Abstract
Sp22D, a modular serine protease encompassing chitin binding, low density lipoprotein receptor, and scavenger receptor cysteine-rich domains, was identified by molecular cloning in the malaria vector, Anopheles gambiae. It is expressed in multiple body parts and during much of development, most intensely in hemocytes. The protein appears to be posttranslationally modified. Its integral, putatively glycosylated form is secreted in the hemolymph, whereas a smaller form potentially generated by proteolytic processing is associated with the tissues. Bacterial challenge or wounding result in low-level RNA induction, but the protein does not bind to bacteria, nor is its processing affected by infection. However, Sp22D binds to chitin with high affinity and undergoes transient changes in processing during pupal to adult metamorphosis; it may respond to exposure to naked chitin during tissue remodeling or damage.
Like all insects, the mosquito Anopheles lacks adaptive immunity but possesses an efficient innate defense system, much reminiscent of vertebrate innate immunity (1). During its life cycle within Anopheles, the malaria parasite Plasmodium is exposed to mosquito defense mechanisms. Indeed immune mechanisms of the Anopheles vector are activated during critical stages of Plasmodium development in the gut, hemocoel, and salivary glands (2, 3); their variations may underlie some cases of natural refractoriness of the vector to the parasite (4).
The molecular mechanisms for early recognition of invading pathogens by the innate immune system of the mosquito are of special interest. In vertebrates, an attractive mechanism for selective recognition has been proposed (5) based on receptors that bind to specific molecular patterns of pathogen surfaces. However, to date rather little is known about recognition and the early events of immune activation in invertebrates. In general, regulated serine protease cascades are implicated, as in the activation of the Toll pathway in Drosophila (6). In the horseshoe crab Limulus, cascades of autocatalytically activated proteases are triggered by lipopolysaccharide or glucans and activate the hemolymph clotting system, which functions both in coagulation and in the defense against pathogens (7). Furthermore, the defensive prophenoloxidase cascade in Manduca sexta is initiated by proteolytic processing (8). Finally, in vertebrates the role of serine proteases in the stimulation of both the alternative and the classical pathways of complement activation is well documented (9). It is important to recognize that immune regulatory pathways may be multifunctional, playing roles in development or tissue remodeling and repair, as well as in molecular immunity per se. The Drosophila Toll pathway is a case in point.
In a search of the Drosophila expressed sequence tag databases for potential pattern recognition modules, we (M.L.) noted an intriguing sequence (CK 0198) that encompassed both a serine protease and the adhesive domain known as SRCR (scavenger receptor cysteine rich). Studies on this gene, named GRAAL, will be reported elsewhere (A. I. Munier, H. Caporilla, R. Lanot, D. Zachary, R. Medzhitov, C. A. Janeway, and M.L., unpublished work). Here we report the cloning and characterization of Sp22D, an Anopheles homologue of GRAAL. The same gene has been cloned independently by others (10) and has been named Sp22D (serine protease from chromosomal division 22D). We have confirmed the reported chromosomal location of this gene and adopted its prior name. It encodes a modular protein with a trypsin-like domain linked to a tandem array of low density lipoprotein receptor-like (LDLr) and SRCR domains, mucin-like repeats and two chitin-binding domains (CBDs). Our immunochemical and immunocytochemical studies revealed that in the adult the protein is mainly produced in the hemocytes and secreted in the hemolymph. Using immunoreactive hemocyte-like cell lines, we initiated biochemical studies on this molecule. Taken together our results suggest putative roles of Sp22D in tissue remodeling and guide our efforts to correlate specific functions to its complex structure.
Materials and Methods
Cell lines were cultured and mosquito colonies were maintained and infected with Plasmodium berghei as described (2, 11). Four-day-old 4a r/r adult females were wounded by a thin sterile glass needle or infected by pricking (2) with an overnight culture of Escherichia coli strain 1106 and Micrococcus luteus strain A270.
cDNA Cloning of Sp22D.
Using degenerate primers for SRCR domains (5′-TGGGGIASTRTKTGYGAYG and 5′-CAGKKSACSCCIRCRTCYTC, I = inosine), a 300-bp PCR product was amplified from a lambda ZAP express (Stratagene) cell line cDNA library, gel-purified, cloned into a Topo-TA vector pCR-2.1 (Invitrogen), confirmed by sequencing, and used to screen that library and one from fourth instar larvae. Four partial cDNA clones were sequenced, and the missing 5′ sequence was obtained by 5′ rapid amplification of cDNA ends with CLONTECH Marathon Kit by using the specific primer 5′-CATCATCGTGCTCAGCTTCGTTGTGTTC.
Expression Analysis by Reverse Transcription–PCR (RT-PCR).
Total RNA from mosquitoes and cell lines was isolated with RNAid PLUS Kit (Bio 101) and used for both standard Northern and RT-PCR analysis. First-strand cDNA synthesis using 2 μg of RNA was performed as described (11). The Sp22D primers 5′-GGAGCTGGACATCTTCATCGAG, 5′-CTTCGGCATACACTGTCC were used at 20 pmol in 50-μl reactions with Amplitaq (Perkin–Elmer), to amplify a 246-bp product through seven cycles (45 s at 94°C, 45 s at 60°C, and 30 s at 72°C). During a pause at 72°C, primers complementary to the ribosomal protein S7 gene were added. The reaction was allowed to proceed for additional cycles (21 for Fig. 2, 18 for Fig. 5). The same thermocyclic program was used for defensin expression analysis with three initial and 18 subsequent amplification cycles. After electrophoresis on 1.5% agarose, gels were stained with the sensitive SYBR green dye (Molecular Probes) for 45 min and analyzed with a fluorimager (Fuji).
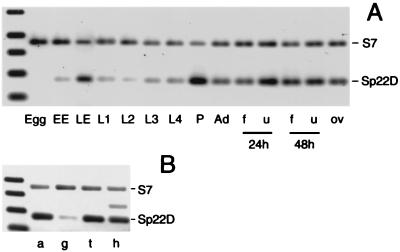
Figure 2.
Developmental and spatial expression profiles of Sp22D mRNA relative to an internal control of ribosomal protein S7 mRNA. (A) RNA of eggs collected within 30 min after oviposition (Egg), early embryos (EE, 18 h), late embryos (LE, 42 h), larvae of first (L1), second (L2), third (L3) and fourth (L4) instar, pupae (P), 4-day-old females (Ad), and females 1 day postoviposition (ov), or 24 and 48 h after feeding on blood (f) or control sugar solution (u). (B) Levels of transcript in dissected abdomens (a), gut (g), thorax carcass with legs (t), and head (h). The extra band in h is caused by genomic DNA contamination.
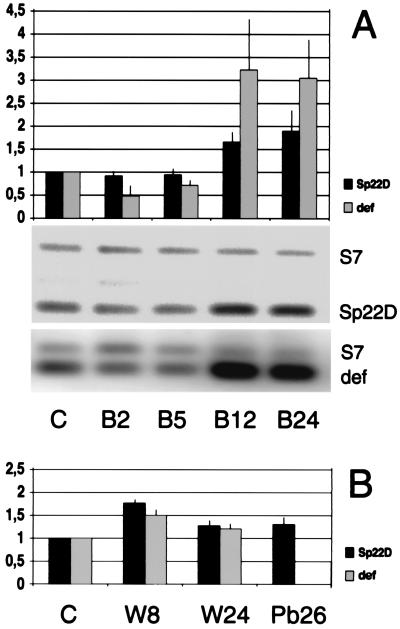
Figure 5.
Expression of Sp22D and the immune marker defensin (def) after bacterial challenge or wounding. RNAs from the indicated sources were used as RT-PCR templates standardized against S7 and normalized against unchallenged controls (C). (A) Females 2, 5, 12, and 24 h after bacterial challenge (B2, B5, B12, B24). (B) Females 8 and 24 h after wounding (W8 and W24) or 26 h after a malarial blood meal (Pb26). The mean and standard error of triplicate experiments are shown.
Generation of Sp22D Antiserum.
The sequence encoding the LDLr and SRCR domains (bp 1175 to 3085) was subcloned into a pQE-31 vector (Qiagen, Chatsworth, CA). The recombinant peptide produced in E. coli was purified on a Ni-nitrilotriacetic acid Agarose column (Qiagen) and run on an SDS/PAGE gel. The protein band was excised, ground to a fine powder, and resuspended in 2 ml of PBS. Two rats were immunized with 100 μg protein in Ribi Adjuvant (RAS, Ribi Immunochem), and boosted every third week with 50–70 μg antigen until final bleeding.
Immunoblotting.
Adults were dissected and/or homogenized in protein extraction buffer (20 mM Hepes, pH 7.5/30 mM NaCl) containing a protease inhibitor mixture (complete, Boehringer Mannheim). After the proboscis of mosquitoes were clipped, a small droplet of hemolymph was promptly collected in a pipette tip filled with the same medium. All extractions were performed on ice. The extracts, or supernatants of cell lines 4a-3A (3A) and 4a-3B (3B) were mixed with SDS-loading buffer and displayed on 6–8% SDS/PAGE gels. Separated polypeptides were transferred to Hybond-P membranes (Amersham Pharmacia), blocked with 5% dry milk, and incubated with the Sp22D antiserum (1:3,000). Bound antibodies were detected by an anti-rat IgG conjugated to horseradish peroxidase (Sigma) by using a chemilluminecence kit (ECL detection system, Amersham Pharmacia).
Immunofluorescence.
Hemolymph perfusion and cell line staining were carried out as described (11). For whole mounts, dissected tissues were fixed for 45 min in ice-cold 4% paraformaldehyde, washed twice, blocked for 1 h (1% BSA, 5% normal goat serum, 0.2% Triton X-100), all in PBS, and subsequently incubated overnight at 4°C (Ab dilutions 1:1,000 Sp22D; 1:8,000 defensin; 1:500 histones MAB052, Chemicon) and treated with secondary antibodies (The Jackson Laboratory, dilution 1:1,000). For sectioning, mosquito bodies were freed of appendages, fixed 3 h in ice-cold PBS, 4% paraformaldehyde, dehydrated, incubated overnight in a 1:1 paraplast/xylene emulsion at 42°C, changed to fresh paraplast (Sigma) at 60°C, and embedded at 4°C. Sagittal sections (10 μm) were taken by using a microtome and stretched on poly-l-lysine-coated slides. All samples were examined with a Zeiss LSM 510 confocal microscope.
Binding Assays.
Chitin beads (NEB 6651L) in PBS were incubated 60 min with four matrix volumes cell line supernatants, packed into a column, and washed with 10 matrix volumes PBS. Fractions were eluted with 2.5 matrix volumes of 50 mM sodium acetate and 50 mM glycine pH 2.0 and subjected to immunoblotting.
Results
Domain Organization of Sp22D.
The Sp22D cDNA encompasses a 3,969-bp ORF, encoding a protein with a predicted molecular mass of 146.8 kDa (Fig. 1). It begins with 24 residues characteristic of a signal peptide and ends with a C-terminal serine protease domain of the trypsin family including the characteristic catalytic triad. In between, no evident transmembrane domain exists (suggesting that the protein is secreted), but several putative adhesive domains are embedded in repetitive sequences. Thus, toward the N terminus two predicted CBDs are embedded in a region rich in Gln residues. Next, a proline-rich region is interrupted by three polythreonine stretches. Between that region and the protease domain, two LDLr and SRCR domains are found in alternating arrangement. Polythreonine stretches are commonly found in mucin-like domains and are known to serve as O-glycosylation sites. Potential N-glycosylation sites also are revealed by profilescan analysis at positions 570, 825, 850, 971, 996, 1201, and 1314 of the deposited protein sequence. Several dibasic potential proteolytic cleavage sites are located in the central half of the protein sequence (see arrowheads in Fig. 3E). Importantly such a site is located between the protease domain and the rest of the molecule.
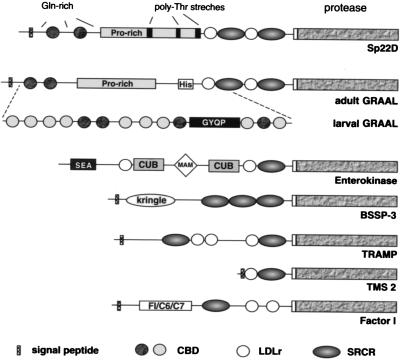
Figure 1.
Domain organization of Sp22D and other serine proteases linked to SRCR domains. Sp22D is the ortholog of the Drosophila adult GRAAL isoform; larval GRAAL is derived from the same gene by alternative splicing in the region indicated (A. I. Munier, H. Caporilla, R. Lanot, D. Zachary, R. Medzhitov, C. A. Janeway, and M.L., unpublished work). CBDs are present only in Sp22D and GRAAL and include a second type in larval GRAAL (lightly shaded circles).
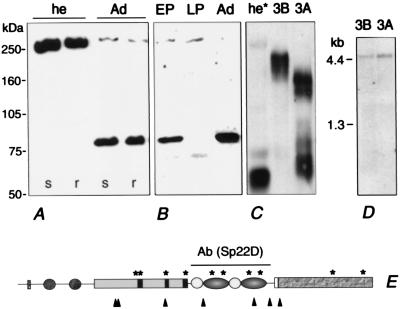
Figure 3.
Sp22D polypeptides detected by immunoblotting. (A) Hemolymph (he) of 4-day-old mosquitoes of both susceptible (s) and refractory (r) strains; (Ad) whole extracts of adult mosquitoes. (B) Early pupal (EP), late pupal (LP), and adult (Ad) extracts. (C) Supernatants (conditioned medium) of cell lines 3A and 3B; (he*) hemolymph of freshly emerged adults. (D) RNA blot displaying a single ca. 4.5-kb transcript in cell lines 3A and 3B. (E) Schematic representation of the Sp22D protein indicating the region against which a specific rat antiserum was raised Ab(Sp22D), the potential proteolytic cleavage sites (arrowheads), and the possible glycosylation sites (*).
The various domains were characterized by similarity values obtained by blast analysis. The protease domain shares significant similarity with the cognate fruitfly homologue GRAAL (GenBank accession no. AJ251803), with mammalian trypsin-like proteases of the coagulation cascade (kallikrein, GenBank accession no. NP036857; factor XII, GenBank accession no. Q04962) and the inflammatory responses (human airway trypsin-like protease, GenBank accession no. NP004253), and with proteases associated with the nervous system (BSSP-3 GenBank accession no. 008762; neurotrypsin, GenBank accession no. NP003610).
The SRCR domains of Sp22D belong to group A, characterized by positional conservation of six cysteines (12), and are remarkably conserved (data not shown). Several proteins bearing SRCR-linked serine protease domains have been reported (Fig. 1). These are either secreted or membrane-bound molecules with diverse biological roles in development (13) and immunity (14). Intriguing features of Sp22D are the CBD motifs that share maximum similarity with AgAper-1, a mosquito midgut-specific molecule with peritrophic matrix binding properties (15). They are also similar to the motifs of other proteins that interact with chitin, including GASP, a molecule expressed in the embryonic tracheae of Drosophila (16), and macroparasite chitinases (e.g., Brugia).
Temporal and Spatial RNA Expression Profiles.
An RT-PCR-based approach assisted by fluorimaging has been used to determine the expression levels of Sp22D throughout development (Fig. 2A). Sp22D transcripts are absent from freshly laid eggs, appear in early embryos, and increase substantially at later embryonic stages. Expression levels are weaker throughout all four larval stages, but a second transient and robust peak becomes evident during the pupal stage. Moderately high RNA levels are detected in the adults, with a slight decrease when females feed on blood.
The distribution of Sp22D transcripts in dissected adult body parts was similarly analyzed. As shown in Fig. 2B, the transcripts are abundant in the head (h), abdomen (a), and thorax (t), but rare in midguts (g). This spatial pattern could be attributed to hemocytes, which are distributed throughout the body but are rarely seen attached to the gut.
Alternative Forms of Sp22D Protein Attributable to Postranslational Modifications.
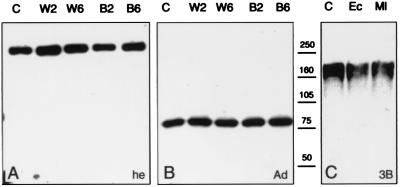
To monitor the protein itself, a polyclonal antiserum was raised against a His-tagged recombinant polypeptide encopassing the tandem array of LDLr and SRCR domains (highlighted in Fig. 3E). The specificity of the antiserum was established in immunoblots of recombinant polypeptide and unrelated His-tagged proteins (data not shown). Immunoblot analysis of two different parasite-susceptible and refractory mosquito strains (Fig. 3A, s and r) reveal the presence of a single 250-kDa band in the hemolymph of adults, further supporting the specificity of the antiserum (Fig. 3A, he). This apparent size is higher than predicted for the integral polypeptide; however, glycosylation (Fig. 3E, *) could well explain this disparity. In adult protein extracts (which include hemolymph), a strong band of 80 kDa appears together with the 250-kDa component that is much fainter (Fig. 3A, Ad). In extracts derived from dissected and rinsed adult tissues, the 80-kDa band is ubiquitous, whereas the 250-kDa band is absent (data not shown). These results suggest that the 250-kDa band, presumably authentic posttranslationally modified full-length protein, is abundantly secreted in the hemolymph and that tissues throughout the body contain a derivative, presumably generated by limited proteolysis.
One potential origin of a circulating hemolymph protein is hemocytes. Therefore we checked for the presence of Sp22D protein in two A. gambiae cell lines (4a-3A and 4a-3B, henceforth 3A and 3B) which have characteristics of hemocytes (11). Using the same antiserum, a complex pattern of polypeptides was associated with the lines (Fig. 3C). In supernatants (conditioned medium) of line 3B, Sp22D is represented by multiple components corresponding to ca. 210 kDa, whereas in supernatants of line 3A smaller products are seen at two size ranges, approximately 160–170 and 70–80 kDa, respectively. The 250-kDa protein is detected only in the 3A and 3B cell pellets (data not shown). Because RNA blot analysis demonstrates the presence of only one transcript of the expected size, identical in both cell lines (Fig. 3D), and recalling the presence of widely distributed dibasic potential cleavage sites (Fig. 3E) in Sp22D, we suggest that different stages of proteolytic processing account for these differences. Indeed in supernatants of prolonged cultures of both cell lines the relative concentration of smaller polypeptides increases at the expense of larger ones (data not shown). Although we cannot exclude the possibility of differences in glycosylation in the two cell lines, we are confident that at least moieties smaller than 147 kDa (the size of the ORF) are products of limited proteolysis.
Enhanced Processing During Metamorphosis.
In extracts of early pupae the presumed proteolytic pattern of Sp22D, with a dominant 80-kDa component, is similar to that observed in adults (Fig. 3B). Interestingly, in extracts derived from late pupae the 80-kDa band is absent, whereas a new band approximately 65 kDa in size and the 250-kDa protein are detected; the total amount of Sp22D is much lower than in early pupae or adults (Fig. 3B). Enhanced processing of Sp22D also is seen in the hemolymph of freshly emerged adults (Fig. 3C, he*). Here, in addition to a faint 250-kDa band and a 65-kDa band, a strong diffuse band of lower size is present. These observations support the notion of enhanced dynamics and suggest some role for Sp22D during the metamorphic transition from the pupal to the adult stage.
The Main Expression Sites in the Adult Mosquito Are Hemocytes.
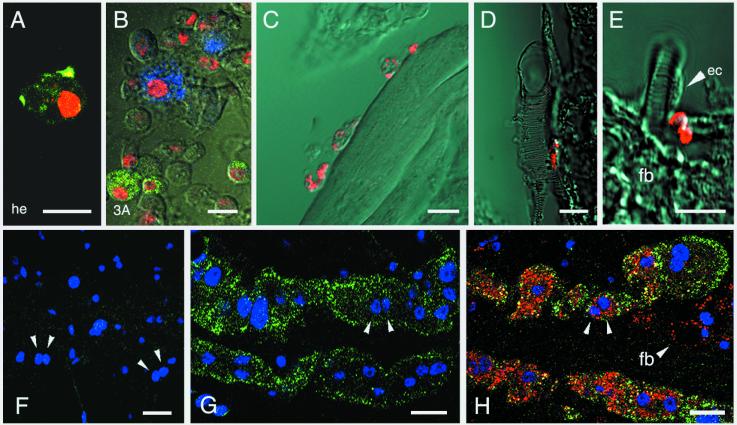
Hypothesis that hemocytes are a major site of Sp22D production is supported by the evidence that the authentic protein is abundant in hemolymph but not in tissues, and that Sp22D is produced and secreted by hemocyte-like cell lines. To test the hypothesis directly, we tested for Sp22D protein in hemocytes circulating in the hemolymph of perfused adult mosquitoes. Loosely attached fat body cells, accidentally present in the perfusate, could be discriminated by their characteristic presence of lipid vacuoles (11). We were able to detect granular staining in individual hemocytes, typical for a secreted protein (Fig. 4A). The same granular pattern was observed in up to 5% of the cells of the hemocyte-like lines 3A and 3B (Fig. 4B). In the 3A line, in which high levels of constitutive expression of the immune marker defensin have been reported (11), a comparable fraction of the cells stained with a specific antibody directed against the antibacterial peptide, defensin. However, the two cell populations were completely distinct: we were not able to detect a single cell in which Sp22D and defensin colocalized (Fig. 4B). Upon bacterial challenge, both lines 3A and 3B responded with a massive degranulation of defensin but not Sp22D (data not shown). Thus Sp22D is expressed by a distinct population of hemocyte-like cells and is not affected by immune challenge.
Figure 4.
Immunolocalization of Sp22D protein in adult tissues and cell lines. Nuclei are counterstained with anti-histone mAb (red in A and B, blue in F–H). (A) Sp22D staining (green) in a perfused hemocyte from a female mosquito. (B) Two distinct hemocyte-like subpopulations of cell line 3A expressing Sp22D (green) and defensin (blue). (C–E) Sections of paraffin-embedded adult mosquitoes showing Sp22D (red) (C) in hemocytes attached to muscle, (D) in a single hemocyte attached to a trachea, (E) in minute tracheal associated cells but not in tracheal epithelial cells (ec) or in fat body (fb). (F–H) Whole-mount triple staining of dissected abdomens showing two characteristic rows of binucleated pericardial cells. Arrows highlight pairs of nuclei, counterstained in blue; Sp22D stains green and defensin red. (F) Control treated with Sp22D and defensin preimmune sera. (G) An unchallenged mosquito showing constitutive presence of Sp22D and absence of defensin. (H) Accumulation of defensin as well as Sp22D in a bacterially challenged mosquito; defensin but not Sp22D is also detected in fat body cells (fb, arrowhead; note the unstained large lipid vacuole characteristic of this tissue). In B–E, the fluorescence channel and Nomarski optics of a Zeiss LSM 510 confocal microscope were combined. (Scale bars = 10 μm.)
To extend our studies on tissue localization of the Sp22D protein, whole-mount and paraffin-embedded sections of adult mosquitoes were tested by immunostaining. We detected strongly stained hemocytes, circulating or attached to tissues such as muscles (Fig. 4C) and tracheae (Fig. 4D). Whenever hemocytes occurred in clusters, not all of them were Sp22D positive (data not shown). We also noticed staining on tracheae and tracheoles but could not attribute it to hemocytes or epithelial cells lining the tracheae, some of which were definitely unstained (Fig. 4E, ec). We were not able to observe midgut and fat body staining (Fig. 4E, fb and data not shown). However, the strongest staining apart the hemocytes was observed in pericardial cells (Fig. 4 F–H). These are large binucleated cells found along the dorsal sinus, which line up in two rows along the outer surface of the tubular heart, and are in direct contact with hemolymph (17, 18). Sp22D protein is present in the cytoplasm of pericardial cells both in naïve and bacterially challenged mosquitoes, in contrast with defensin, which starts to accumulate in these cells several hours after the bacterial challenge (Fig. 4 G and H).
Expression upon Bacterial Challenge, Wounding, and Parasite Infection.
Because of the suggestive sequence features of the protein and its association with hemocytes, we investigated its potential involvement in immunity by assaying through RT-PCR changes in the levels of Sp22D transcript upon immune challenge. Mosquitoes were pricked with a mixture of Gram-positive and negative bacteria or were infected with the rodent malaria parasite P. berghei. A 1.7-fold induction was observed by 12 h after bacterial challenge, concomitant with robust induction of defensin (Fig. 5A). Aseptic wounding yielded similar, albeit more transient, induction (Fig. 5B). Induction rates of Sp22D upon midgut invasion by the Plasmodium parasite were assayed 26 h after an infective blood meal. Relative to control mosquitoes fed on uninfected mice, up-regulation was at best marginal (Fig. 5B, Pb26).
We considered that altered processing might be a more vigorous and immediate mode of response of Sp22D. However, neither wounding nor bacterial challenge significantly affected the processing of Sp22D in vivo (Fig. 6 A and B). Absence of processing also was observed when cell line supernatants were incubated with heat-inactivated bacteria (Fig. 6C, Ec and Ml).
Figure 6.
Lack of Sp22D processing after wounding (W) and bacterial challenge (B) relative to controls (C). Immunoblots were performed with protein extracts from hemolymph (A) and adults (B) at 2 and 6 h after treatment, and from cell-free conditioned supernatants of cell line 3B (C) incubated with Gram-positive or -negative bacteria (Ec and Ml, respectively).
Sp22D Binds to Chitin but Not to Bacteria.
Because binding to bacteria has been reported for other SRCR domains (19), we tested whether Sp22D shows similar binding. Both Gram-positive (M. luteus) and negative (E. coli) bacteria were incubated with supernatants of cell lines 3A and 3B and were washed at increasing salt concentration and decreasing pH. Immunoblotting showed that none of the eluates contained Sp22D (data not shown).
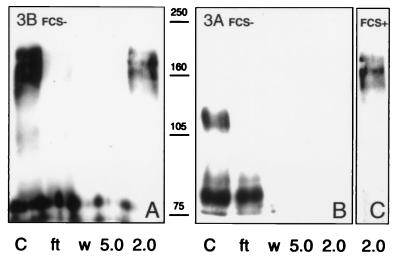
To test whether the two CBDs confer chitin binding activity, chitin beads were incubated with supernatants of cell line 3B cultured in the absence of FCS to avoid the presence of BSA. The beads were washed with PBS, and adsorbed material then was eluted by pH reduction and analyzed by immunoblotting (Fig. 7A). The larger Sp22D polypeptides present in the 3B/FCS− supernatants were in the range of 160 kDa, and they were found to adhere to the beads tightly, eluted only at pH 2.0 (Fig. 7A). Absence of FCS further reduced the apparent size of Sp22D components in the 3A supernatants. Neither the largest component of ca. 110 kDa nor other smaller products bind to chitin (Fig. 7B). However, supernatants of 3A cultured in the presence of FCS showed Sp22D polypeptides comparable in size to those of 3B/FCS−, and these were able to bind to the chitin beads (Fig. 7C). In summary, high, but not low, molecular mass Sp22D forms contain functional, high affinity CBDs.
Figure 7.
Immunoblots showing chitin binding of Sp22D from cell lines. 3B/FCS− (A), 3A/FCS− (B), and 3A/FCS+ (C), conditioned medium supernatants (lanes C) were mixed with chitin beads, which were washed and eluted at progressively lower pH (5.0 and 2.0); flow-through (ft) and PBS washes (w) also are shown. The 160- to 170-kDa polypeptides of 3B/FCS− and 3A/FCS+ samples bind tightly and are eluted from chitin only at pH 2.0. Smaller polypeptides, including all components of the 3A/FCS− samples, do not bind.
Discussion
Hemolymph is an important fluid that integrates the insect body plan and mediates many of its physiological responses, including systemic immunity. Sp22D, a modular adhesive protease produced by hemocytes and secreted in the hemolymph, is an attractive candidate to link proteolytic pathways that activate immune responses with the recognition of invading microorganisms. Although its transcript increases somewhat upon bacterial challenge, we observed that Sp22D does not bind to Gram-positive or -negative bacteria. However, we did detect an intriguing chitin binding property. Insect chitin is found in the exoskeleton, respiratory tracheal system, and peritrophic matrix but is insulated from the hemolymph by epithelial layers. Exposure of chitin to the hemolymph may signal unscheduled mechanical disruption of protective barriers during injury and pathogen intrusion or extensive tissue remodeling during development. Direct contact of the hemolymph with chitin has been reported in the pupa, because of degradation of the tracheal epithelium and the larval hypodermis (20). The demonstrated chitin binding activity of Sp22D, together with its RNA induction in the pupa and during wounding or immune challenge, lead us to suggest that constitutive Sp22D may serve as sentinel to detect exposed chitin, and then trigger appropriate physiological or developmental responses, including its own up-regulation. Chitin also is found on the surface of several invading agents, so it is therefore worth exploring whether Sp22D can bind to entomopathogenic fungi or nematodes.
Although most of the Sp22D protein is seen in hemocytes, only a fraction of these cells are positive, both within in vivo clusters and in the cell lines. Moreover, in the cell lines the Sp22D-positive cells are distinct from those that produce defensin. Heterogeneity in the circulating hemocytes has been reported in Drosophila (21, 22) and deserves further molecular analysis. An intriguing finding is the presence of both Sp22D and defensin in the pericardial cells, which are sessile and thought to play a role in homeostasis and detoxification of the insect hemolymph (17, 18). This raises the question whether their content of Sp22D and defensin protein reflects endogenous production or the uptake of these molecules from the hemolymph: ongoing in the case of Sp22D and acute for defensin when antimicrobial peptides exceed desirable levels.
Our biochemical studies on the Sp22D protein unveiled extensive processing of the molecule, both in vivo and in the mosquito cell lines. A complex pattern of processing of Sp22D was predicted from the prevalence of putative proteolytic cleavage and glycosylation sites in the sequence and was detected by immunoblotting. Although processing is inhibited by FCS in the culture medium of cell lines, it is not yet clear if proteolysis is caused by autocatalysis or by proteases present in the adult tissues where Sp22D is extensively processed. We have not observed processing upon exposure to bacteria or upon chitin binding in vitro. Whatever the mechanism and mode of regulation, processing could generate a panel of polypeptides with different protease/adhesive domain combinations and physiological roles. The observed changes in the profile of Sp22D polypeptides during pupal to adult metamorphosis suggest that specific products may play various roles in the extensive tissue remodeling that takes place during this phase. Insect molecules combining adhesive and enzymatic motifs have been proposed to help generate the extracellular matrix (23). Detailed biochemical analysis of the mechanisms and pathways of Sp22D processing will be essential to unravel the function(s) of this intriguing molecule.
Acknowledgments
We are grateful to E. Levashina for sharing materials and for discussion. We thank R. Cantera for offering his expertise in histology and M. Spiegel for mosquito rearing. This research was supported by a Network Grant from the Training and Mobility of Researchers Program of the European Community and Grant SFB 544 from the Deutsche Forschungsgemeinschaft.
Abbreviations
- CBD
chitin binding domain
- SRCR
scavenger receptor cysteine rich
- LDLr
low density lipoprotein receptor-like
- RT-PCR
reverse transcription–PCR
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ276428).
References
- 1.Hoffmann J A, Kafatos F C, Janeway C A, Ezekowitz R A. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 2.Richman A M, Dimopoulos G, Seeley D, Kafatos F C. EMBO J. 1997;16:6114–6119. doi: 10.1093/emboj/16.20.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dimopoulos G, Seeley D, Wolf A, Kafatos F C. EMBO J. 1998;17:6115–6123. doi: 10.1093/emboj/17.21.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins F H, Sakai R K, Vernick K D, Paskewitz S, Seeley D C, Miller L H, Collins W E, Campbell C C, Gwadz R W. Science. 1986;234:607–610. doi: 10.1126/science.3532325. [DOI] [PubMed] [Google Scholar]
- 5.Janeway C A. Immunol Today. 1992;13:11–16. doi: 10.1016/0167-5699(92)90198-G. [DOI] [PubMed] [Google Scholar]
- 6.Levashina E A, Langley E, Green C, Gubb D, Ashburner M, Hoffmann J A, Reichhart J M. Science. 1999;285:1917–1919. doi: 10.1126/science.285.5435.1917. [DOI] [PubMed] [Google Scholar]
- 7.Iwanaga S, Kawabata S, Muta T. J Biochem. 1998;123:1–15. doi: 10.1093/oxfordjournals.jbchem.a021894. [DOI] [PubMed] [Google Scholar]
- 8.Jiang H, Wang Y, Kanost M R. Proc Natl Acad Sci USA. 1998;95:12220–12225. doi: 10.1073/pnas.95.21.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liszewski M K, Atkinson J P. In: Fundamental Immunology. 3rd Ed. Paul W E, editor. New York: Raven; 1993. pp. 917–939. [Google Scholar]
- 10.Gorman M J, Andreeva O V, Paskewitz S M. Insect Biochem Mol Biol. 2000;30:35–46. doi: 10.1016/s0965-1748(99)00095-8. [DOI] [PubMed] [Google Scholar]
- 11.Müller H-M, Dimopoulos G, Blass C, Kafatos F C. J Biol Chem. 1999;274:11727–11735. doi: 10.1074/jbc.274.17.11727. [DOI] [PubMed] [Google Scholar]
- 12.Resnick D, Pearson A, Krieger M. Trends Biochem Sci. 1994;19:5–8. doi: 10.1016/0968-0004(94)90165-1. [DOI] [PubMed] [Google Scholar]
- 13.Ohashi M, Kawamura K, Fujii N, Yubisui T, Fujiwara S. Dev Biol. 1999;214:38–45. doi: 10.1006/dbio.1999.9400. [DOI] [PubMed] [Google Scholar]
- 14.Vyse T J, Bates G P, Walport M J, Morley B J. Genomics. 1994;24:90–98. doi: 10.1006/geno.1994.1585. [DOI] [PubMed] [Google Scholar]
- 15.Shen Z, Jacobs-Lorena M. J Biol Chem. 1998;273:17665–17670. doi: 10.1074/jbc.273.28.17665. [DOI] [PubMed] [Google Scholar]
- 16.Barry M K, Triplett A A, Christensen A C. Insect Biochem Mol Biol. 1999;29:319–327. doi: 10.1016/s0965-1748(99)00004-1. [DOI] [PubMed] [Google Scholar]
- 17.Bowers B. Protoplasma. 1964;59:351–367. [Google Scholar]
- 18.Smith D S. In: Insects Cells. Smith D S, editor. Edinburgh: Oliver and Boyd; 1968. pp. 171–180. [Google Scholar]
- 19.Elomaa O, Sankala M, Pikkarainen T, Bergmann U, Tuuttila A, Raatikainen-Ahokas A, Sariola H, Tryggvason K. J Biol Chem. 1998;273:4530–4538. doi: 10.1074/jbc.273.8.4530. [DOI] [PubMed] [Google Scholar]
- 20.Manning G, Krasnow M A. In: The Development of Drosophila melanogaster. Bate M, Martinez Arias A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 609–685. [Google Scholar]
- 21.Meister M, Braun A, Kappler C, Reichhart J M, Hoffmann J A. EMBO J. 1994;13:5958–5966. doi: 10.1002/j.1460-2075.1994.tb06941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samakovlis C, Kimbrell D A, Kylsten P, Engstrom A, Hultmark D. EMBO J. 1990;9:2969–2976. doi: 10.1002/j.1460-2075.1990.tb07489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson R E, Fessler L I, Takagi Y, Blumberg B, Keene D R, Olson P F, Parker C G, Fessler J H. EMBO J. 1994;13:3438–3447. doi: 10.1002/j.1460-2075.1994.tb06649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]