mechanism for differential responses to Ca2+ stimulation among NFAT isoforms.
There exists a great deal of interest in small-molecule agents that selectively target the Cn⋅NFAT interaction, while leaving unaffected the interaction of Cn with its other substrates. The wild-type NFATp CnBP-A peptide and an affinity-optimized derivative of it potently inhibit the Cn-dependent functions of NFAT, but the Cn activity on other proteins and non-NFAT-dependent promoters (30, 36). These results have raised the prospect that drugs targeting the CnBP-A docking site on Cn might act as selective immunosuppressants. In a similar vein, it is to be expected that effective competition at the CnBP-B docking site also will result in immunosuppression. However, given that CnBP-B and CnBP-A docking sites on Cn are distinct, targeting of each should produce distinct biological and pharmacologic responses.
Acknowledgments
We thank Susan Taylor and Peter Roach for gifts of kinases and kinase expression constructs, Jerry Crabtree for sharing results on the identity of the NFAT kinases before publication and for the SEAP reporter construct, and members of the Verdine lab, especially Li Jing Sun and Pei Zhou, for advice and assistance. This work was supported by the Hoffmann-LaRoche/Harvard Institute for Chemistry and Medicine.
Abbreviations
- NFAT
nuclear factor of activated T cells
- NFAT-h
NFAT homology region
- Cn
calcineurin
- NLS
nuclear localization signal
- NES
nuclear export signal
- CnBP-A
Cn binding peptide A
- CnBP-B
Cn binding peptide B
- PKA
protein kinase A
- GSK-3β
glycogen synthase kinase-3β
- GST
glutathione S-transferase
- HSQC
heteronuclear single quantum correlation
- SEAP
secreted alkaline phosphatase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Crabtree G R, Clipstone N A. Annu Rev Biochem. 1994;63:1045–1083. doi: 10.1146/annurev.bi.63.070194.005145. [DOI] [PubMed] [Google Scholar]
- 2.Crabtree G R. Cell. 1999;96:611–614. doi: 10.1016/s0092-8674(00)80571-1. [DOI] [PubMed] [Google Scholar]
- 3.Rao A, Luo C, Hogan P G. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 4.Northrop J P, Ho S N, Chen L, Thomas D J, Timmerman L A, Nolan G P, Admon A, Crabtree G R. Nature (London) 1994;369:497–502. doi: 10.1038/369497a0. [DOI] [PubMed] [Google Scholar]
- 5.McCaffrey P G, Luo C, Kerppola T K, Jain J, Badalian T M, Ho A M, Burgeon E, Lane W S, Lambert J N, Curran T, et al. Science. 1993;262:750–754. doi: 10.1126/science.8235597. [DOI] [PubMed] [Google Scholar]
- 6.Hoey T, Sun Y L, Williamson K, Xu X. Immunity. 1995;2:461–472. doi: 10.1016/1074-7613(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 7.Masuda E S, Naito Y, Tokumitsu H, Campbell D, Saito F, Hannum C, Arai K I, Arai N. Mol Cell Biol. 1995;15:2697–2706. doi: 10.1128/mcb.15.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez-Rodriguez C, Aramburu J, Rakeman A S, Rao A. Proc Natl Acad Sci USA. 1999;96:7214–7219. doi: 10.1073/pnas.96.13.7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xanthoudakis S, Viola J P, Shaw K T, Luo C, Wallace J D, Bozza P T, Luk D C, Curran T, Rao A. Science. 1996;272:892–895. doi: 10.1126/science.272.5263.892. [DOI] [PubMed] [Google Scholar]
- 10.de la Pompa J L, Timmerman L A, Takimoto H, Yoshida H, Elia A J, Samper E, Potter J, Wakeham A, Marengere L, Langille B L, et al. Nature (London) 1998;392:182–186. doi: 10.1038/32419. [DOI] [PubMed] [Google Scholar]
- 11.Ranger A M, Grusby M J, Hodge M R, Gravallese E M, de la Brousse F C, Hoey T, Mickanin C, Baldwin H S, Glimcher L H. Nature (London) 1998;392:186–190. doi: 10.1038/32426. [DOI] [PubMed] [Google Scholar]
- 12.Oukka M, Ho I C, de la Brousse F C, Hoey T, Grusby M J, Glimcher L H. Immunity. 1998;9:295–304. doi: 10.1016/s1074-7613(00)80612-3. [DOI] [PubMed] [Google Scholar]
- 13.Jain J, Burgeon E, Badalian T M, Hogan P G, Rao A. J Biol Chem. 1995;270:4138–4145. [PubMed] [Google Scholar]
- 14.Jain J, McCaffrey P, Miner Z, Kerppola T K, Lambert J N, Verdine G L, Curran T, Rao A. Nature (London) 1993;365:352–355. doi: 10.1038/365352a0. [DOI] [PubMed] [Google Scholar]
- 15.Molkentin J D, Lu J R, Antos C L, Markham B, Richardson J, Robbins J, Grant S R, Olson E N. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musaro A, McCullagh K J, Naya F J, Olson E N, Rosenthal N. Nature (London) 1999;400:581–585. doi: 10.1038/23060. [DOI] [PubMed] [Google Scholar]
- 17.Shibasaki F, Price E R, Milan D, McKeon F. Nature (London) 1996;382:370–373. doi: 10.1038/382370a0. [DOI] [PubMed] [Google Scholar]
- 18.Luo C, Shaw K T, Raghavan A, Aramburu J, Garcia-Cozar F, Perrino B A, Hogan P G, Rao A. Proc Natl Acad Sci USA. 1996;93:8907–8912. doi: 10.1073/pnas.93.17.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loh C, Shaw K T, Carew J, Viola J P, Luo C, Perrino B A, Rao A. J Biol Chem. 1996;271:10884–10891. doi: 10.1074/jbc.271.18.10884. [DOI] [PubMed] [Google Scholar]
- 20.Beals C R, Clipstone N A, Ho S N, Crabtree G R. Genes Dev. 1997;11:824–834. doi: 10.1101/gad.11.7.824. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Farmer J D, Lane W S, Friedman J, Weissman I, Schreiber S L. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 22.Griffith J P, Kim J L, Kim E E, Sintchak M D, Thomson J A, Fitzgibbon M J, Fleming M A, Caron P R, Hsiao K, Navia M A. Cell. 1995;82:507–522. doi: 10.1016/0092-8674(95)90439-5. [DOI] [PubMed] [Google Scholar]
- 23.Kissinger C R, Parge H E, Knighton D R, Lewis C T, Pelletier L A, Tempczyk A, Kalish V J, Tucker K D, Showalter R E, Moomaw E W, et al. Nature (London) 1995;378:641–644. doi: 10.1038/378641a0. [DOI] [PubMed] [Google Scholar]
- 24.Hemenway C S, Heitman J. Cell Biochem Biophys. 1999;30:115–151. doi: 10.1007/BF02737887. [DOI] [PubMed] [Google Scholar]
- 25.Beals C R, Sheridan C M, Turck C W, Gardner P, Crabtree G R. Science. 1997;275:1930–1934. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- 26.Zhu J, Shibasaki F, Price R, Guillemot J C, Yano T, Dotsch V, Wagner G, Ferrara P, McKeon F. Cell. 1998;93:851–861. doi: 10.1016/s0092-8674(00)81445-2. [DOI] [PubMed] [Google Scholar]
- 27.Klemm J D, Beals C R, Crabtree G R. Curr Biol. 1997;7:638–644. doi: 10.1016/s0960-9822(06)00290-9. [DOI] [PubMed] [Google Scholar]
- 28.Zhu J, McKeon F. Nature (London) 1999;398:256–260. doi: 10.1038/18473. [DOI] [PubMed] [Google Scholar]
- 29.Wesselborg S, Fruman D A, Sagoo J K, Bierer B E, Burakoff S J. J Biol Chem. 1996;271:1274–1277. doi: 10.1074/jbc.271.3.1274. [DOI] [PubMed] [Google Scholar]
- 30.Aramburu J, Garcia-Cozar F, Raghavan A, Okamura H, Rao A, Hogan P G. Mol Cell. 1998;1:627–637. doi: 10.1016/s1097-2765(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 31.Mondragon A, Griffith E C, Sun L, Xiong F, Armstrong C, Liu J O. Biochemistry. 1997;36:4934–4942. doi: 10.1021/bi9631935. [DOI] [PubMed] [Google Scholar]
- 32.Dadlez M, Kim P S. Biochemistry. 1996;35:16153–16164. doi: 10.1021/bi9616054. [DOI] [PubMed] [Google Scholar]
- 33.Belshaw P J, Spencer D M, Crabtree G R, Schreiber S L. Chem Biol. 1996;3:731–738. doi: 10.1016/s1074-5521(96)90249-5. [DOI] [PubMed] [Google Scholar]
- 34.Crowley M P, Reich Z, Mavaddat N, Altman J D, Chien Y. J Exp Med. 1997;185:1223–1230. doi: 10.1084/jem.185.7.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fiol C J, Williams J S, Chou C H, Wang Q M, Roach P J, Andrisani O M. J Biol Chem. 1994;269:32187–32193. [PubMed] [Google Scholar]
- 36.Aramburu J, Yaffe M B, Lopez-Rodriguez C, Cantley L C, Hogan P G, Rao A. Science. 1999;285:2129–2133. doi: 10.1126/science.285.5436.2129. [DOI] [PubMed] [Google Scholar]
- 37.Coghlan V M, Perrino B A, Howard M, Langeberg L K, Hicks J B, Gallatin W M, Scott J D. Science. 1995;267:108–111. doi: 10.1126/science.7528941. [DOI] [PubMed] [Google Scholar]
- 38.Shibasaki F, Kondo E, Akagi T, McKeon F. Nature (London) 1997;386:728–731. doi: 10.1038/386728a0. [DOI] [PubMed] [Google Scholar]
- 39.Masuda E S, Liu J, Imamura R, Imai S I, Arai K I, Arai N. Mol Cell Biol. 1997;17:2066–2075. doi: 10.1128/mcb.17.4.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J O, Russo A A, Pavletich N P. Nature (London) 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- 41.Jeon Y H, Negishi T, Shirakawa M, Yamazaki T, Fujita N, Ishihama A, Kyogoku Y. Science. 1995;270:1495–1497. doi: 10.1126/science.270.5241.1495. [DOI] [PubMed] [Google Scholar]
- 42.Gorlich D. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ullman K S, Powers M A, Forbes D J. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 44.Garcia-Cozar F J, Okamura H, Aramburu J F, Shaw K T Y, Pelletier L, Showalter R, Villafranca E, Rao A. J Biol Chem. 1998;273:23877–23883. doi: 10.1074/jbc.273.37.23877. [DOI] [PubMed] [Google Scholar]
- 45.Kehlenbach R H, Dickmanns A, Gerace L. J Cell Biol. 1998;141:863–874. doi: 10.1083/jcb.141.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]