Abstract
The degradation of 7,12-dimethylbenz[a]anthracene (DMBA), a carcinogenic polycyclic aromatic hydrocarbon, by cultures of Mycobacterium vanbaalenii PYR-1 was studied. When M. vanbaalenii PYR-1 was grown in the presence of DMBA for 136 h, high-pressure liquid chromatography (HPLC) analysis showed the presence of four ethyl acetate-extractable compounds and unutilized substrate. Characterization of the metabolites by mass and nuclear magnetic resonance spectrometry indicated initial attack at the C-5 and C-6 positions and on the methyl group attached to C-7 of DMBA. The metabolites were identified as cis-5,6-dihydro-5,6-dihydroxy-7,12-dimethylbenz[a]anthracene (DMBA cis-5,6-dihydrodiol), trans-5,6-dihydro-5,6-dihydroxy-7,12-dimethylbenz[a]anthracene (DMBA trans-5,6-dihydrodiol), and 7-hydroxymethyl-12-methylbenz[a]anthracene, suggesting dioxygenation and monooxygenation reactions. Chiral stationary-phase HPLC analysis of the dihydrodiols showed that DMBA cis-5,6-dihydrodiol had 95% 5S,6R and 5% 5R,6S absolute stereochemistry. On the other hand, the DMBA trans-5,6-dihydrodiol was a 100% 5S,6S enantiomer. A minor photooxidation product, 7,12-epidioxy-7,12-dimethylbenz[a]anthracene, was also formed. The results demonstrate that M. vanbaalenii PYR-1 is highly regio- and stereoselective in the degradation of DMBA.
7,12-Dimethylbenz[a]anthracene (DMBA) is a potent carcinogen in rodent skin and mammary cells (7). Alkylated benz[a]-anthracenes are formed in sediments by thermal processes and have also been identified in cigarette smoke condensate, coal gasification and stock gas particulates, and roofing tar extracts (1, 10). Studies from several laboratories have indicated that DMBA is bioactivated by various isoforms of mammalian cytochrome P-450 and epoxide hydrolase to form DMBA-3,4-dihydrodiol-syn- and anti-1,2-epoxides that react with DNA to form adducts that lead to tumor initiation (22, 32, 33). An alternative mechanism for metabolic activation is hydroxylation of the alkyl side chain to form hydroxymethyl derivatives, followed by sulfotransferase-catalyzed activation of DMBA to form electrophilic sulfuric acid esters that bind to DNA (6). 7,12-Dimethylbenz[a]anthracene has been used as a model polycyclic aromatic hydrocarbon (PAH) in the following ways: (i) as a tumor initiator or as a complete carcinogen in rodent skin and mammary gland models of carcinogenesis (15, 22), (ii) as a prototype methyl-substituted PAH in mutation and cancer research (15), (iii) to determine whether metabolic activation occurs via bay-region dihydrodiol epoxide metabolites or hydroxylation of the methyl substituent with subsequent formation of electrophilic sulfuric acid esters (6), and (iv) to determine the environmental fate of methyl-substituted PAHs.
In contrast to the numerous studies of the metabolism, DNA adduct formation, and tumor induction by DMBA in mammalian cells, little is known about the microbial metabolism of this potent carcinogen. Several strains of fungi metabolize methylbenz[a]anthracenes at the methyl group, with further oxidation of the aromatic ring to form hydroxymethylbenz[a]anthracene-trans-dihydrodiols. The results from these studies indicate that the fungal cytochrome P-450 monooxygenase and epoxide hydrolase enzyme systems were highly regio- and stereoselective in the metabolism of methylbenz[a]anthracenes (3, 4, 24, 25). The mutagenic activity of ethyl acetate extracts from two filamentous fungi, Cunninghamella elegans and Syncephalastrum racemosum, grown in the presence of 7,12-dimethylbenz[a]anthracene decreased with time of incubation and metabolism (5, 25). Pseudomonas aeruginosa was able to oxidize the methyl groups of DMBA to form hydroxymethyl derivatives (36).
Recently, we reported that Mycobacterium vanbaalenii PYR-1 can metabolize a wide variety of PAHs, including naphthalene, biphenyl, anthracene, phenanthrene, pyrene, fluoranthene, and benzo[a]pyrene (11-14, 16, 17-21, 26, 27). In the present paper, we provide the first detailed report on the bacterial metabolism of DMBA by M. vanbaalenii PYR-1. Enantiomeric resolution of the initial dioxygenation and monooxygenation metabolites is described.
MATERIALS AND METHODS
Chemicals.
[7,12-14C]DMBA (specific activity, 97.4 mCi/mmol) was purchased from New England Nuclear (Boston, Mass.). Unlabeled DMBA (95% pure) was purchased from Sigma Chemical Company (St. Louis, Mo.). Bacteriological media and reagents were purchased from BD Biosciences, Difco Laboratories (Detroit, Mich.). Deuterated acetone (99.96% pure), used as the solvent for nuclear magnetic resonance (NMR) analysis, was purchased from Isotec, Inc. (Miamisburg, Ohio). Other solvents were purchased from J. T. Baker, Inc. (Phillipsburg, N.J.), and were of the highest purity available. Racemic DMBA cis-5,6-dihydrodiol and DMBA trans-5,6-dihydrodiol were prepared following published procedures (8).
Cultural conditions.
Cultures of M. vanbaalenii PYR-1 DSM 7251 were grown in 1-liter Erlenmeyer flasks containing 500 ml of minimal basal salts medium supplemented with 0.38 g/ml each of peptone, yeast extract, and soluble starch. A 100-μl aliquot of 70 mM phenanthrene in N,N-dimethylformamide was added to each flask for enzyme induction. The cultures were grown for 4 days in the dark at 28°C with shaking at 110 rpm. The cells from the flasks were harvested by centrifugation (7,000 × g for 10 min) and resuspended in 50 ml of the medium in 250-ml Erlenmeyer flasks. DMBA was dissolved in N,N-dimethylformamide and added to the cultures at a final concentration of 0.47 mM. Flasks were harvested at 6, 24, 48, 72, 96, 129, 136, 160, 184, and 256 h to determine the time of optimum metabolite production. A larger group of cultures was grown, treated with DMBA in the same manner, and incubated for 136 h. Flasks containing only the culture and noninoculated flasks containing only DMBA and medium were used as controls.
The contents of each flask, including the controls, were extracted with 3 equal volumes of ethyl acetate. The organic extracts were dried over sodium sulfate and evaporated to dryness with a Buchi 011 rotary evaporator (Brinkmann Instruments, Westbury, N.Y.). The pH of the remaining aqueous fraction was lowered to 2.5 with 6 M hydrochloric acid, and the contents were extracted again with ethyl acetate to isolate the acidic metabolites. The residues were dissolved in 3 ml of methanol and concentrated to approximately 100 μl with a model SS21 Savant Speed-vac system (Savant Instruments, Holbrook, N.Y.) for analysis by reversed-phase high-pressure liquid chromatography (HPLC).
Radiolabeling experiments.
Mineralization experiments, as evidenced by CO2 evolution, were carried out with a CO2 trap containing 15 ml of 5.0 M NaOH. Phenanthrene-induced Mycobacterium cells were added to 50 ml of minimal basal salts medium supplemented with 0.38 g/ml each of peptone, yeast extract, and soluble starch and treated with 1.0 μCi of 7,12-[dimethyl-14C]benz[a]anthracene and 6 mg of unlabeled DMBA. A flask containing only DMBA and one with only M. vanbaalenii PYR-1 were used as controls. Each flask was immediately sampled for CO2 production by removing 1.0 ml of the trapping solution. This sample was added to 14 ml of Ultima Gold liquid scintillation fluid (Packard Instruments, Downers Grove, Ill.) and counted in a Packard Tri-Carb 2000A scintillation analyzer. Flasks were sampled and counted at 24, 48, 72, 96, 144, 168, 192, 216, 240, 312, 336, and 360 h.
Physical and chemical analysis.
DMBA and its metabolites were separated by HPLC with a Hewlett-Packard model 1050 chromatograph (Hewlett-Packard, Palo Alto, Calif.) with a 5-μm C18 Inertsil ODS-3 column (4.6 by 250 mm; MetaChem Technologies, Torrance, Calif.) at a flow rate of 1 ml/min. UV absorbance spectra were obtained on line with a diode array model 1040A detector at 254 nm. The compounds were eluted with a linear gradient of 40 to 95% methanol-water over 40 min. For collection of larger amounts of metabolites, a Beckman model 100A chromatograph (Beckman Instruments, Fullerton, Calif.) with an Inertsil ODS-3 column (10 by 250 mm; MetaChem) was used. The mobile phase and gradient were the same as above, but with a 5-ml/min flow rate.
The compounds were analyzed by gas chromatography-mass spectrometry (GC-MS) on a ThermoFinnigan TSQ 700 gas chromatograph-mass spectrometer (Finnigan Corp., San Jose, Calif.) in the electron ionization, single-quadrupole mode. The ion source temperature was 150°C, and the electron energy was 70 eV (uncorrected). The first quadrupole analyzer was scanned from m/z 50 to 450 in 0.5 s. The Varian model 3400 gas chromatograph (Varian, Inc., Palo Alto, Calif.) employed a septum-equipped, temperature-programmable injector and a J&W (Agilent Technologies, Palo Alto, Calif.) DB-5ms capillary column (30 m by 0.25 mm by 0.25 μm) that was coupled directly to the ion source through a heated interface.
NMR spectra were recorded at 500.13 MHz on a Bruker AM500 spectrometer (Bruker Instruments, Billerica, Mass.). The metabolites were dissolved in 0.5 ml of deuterated acetone. 1H chemical shifts are reported on the δ scale (in parts per million) by assigning the residual solvent peak to 2.04 ppm. Typical 1H data acquisition parameters were: data size, 32,000; sweep width, 7,042 Hz; filter width, 8,900 Hz; acquisition time, 2.33 s; flip angle, 90°; relaxation delay, 0 s; temperature, 301 K. For spectra recorded under quantitative conditions, a 10-s relaxation delay was used. For measurement of coupling constants, the free induction decay was zero-filled to 64,000, resulting in a final data point resolution of 0.215 Hz per point. Only first-order coupling constants are reported.
Direct resolution of DMBA cis-5,6-dihydrodiol and DMBA trans-5,6-dihydrodiol enantiomers by chiral stationary-phase (CSP) HPLC was performed with an (R,R) Whelk-O1 column (4.6 by 250 mm; Regis Technologies, Morton Grove, Ill.). The dihydrodiol enantiomers were eluted isocratically with hexane-isopropanol (80%:20%, vol/vol) at a flow rate of 1 ml/min. The ratios of the enantiomers were calculated based on comparison of peak areas of the two enantiomers.
Circular dichroism (CD) spectra of DMBA cis-5,6-dihydrodiol and DMBA trans-5,6-dihydrodiol dissolved in methanol were measured in a quartz cell of 1-cm path length on a Jasco model 500A spectropolarimeter. CD spectra are expressed as ellipticity (φλ in millidegrees) as described by Fu and Yang (8).
RESULTS
Identification of DMBA metabolites.
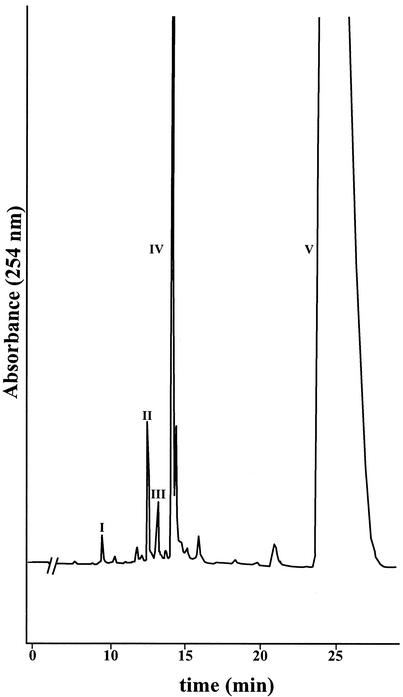
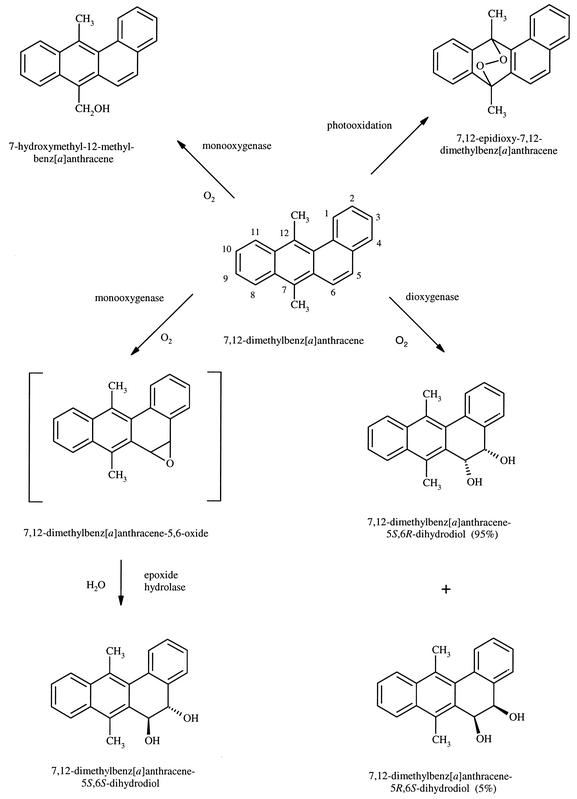
After 136 h of incubation of M. vanbaalenii PYR-1 in the presence of DMBA, HPLC analysis of the ethyl acetate extracts showed compounds with retention times (tR) of 9.4, 12.5, 13.1, 14.1, and 26.0 min (Fig. 1). Radiolabel experiments indicated that less than 5% and 1% of the DMBA was metabolized and mineralized, respectively. The metabolites, compounds I through IV, were found in a ratio of 1:4.2:2.5:20.
FIG. 1.
Reversed-phase HPLC chromatogram showing DMBA and the metabolites formed by M. vanbaalenii PYR-1. The positions of compounds I to V are shown.
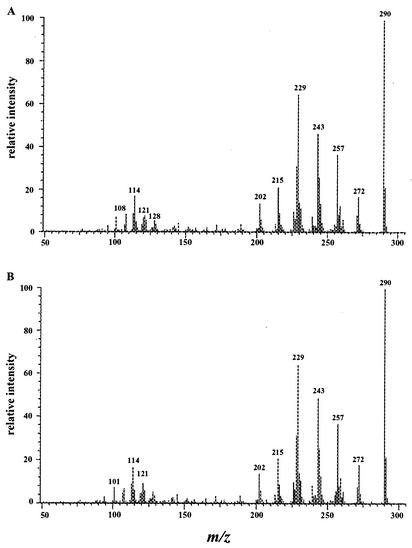
The compounds were further analyzed by GC-MS. Compounds I (tR = 9.4 min) and II (tR = 12.5 min) had GC-MS retention times of 16.70 min and 17.13 min, respectively. They had essentially the same mass spectra, with base peak molecular ions at m/z 290 [M+.] and significant fragment ions at m/z 272 [M-18]+, 257 [M-18-15]+, 243 [M-18-29]+, 229 [M-18-15-28]+, 215 [M-18-29-28]+, and 202 (Fig. 2). The molecular weight indicates the addition of two hydrogen and two oxygen atoms. Losses of water (18 Da) and carbonyl (29 and 28 Da) are characteristic of dihydrodiols. The loss of methyl (15 Da) is characteristic of methyl substituents.
FIG. 2.
Electron ionization mass spectra of DMBA trans-5,6-dihydrodiol (A) and DMBA cis-5,6-dihydrodiol (B).
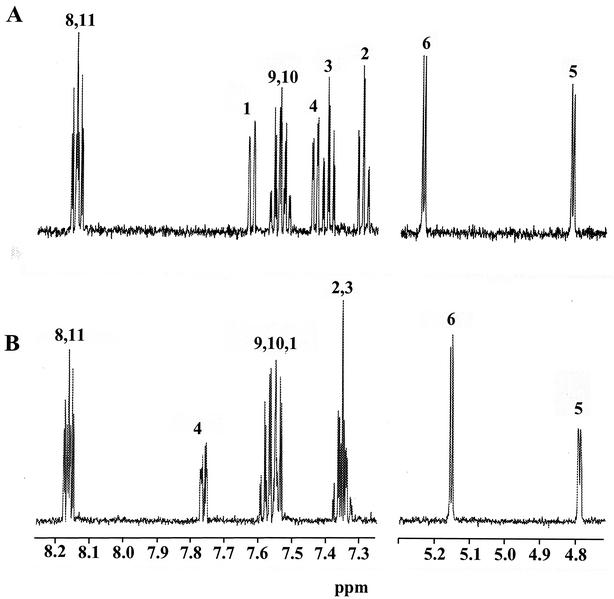
Compound I (tR = 9.4 min) was identified as DMBA trans-5,6-dihydrodiol. The NMR spectrum [7.62 (H1, J1,2 = 7.7 Hz), 7.29 (H2, J2,3 = 7.3 Hz, J2,4 = 1.3 Hz), 7.40 (H3, J3,4 = 7.5 Hz), 7.44 (H4), 4.82 (H5, J5,6 = 3.4 Hz), 5.24 (H6), 2.79 (H7), 8.14 (H8, H11), 7.54 (H9, H10), 2.89 (H12)] was identical to that reported by McMillan et al. (24), with the hydroxyl groups in a quasi-diaxial conformation (Fig. 3). Compound II eluted at 12.5 min and had the same UV absorbance spectrum as compound I (λmax = 227, 259, 266, and 298 nm). The chemical shifts of the resonances in the NMR spectrum were within 0.08 ppm of those of compound I, with the exception of the H4 resonance, which was shifted downfield 0.37 ppm (Fig. 3). The H5 and H6 coupling constants were also 3.4 Hz. NMR assignments are: [7.54 (H1), 7.35 (H2, H3), 7.77 (H4), 4.79 (H5), 5.16 (H6), 2.79 (H7), 8.16 (H8, H11), 7.57 (H9, H10), 2.89 (H12)]. Comparison of the retention time, UV absorption spectrum, mass spectrum, and NMR spectrum of the metabolite to those of a synthetic standard identified it as DMBA cis-5,6-dihydrodiol. The 5- and 6-hydroxyl groups were in quasi-equatorial and quasi-axial conformations, respectively (40). The ratio of DMBA cis-5,6-dihydrodiol to the trans enantiomer was 4.2:1.
FIG. 3.
Proton NMR spectra of DMBA trans-5,6-dihydrodiol (A) and DMBA cis-5,6-dihydrodiol (B).
Compound III (tR = 13.1 min, UV λmax = 212, 229, and 292 nm) had a GC-MS retention time of 12.19 min. The mass spectrum consisted of a molecular ion at m/z 288 [M+.] and significant fragment ions at m/z 270 [M-18]+, 255 [M-18-15]+, 245 [M-43]+, 215, and 202. The base peak ion was m/z 245. The molecular weight indicates the addition of 2 oxygen atoms. Losses of water (18 Da) and methyl (15 Da) could be characteristic of the dihydrodiols above, but the loss of 43 Da to form the base peak ion at m/z 245 may be due to the loss of CH3—C=O from the endoperoxide portion of the molecule. The NMR spectrum revealed that all the aromatic protons were still attached to their respective carbons. Some of those protons were shifted upfield and some downfield from the corresponding ones in DMBA. However, the methyl proton resonances of compound III were shifted upfield by 0.84 ppm (H7) and 0.71 ppm (H12). NMR assignments and coupling constants are: 8.62 (H1, J1,2 = 8.4 Hz, J1,3 = 1.3 Hz), 7.57 (H2, J2,3 = 7.5 Hz, J3,4 = 8.4 Hz), 7.48 (H3), 7.95 (H4), 7.91 (H5, J5,6 = 8.4 Hz), 7.71 (H6), 2.22 (H7), 7.50 (H8), 7.29 (H9,10), 7.60 (H11), 2.62 (H12). A 7,12-endoperoxide structure was proposed, so a standard of 7,12-epidioxy-7,12-dimethylbenz[a]anthracene was prepared according to Wood et al. (35). The resulting compound had the same UV, mass, and NMR spectra and HPLC retention time as the peak at 13.1 min. The compound, identified as 7,12-epidioxy-7,12-dimethylbenz[a]anthracene, was actually a photooxidation product (35) and not a true metabolite, since it was formed in the control extract as well as the experimental extract.
Compound IV (tR = 14.1 min, UV λmax = 261, 280, 290, 355 nm) had a GC-MS retention time of 18.73 min. The mass spectrum consisted of a molecular ion at m/z 272 [M+.] and significant fragment ions at m/z 255 [M-17]+, 243 [M-29]+, 239 [M-18-15]+, and 228 [M-44]+. The molecular weight indicates the addition of one oxygen atom. The NMR spectrum [8.54 (H1), 7.61 (H2, H3), 7.90 (H4), 7.67 (H5, J5,6 = 9.5 Hz), 8.25 (H6), 5.56 (H7), 8.57 (H8), 7.66 (H9, H10), 8.42 (H11), 3.37 (H12)] showed one methyl resonance that integrated as three, ten aromatic resonances, and a singlet at 5.56 ppm that integrated as two. The metabolite was identified as 7-hydroxymethyl-12-methylbenz[a]anthracene.
Unmetabolized DMBA, contained in peak V, eluted at 26.0 min and was identified by comparison of its retention time and UV, mass, and NMR spectra to those of the standard. NMR assignments and coupling constants were: 8.53 (H1), 7.59 (H2, H3), 7.88 (H4), 7.64 (H5, J5,6 = 9.5 Hz), 8.09 (H6), 3.06 (H7), 8.40 (H8, H11), 7.65 (H9, H10), 3.33 (H12).
Optical purity and absolute configuration of DMBA 5,6-dihydrodiol metabolites.
The optical purity and absolute configuration of the DMBA trans-5,6-dihydrodiol metabolite were determined by CSP HPLC and CD spectral analyses. To develop optimal conditions for direct resolution of this metabolite by CSP HPLC, direct separation of the synthetically prepared racemic DMBA trans-5,6-dihydrodiols was required. Several new types of CSP HPLC Pirkle columns, including the π-acceptor type β-Gem 1 CSP column, the π-acceptor type α-Burke 2 CSP column, and the π-electron acceptor/π-electron donor type (R,R) Whelk-O1 CSP column, as well as different solvent systems were used for comparison. The (R,R) Whelk-O1 CSP HPLC column with a hexane-isopropanol (80:20; vol/vol) isocratic mixture provided the best resolution. It resulted in a baseline separation of the two DMBA trans-5,6-dihydrodiol enantiomers, with retention times of 8.35 and 9.60 min, respectively. Both of their UV-visible absorption spectra were identical to that of the DMBA trans-5,6-dihydrodiol metabolite, compound I.
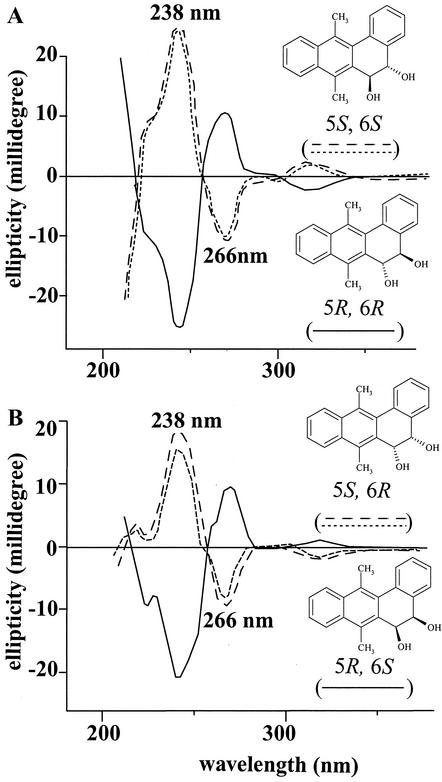
The CD spectra of the two pure enantiomers were measured (Fig. 4). The enantiomer that eluted first by CSP HPLC, 8.35 min, had a CD spectrum that was a mirror image of that of the enantiomer that eluted at 9.60 min. Both had identical Cotton effects at 238 and 266 nm, but had ellipticities of opposite value. Comparison with the CD spectrum of DMBA 5R,6R-dihydrodiol published by Yang and Fu (37) and by McMillan et al. (24) revealed that the DMBA trans-5,6-dihydrodiol enantiomers eluting at 8.35 and 9.60 min had the 5R,6R and 5S,6S absolute configurations, respectively (Fig. 4A).
FIG. 4.
CD spectra of the two enantiomers of synthetically prepared racemic DMBA trans-5,6-dihydrodiol (— and –––) and DMBA trans-5,6-dihydrodiol from M. vanbaalenii PYR-1 (----) (A) and synthetically prepared racemic DMBA cis-5,6-dihydrodiol (— and –––) and DMBA cis-5,6-dihydrodiol from M. vanbaalenii PYR-1 (----) (B).
After the optimal conditions for direct resolution of the synthetically prepared racemic DMBA trans-5,6-dihydrodiol by CSP HPLC were obtained, the DMBA trans-5,6-dihydrodiol metabolite, compound I, was separated under identical conditions. Within the detection limit of the CSP HPLC separation conditions employed, this DMBA trans-5,6-dihydrodiol metabolite contained only the DMBA 5S,6S-dihydrodiol enantiomer, since it eluted at 9.60 min. This was further confirmed by analysis of its CD spectrum. As shown in Fig. 4A, the CD spectrum was similar to that of the DMBA 5S,6S-dihydrodiol enantiomer, having the identical Cotton effects shown at 238 and 266 nm and the same ellipticities. This CD spectral comparison confirmed that the DMBA trans-5,6-dihydrodiol metabolite obtained from metabolism of DMBA by M. vanbaalenii PYR-1 was a 100% pure 5S,6S enantiomer.
The optical purity and absolute configuration of the DMBA cis-5,6-dihydrodiol metabolite obtained from metabolism of DMBA by M. vanbaalenii PYR-1 were similarly determined. CSP HPLC directly resolved the synthetically prepared racemic DMBA cis-5,6-dihydrodiol into the two enantiomers. They both had UV-visible absorption spectra identical to that of the DMBA cis-5,6-dihydrodiol metabolite.
The CD spectra of the two enantiomers were measured (Fig. 4B). The enantiomer that eluted first, 8.30 min, had a CD spectrum that was a mirror image to that of the enantiomer that eluted at 9.61 min. Both had the same Cotton effects at 238 and 266 nm, but with ellipticities of opposite value. After comparison of the spectra with the CD spectrum (Fig. 4B) of DMBA 5R,6S-dihydrodiol published by Yang et al. (39), we concluded that the DMBA cis-5,6-dihydrodiol enantiomers eluting at 8.30 and 9.61 min had the 5S,6R and 5R,6S absolute configurations, respectively.
CSP HPLC was then used to separate the DMBA cis-5,6-dihydrodiol metabolite, compound II. The resulting chromatogram showed two peaks that had UV-visible absorption spectra identical to that of the DMBA cis-5,6-dihydrodiol metabolite. The ratio of the two enantiomers was determined based on comparison of peak areas of the two enantiomers. It was determined that compound II was 95% DMBA 5S,6R-dihydrodiol and 5% DMBA 5R,6S-dihydrodiol. Thus, the optical purity of the DMBA cis-5,6-dihydrodiol metabolite was 90%.
DISCUSSION
M. vanbaalenii PYR-1 initially oxidized DMBA at the K-region (C-5 and C-6) to form cis- and trans-5,6 dihydrodiols, indicating aromatic ring dioxygenation and monooxygenation, respectively. More of the cis than the trans enantiomer was formed, with a ratio of 4.2:1, indicating that the dioxygenation mechanism is the preferred one for the formation of dihydrodiols. These results support earlier observations in our laboratory that this organism is highly regio- and stereoselective in the initial oxidation of high-molecular-weight PAHs. For example, M. vanbaalenii PYR-1 initially attacked the pyrene and phenanthrene nucleus to form pyrene and phenanthrene cis- and trans-dihydrodiols in the 4,5 and 9,10 positions, the K-region of each PAH.
Based on 18O2 incorporation experiments and rigorous structure elucidation analyses, we have shown that M. vanbaalenii has both a cytochrome P-450 monooxygenase/epoxide hydrolase and dioxygenases to catalyze the initial attack on the PAH (13, 16, 26). The preferred regioselective attack at the K-region bonds and the mechanism of oxidation seem to be unique to Mycobacterium species. Recently, the PAH catabolic genes coding for the α- and β-subunits of the terminal dioxygenase for pyrene degradation have been characterized for this strain (21). The nidA and nidB genes, encoding the α- and β-subunits of aromatic ring-hydroxylating dioxygenase, respectively, were novel compared to sequences of this enzyme from gram-negative bacteria that degrade PAHs. Although we have not attempted to characterize the cytochrome P-450 in M. vanbaalenii PYR-1, a soluble cytochrome P-450 monooxygenase has been detected in several mycobacterial strains involved in the degradation of morpholine, piperidine, and pyrrolidine (28, 29, 30). The genes encoding the piperidine-inducible cytochrome P-450 have been characterized in Mycobacterium smegmatis strain mc2155 (30).
The DMBA trans-5,6-dihydrodiol formed by Mycobacterium vanbaalenii PYR-1 has similarities to and differences from those produced from mammalian and fungal systems. It is known that the metabolism of PAHs by mammalian enzymatic systems produces only the trans-dihydrodiol metabolites, never a cis-dihydrodiol (37, 38). This is because a PAH is biotransformed into a trans-dihydrodiol in two steps: epoxidation of the PAH to form an arene oxide and subsequent hydrolysis of the arene oxide. The first reaction is catalyzed by cytochrome P-450 and the second by epoxide hydrolase (Fig. 5). The fungi Syncephalastrum racemosum and Cunninghamella elegans both formed 7-hydroxy-trans-5,6-dihydrodiol DMBA that was predominantly 5R,6R (24) (Table 1). In mammalian systems, metabolism of DMBA produces a trans-5,6-dihydrodiol as one of the major metabolites that has a predominantly S,S absolute stereochemistry (37). The R,R conformation is formed when epoxide hydrolase causes hydration at the least hindered epoxide carbon atom, C-5. However, when DMBA is metabolized in the same manner, the more sterically hindered site (C-6) is attacked, leading to a 5S,6S-dihydrodiol (24). This mechanism of bond attack may be similar in M. vanbaalenii PYR-1, and like that produced by rat liver microsomes, the trans-5,6-dihydrodiol adopts the 5S,6S conformation with 100% optical purity. Table 1 compares the enantiomeric composition of trans-5,6-dihydrodiols of benz[a]anthracene and methylated benz[a]anthracenes from various microbial and mammalian sources.
FIG. 5.
Proposed pathway for the transformation of DMBA by M. vanbaalenii PYR-1.
TABLE 1.
Comparison of the enantiomeric composition of the trans-5,6-dihydrodiol metabolites formed from the metabolism of benz[a]anthracene and methyl-substituted benz[a]anthracenes by rat liver microsomes and microorganisms
| PAHa | Microsomes | Enantiomer (%)
|
Optical purity (%) | Refer- ence(s) | |
|---|---|---|---|---|---|
| 5R,6R | 5S,6S | ||||
| BA | PB microsomes | 68 | 32 | 36 | 31 |
| MC microsomes | 81 | 19 | 62 | ||
| 7-MBA | MC microsomes | 53 | 47 | 6 | 38 |
| 12-MBA | MC microsomes | 5 | 95 | 90 | 37 |
| DMBA | Control microsomes | 11 | 89 | 78 | 37, 38 |
| PB microsomes | 5 | 95 | 90 | ||
| MC microsomes | 6 | 94 | 88 | ||
| M. vanbaalenii PYR-1 | 0 | 100 | 100 | This study | |
| 7-OH-12-MBA | Syncephalastrum racemosum | 88 | 12 | 76 | 24, 25 |
| Cunninghamella elegans | 77 | 23 | 54 | ||
BA, benz[a]anthracene; PB, phenobarbital; MC, 3-methylcholanthrene; OH, hydroxy; MBA, methylbenz[a]anthracene.
M. vanbaalenii PYR-1 oxidized the 7-methyl substituent of DMBA to 7-hydroxymethyl-12-methylbenz[a]anthracene, as did Pseudomonas aeruginosa and the fungus Penicillium notatum (36). Since a triple mutant of cytochrome P-450 BM-3 from Bacillus megaterium hydroxylated several PAHs and transformed 9-methylanthracene to 9-hydroxymethylanthracene (23), we are investigating the role of cytochrome P-450 in the hydroxylation of PAHs by M. vanbaalenii PYR-1.
Since K-region dihydrodiols of PAHs do not form carcinogenic vicinal diol epoxides, these reactions formed by M. vanbaalenii PYR-1 can be considered detoxification processes. However, 7-hydroxymethyl-12-methylbenz[a]anthracene has been shown to cause adrenal apoplexy and mammary cancer in rats (2) and so is not considered a detoxification intermediate. 7,12-Epidioxy-7,12-dimethylbenz[a]anthracene, a photooxidation product of DMBA (35), has been shown to be toxic to chicken fibroblast cells (34). While mass spectrometric analysis showed no 7,12-epidioxy-7,12-dimethylbenz[a]anthracene present in the DMBA used in treating the Mycobacterium cells, the compound did form in the control flask. Since the compound can be formed upon exposure to light, even more would have been formed if the cultures had not been incubated in the dark.
This study is the first example of the ability of M. vanbaalenii PYR-1 to degrade methyl-substituted high-molecular-weight PAHs in a regio- and stereoselective manner to form known as well as novel metabolites. This study provides evidence for the potential application of this organism for improved PAH bioremediation and enantioselective production of dihydroxylated synthons.
Acknowledgments
We thank John B. Sutherland and Thomas M. Heinze for critical review of the manuscript and Barbara Jacks for illustrations.
REFERENCES
- 1.Blumer, M. 1976. Polycyclic aromatic compounds in nature. Scientific American 234:35-45. [PubMed] [Google Scholar]
- 2.Boyland, E., P. Sims, and C. Huggins. 1969. Induction of adrenal damage and cancer with metabolites of 7,12-dimethylbenz[a]anthracene. Nature 207:816-817. [DOI] [PubMed] [Google Scholar]
- 3.Cerniglia, C. E., P. P. Fu, and S. K. Yang. 1982. Metabolism of 7-methylbenz[a]anthracene and 7-hydroxymethylbenz[a]anthracene by Cunninghamella elegans. Appl. Environ. Microbiol. 49:682-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerniglia, C. E., P. P. Fu, and S. K. Yang. 1983. Regio- and stereoselective metabolism of 4-methylbenz[a]anthracene by the fungus Cunninghamella elegans. Biochem. J. 216:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerniglia, C. E., G. L. White, and R. H. Heflich. 1985. Fungal metabolism and detoxification of polycyclic aromatic hydrocarbons. Arch. Microbiol. 50:105-110. [DOI] [PubMed] [Google Scholar]
- 6.Chou, H.-C., S. Ozawa, P. P. Fu, N. P. Lang, and F. F. Kadlubar. 1998. Metabolic activation of methyl-hydroxylated derivatives of 7,12-dimethylbenz[a]anthracene by human liver dehydroepiandrosterone-steroid sulfotransferase. Carcinogenesis 19:1071-1076. [DOI] [PubMed] [Google Scholar]
- 7.DiGiovanni, J. 1992. Multi stage carcinogenesis in mouse skin. Pharmacol. Ther. 54:63-128. [DOI] [PubMed] [Google Scholar]
- 8.Fu, P. P., and S. K. Yang. 1982. In vitro metabolism of 12-methylbenz[a]anthracene: effect of the methyl group on the stereochemistry of a 5,6-dihydrodiol metabolite. Biochem. Biophys. Res. Commun. 106:940-946. [DOI] [PubMed] [Google Scholar]
- 9.Harvey, R. G., S. H. Goh, and C. Cortez. 1975. “K-region” oxides and related oxidized metabolites of carcinogenic aromatic hydrocarbons. J. Am. Chem. Soc. 97:3468-3479. [DOI] [PubMed] [Google Scholar]
- 10.Haugen, D. A., and V. C. Stamoudes. 1986. Isolation and identification of mutagenic polycyclic aromatic hydrocarbons from a coal gasifier condensate. Environ. Res. 4l:400-419. [DOI] [PubMed] [Google Scholar]
- 11.Heitkamp, M. A., and C. E. Cerniglia. 1988. Mineralization of polycyclic aromatic hydrocarbons by a bacterium isolated from sediment below an oil field. Appl. Environ. Microbiol. 54:1612-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heitkamp, M. A., and C. E. Cerniglia. 1989. Polycyclic aromatic hydrocarbon degradation by a Mycobacterium sp. in microcosms containing sediment and water from a pristine ecosystem. Appl. Environ. Microbiol. 55:1968-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heitkamp, M. A., W. Franklin, and C. E. Cerniglia. 1988. Microbial metabolism of polycyclic aromatic hydrocarbons: Isolation and characterization of pyrene-degrading bacterium. Appl. Environ. Microbiol. 54:2549-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heitkamp, M. A., J. P. Freeman, D. W. Miller, and C. W. Cerniglia. 1988. Pyrene degradation by a Mycobacterium sp.: identification of ring oxidation and ring fission products. Appl. Environ. Microbiol. 54:2556-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izzotti, A., A. Camoirano, C. Cartiglia, C. J. Grubbs, R. A. Lubet, G. J. Kelloff, and S. De Flora. 1999. Patterns of DNA adduct formation in liver and mammary epithelial cells of rats treated with 7,12-dimethylbenz[a]anthracene, and selective effects of chemopreventive agents. Cancer Res. 59:4285-4290. [PubMed] [Google Scholar]
- 16.Kelley, I., J. P. Freeman, and C. E. Cerniglia. 1990. Identification of metabolites from degradation of naphthalene by a Mycobacterium sp. Biodegradation 1:283-290. [DOI] [PubMed] [Google Scholar]
- 17.Kelley, I., and C. E. Cerniglia. 1991. The metabolism of fluoranthene by a species of Mycobacterium. J. Ind. Microbiol. 7:19-26. [Google Scholar]
- 18.Kelley, I., J. P. Freeman, F. E. Evans, and C. E. Cerniglia. 1991. Identification of a carboxylic acid metabolite from the catabolism of fluoranthene by a Mycobacterium sp. Appl. Environ. Microbiol. 57:636-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelley, I., J. P. Freeman, F. E. Evans, and C. E. Cerniglia. 1993. Identification of metabolites from the degradation of fluoranthene by Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 59:800-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelley, I., and C. E. Cerniglia. 1995. Degradation of a mixture of high-molecular-weight polycyclic aromatic hydrocarbons by Mycobacterium strain PYR-1. J. Soil Contam. 4:44-91. [Google Scholar]
- 21.Khan, A. A., S.-J. Kim, D. D. Paine, and C. E. Cerniglia. 2002. Classification of a polycyclic aromatic hydrocarbon-metabolizing bacterium, Mycobacterium sp. strain PYR-1, as Mycobacterium vanbaalenii sp. nov. Int. J. Syst. E vol. Microbiol. 52:1997-2002. [DOI] [PubMed] [Google Scholar]
- 22.Kleiner, H. E., S. V. Vulimiri, M. J. Reed, A. Uberecken, and J. DiGiovanni. 2002. Role of cytochrome P-450 1a1 and 1b1 in the metabolic activation of 7,12-dimethylbenz[a]anthracene and the effects of naturally occurring furanocoumarins on skin tumor initiation. Chem. Res. Toxicol. 15:226-235. [DOI] [PubMed] [Google Scholar]
- 23.Li, Q.-S., J. Ogawa, R. D. Schmid, and S. Shimizu. 2001. Engineering cytochrome P-450 BM-3 for oxidation of polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 67:5735-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMillan, D. C., P. P. Fu, and C. E. Cerniglia. 1987. Stereo-selective fungal metabolism of 7,12-dimethylbenz[a]anthracene: identification and enantiomeric resolution of a K-region dihydrodiol. Appl. Environ. Microbiol. 53:2560-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMillan, D. C., P. P. Fu, J. P. Freeman, D. W. Miller, and C. E. Cerniglia. 1988. Microbial metabolism and detoxification of 7,12-dimethylbenz[a]anthracene. J. Ind. Microbiol. 3:211-225. [Google Scholar]
- 26.Moody, J. D., J. P. Freeman, D. R. Doerge, and C. E. Cerniglia. 2001. Degradation of phenanthrene and anthracene by cell suspensions of Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 67:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moody, J. D., D. R. Doerge, J. P. Freeman, and C. E. Cerniglia. 2002. Degradation of biphenyl by Mycobacterium sp. strain PYR-1. Appl. Microbiol. Biotechnol. 58:364-369. [DOI] [PubMed] [Google Scholar]
- 28.Poupin, P., N. Truffant, B. Combourieu, P. Besse, M. Sancelme, H. Veschambre, and A. M. Delort. 1998. Degradation of morpholine by an environmental Mycobacterium strain involves a cytochrome P-450. Appl. Environ. Microbiol. 64:159-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poupin, P., V. Ducrocq, S. Hallier-Souliee, and N. Truffant. 1999. Degradation of morpholine, piperidine, and pyrrolidine by mycobacteria: evidence for the involvement of a cytochrome P-450. Can. J. Microbiol. 45:209-216. [PubMed] [Google Scholar]
- 30.Poupin, P., J. J. Godon, E. Zumstein, and N. Truffant. 1999. Cloning and characterization of genes encoding a cytochrome P-450 (pipA) involved in piperidine and pyrrolidine utilization and its regulatory protein (pipR) in Mycobacterium smegmatis mc2155. J. Bacteriol. 181:3419-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thakker, D. R., W. Levin, H. Yagi, M. S. Thrujman, D. Kapadia, A. H. Conney, and D. M. Jerina. 1979. Absolute stereochemistry of the trans-dihydrodiols formed from benz[a]anthracene by liver microsomes. Chem. Biol. Interact. 27:145-161. [DOI] [PubMed] [Google Scholar]
- 32.Vericat, J. A., S. C. Cheng, and A. Dipple. 1989. Absolute stereochemistry of the major 7,12-dimethylbenz[a]anthracene-DNA adducts formed in mouse cells. Carcinogenesis 10:567-570. [DOI] [PubMed] [Google Scholar]
- 33.Vericat, J. A., S. C. Cheng, and A. Dipple. 1991. Absolute configuration of 7,12-dimethylbenz[a]anthracene-DNA adducts in mouse epidermis. Cancer Lett. 57:237-242. [DOI] [PubMed] [Google Scholar]
- 34.Warshawsky, D., E. Kerns, M. J. Bissell, and M. Calvin. 1977. Characterization of a photoproduct of 7,12-dimethylbenz[a]anthracene and its effects on chick-embryo cells in culture. Biochem. J. 164:481-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood, J., L., C. L. Barker, and C. J. Grubbs. 1979. Photooxidation products of 7,12-dimethylbenz[a]anthracene. Chem.-Biol. Interact. 26:339-347. [DOI] [PubMed] [Google Scholar]
- 36.Wu, J., and L. K. Wong. 1981. Microbial transformation of 7,12-dimethylbenz[a]anthracene. Appl. Environ. Microbiol. 41:843-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, S. K., and P. P. Fu. 1984. The effect of the bay-region 12-methyl group on the stereoselective metabolism at the K-region of 7,12-dimethylbenz[a]anthracene by rat liver microsomes. Biochem. J. 223:775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, S. K., and P. P. Fu. 1984. Stereoselective metabolism of 7-methylbenz[a]anthracene: absolute configurations of five dihydrodiol metabolites and the effect of dihydrodiol conformation on circular dichroism spectra. Chem. Biol. Interact. 49:71-88. [DOI] [PubMed] [Google Scholar]
- 39.Yang, S. K., M. Mushtaq, and P. P. Fu. 1990. Absolute configuration of cis-5,6-dihydrodiol enantiomers derived from helical conformers of 7,12-dimethylbenz[a]anthracene. Chirality 2:58-64. [DOI] [PubMed] [Google Scholar]
- 40.Zacharias, D. E., J. P. Glusker, R. G. Harvey, and P. P Fu. 1977. Molecular structure of the K-region cis-dihydrodiol of 7,12-dimethylbenz[a]anthracene. Cancer Res. 37:775-782. [PubMed] [Google Scholar]