Abstract
The possible roles of 13 Msn2p- and Msn4p-regulated genes in the adaptation of Saccharomyces cerevisiae to the herbicide 2,4-d-dichlorophenoxyacetic acid (2,4-D) were examined. Single deletion of genes involved in defense against oxidizing agents (CTT1, GRX1, and GRX2/TTR1) or encoding chaperones of the HSP70 family (SSA1, SSA4, and SSE2) showed a slight effect. A more significant role was observed for the heat shock genes HSP78, HSP26, HSP104, HSP12, and HSP42, most of which encode molecular chaperones. However, the SPI1 gene, encoding a member of the glycosylphosphatidylinositol-anchored cell wall protein family, emerged as the major determinant of 2,4-D resistance. SPI1 expression reduced the loss of viability of an unadapted yeast population suddenly exposed to the herbicide, allowing earlier growth resumption. Significantly, yeast adaptation to 2,4-D involves the rapid and transient Msn2p- and Msn4p-mediated activation (fivefold) of SPI1 transcription. SPI1 mRNA levels were reduced to values slightly above those in unstressed cells when the adapted population started duplication in the presence of 2,4-D. Since SPI1 deletion leads to the higher β-1,3-glucanase sensitivity of 2,4-D-stressed cells, it was hypothesized that adaptation may involve an Spi1p-mediated increase in the diffusional restriction of the liposoluble acid form of the herbicide across the cell envelope. Such a cell response would avoid a futile cycle due to acid reentry into the cell counteracting the active export of the anionic form, presumably through an inducible plasma membrane transporter(s). Consistent with this concept, the concentration of 14C-labeled 2,4-D in 2,4-D-energized adapted Δspi1 mutant cells and the consequent intracellular acidification are higher than in wild-type cells.
The widespread and intensive use and misuse of herbicides may give rise to a number of toxicological and environmental problems and have led to a growing number of resistant weed species and classes of herbicides to which resistance has evolved (3, 30). Saccharomyces cerevisiae is a very useful experimental model for studies of the toxic effects of herbicides and other xenobiotics and of mechanisms of adaptation and resistance to environmental stress in more-complex and less genetically accessible eukaryotes. Seven years of postgenomic research focused on yeast have facilitated these studies by making available thousands of mutant forms of all nonessential genes (15) and a large amount of information from genome approaches. In particular, microarray analysis has given crucial insights into the complex gene regulation occurring in yeast cells exposed to stress factors, leading to the identification of a set of genes whose transcription is up-regulated under every stress condition studied. These genes include those with functions in carbohydrate metabolism, cell stress, and energy generation (6, 9). The majority of these genes are regulated by the Msn2p and Msn4p transcription factors. In response to various stresses, these homologous regulators are translocated from the cytosol to the nucleus, activating the transcription of target genes, in general through the stress responsive cis element (CCCCT/AGGGG) (10, 16, 17).
Although the auxin analogue herbicide 2,4-dichlorophenoxyacetic acid (2,4-D) acts in plants via physiological mechanisms that do not exist in yeast, many of the mechanisms underlying resistance to drugs and other chemicals are, apparently, conserved among phylogenetically distant organisms (12, 30). 2,4-D is a highly lipophilic weak acid (log P = 2.8 [25], where P is the octanol-water partition coefficient; pKa = 2.73 [29]), and, consistent with the concept that the toxic form is the liposoluble undissociated form, at values close to its pKa, 2,4-D toxicity is very high, decreasing with the increase of external pH (4). It is likely that this highly liposoluble weak acid significantly affects the spatial organization of plasma membrane, affecting its function as a matrix for enzymes and as a selective barrier, thereby leading to the dissipation of the proton motive force across this membrane (23). As for other weak acids, following the entrance of undissociated acid into the cell by passive diffusion, its dissociation in the approximately neutral cytoplasm leads to a decrease of intracellular pH (pHi) and accumulation of the anion, which cannot cross plasma membrane lipids but which may be actively exported through a specific inducible transporter(s), presumably Pdr5p (26). Yeast adaptation and resistance to the herbicides 2,4-D and 2-methyl-4-chlorophenoxyacetic acid involve the rapid and transient transcriptional activation of the TPO1 and PDR5 genes, encoding putative multidrug transporters belonging to the major facilitator superfamily and to the ATP-binding cassette (ABC) superfamily, respectively, in a Pdr1p- and Pdr3p-dependent manner (26). In the present work, we searched for additional determinants of resistance to 2,4-D in S. cerevisiae. Following the demonstration that the MSN2 and MSN4 genes are required for adaptation and resistance to 2,4-D in yeast, we have examined the possible participation of 13 Msn2p and Msn4p target genes (Table 1) in this phenomenon. The selected genes are involved in the general stress response, in particular, in adaptation to weak acids (2, 6, 8, 9, 20). Among the genes examined in the present work, the SPI1 gene emerged as the major determinant of 2,4-D resistance. SPI1 encodes a cell wall protein containing a signal sequence for the addition of a glycosylphosphatidylinositol (GPI) anchor (11). It is strongly induced during stationary phase and under different stress conditions (6, 9, 21) and is required for resistance to lyticase in 2,4-D-adapted cells (this work), suggesting that this gene may be essential for reducing cell envelope permeability under stress. Experimental evidence reported in this study indicates that yeast adaptation to 2,4-D involves the Msn2p- and Msn4p-mediated rapid transcriptional activation (fivefold) of the SPI1 gene and that Spi1p is required to reduce the intracellular pool of 2,4-D and the consequent intracellular acidification. Therefore, we submit that the role of Spi1p, as part of the yeast response mechanisms to 2,4-D, involves the restriction of the diffusional entry of the acid form across the cell envelope, thus avoiding the counteraction of the inducible active export of the respective anion.
TABLE 1.
Msn2p-and Msn4p-regulated genes tested in the present work for a possible role in yeast resistance to 2,4-Da
| Gene | Cellular role |
|---|---|
| Cell wall protein gene | |
| SPII | GPI-attached cell wall protein |
| Antioxidant enzyme genes | |
| CTT1 | Cytosolic catalase T |
| GRX1 | Glutaredoxin |
| GRX2/TTR1 | Glutaredoxin |
| SOD2 | Manganese superoxide dismutase |
| HSP genes | |
| HSP12 | HSP of 12 kDa |
| HSP26 | HSP of 26 kDa; protein folding chaperone |
| HSP42 | HSP of 42 kDa; protein folding chaperone |
| HSP78 | HSP of 78 kDa; protein folding chaperone |
| HSP104 | HSP of 104 kDa; protein folding chaperone |
| HSP70 family genes | |
| SSA1 | HSP of 70 kDa; protein folding chaperone |
| SSA4 | HSP of 70 kDa; protein folding chaperone |
| SSE2 | HSP of 70 kDa; protein folding chaperone |
HSP, heat shock protein.
MATERIALS AND METHODS
Strains, growth media, and growth conditions.
S. cerevisiae strains used in this work are described in Table 2. Cells of the parental strain BY4741 and the 15 Euroscarf mutant strains examined were batch cultured at 30°C, with orbital agitation (250 rpm) in MM4 liquid medium (acidified to pH 3.5 with HCl) with the following composition: 1.7 g of yeast nitrogen base without amino acids or NH4+ (Difco Laboratories, Detroit, Mich.)/liter and 20 g of glucose, 2.65 g of (NH4)2SO4, 20 mg of methionine, 20 mg of histidine, 60 mg of leucine, and 20 mg of uracil (all from Sigma Chemical Co., St. Louis, Mo)/liter. Agarized solid medium (acidified to pH 4.5 with HCl) contained, besides the above-indicated ingredients, 20 g of agar (Iberagar S.A., Barreiro, Portugal)/liter.
TABLE 2.
S. cerevisiae strains used in this studya
| Strain | Genotpye |
|---|---|
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| BY4741_Δmsn2 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YMR037c::kanMX4 |
| BY4741_Δmsn4 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YKL062w::kanMX4 |
| BY4741_Δspi1 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YER150w::kanMX4 |
| BY4741_Δctt1 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YGR088w::kanMX4 |
| BY4741_Δgrx1 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YCL035c::kanMX4 |
| BY4741_Δgrx2 | MATahis3Δ1 leu2Δ0 met 15Δ0 ura3Δ0 YDR513w:: kanMX4 |
| BY4741_Δsod2 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YHR008c::kanMX4 |
| BY4741_Δhsp12 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YFL014w::kanMX4 |
| BY4741_Δhsp26 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YBR072w::kanMX4 |
| BY4741_Δhsp42 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YDR171w::kanMX4 |
| BY4741_Δhsp78 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YDR258c::kanMX4 |
| BY4741_Δhsp104 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YLL026w::kanMX4 |
| BY4741_Δssa1 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YAL005c::kanMX4 |
| BY4741_Δssa4 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YER103w::kanMX4 |
| BY4741_Δsse2 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YBR169c::kanMX4 |
All strains were from Euroscarf.
2,4-D susceptibility assays.
Comparisons of the susceptibilities of BY4741 and the deletion mutant strains to inhibitory concentrations of 2,4-D were based on spot assays or growth curves in liquid medium. The cell suspensions used to inoculate the agar plates were prepared by cultivation in MM4 liquid medium, until the standardized culture optical density at 600 nm (OD600) of 0.5 ± 0.05 was reached, followed by dilution to a standardized culture OD600 of 0.05 ± 0.005. These cell suspensions and a subsequent dilution (1:4) were applied as spots (4 μl) onto the surface of the agarized MM4 medium (at pH 4.5) supplemented with adequate 2,4-D concentrations (ranging from 2.5 to 4.5 mM). Plates were incubated at 30°C for 3 to 8 days, depending on the severity of growth inhibition. Susceptibility assays were also carried out by comparing the growth curves in MM4 liquid medium (at pH 3.5) supplemented with adequate 2,4-D concentrations at 30°C and subjected to orbital agitation (250 rpm). 2,4-D was obtained from Sigma, and the stock solution was prepared in dimethyl sulfoxide (DMSO) (Sigma), whose concentration in the growth medium was kept constant and below an inhibitory level (0.07% vol/vol). In control experiments, DMSO was also added to the growth medium at this concentration. The low pH of the liquid medium avoided the need for adding high herbicide concentrations to obtain growth inhibition, but, to avoid agarized medium liquefaction as the result of autoclaving, medium pH was adjusted to 4.5. Growth curves were based on measurements of culture OD600.
Northern blot analysis.
RNA extraction from yeast cells cultivated under herbicide stress was performed according to the hot-phenol method, and Northern blot hybridizations were carried out as reported before (26). Total RNA in each sample used for Northern blotting was approximately constant (20 μg; RNA quantification was based on the measurement of RNA solution absorbance at 260 nm). The specific DNA probe to detect SPI1 transcripts was prepared by PCR amplification with the following primers: 5′ AAACTTCTCGAAGTTCCCAGA 3′ and 5′ TTGCAGTAGCAGTCGAGTTGT 3′. This probe includes 200 bp of the SPI1 promoter region and the first 200 bp of the coding region, and its nucleotide sequence shows no significant homology to the rest of the genome. The specificity of the probe was tested by using total RNA extracted from cells of the Δspi1 deletion mutant that had been exposed to the herbicide under conditions leading to gene transcription activation. The ACT1 mRNA level was used as an internal control. Kodak BIOMAX MS (Amersham Pharmacia Biotech, Carnaxide, Portugal) films were exposed to the nitrocellulose membranes following the hybridization step and incubated with an intensifying screen at −70°C for 1 day to obtain SPI1 hybridization signals. The relative intensities in the autoradiograms were quantified by densitometry (computing densitometer and ImageQuant software, version 3.3; Molecular Dynamics).
β-1,3-Glucanase sensitivity assay.
To monitor structural changes in the yeast cell wall, a lyticase (a β-1,3-glucanase from Micrococcus luteus; Sigma) sensitivity assay was conducted as described by Shimoi and coworkers (24). Yeast cells were cultured in MM4 liquid medium, either unsupplemented or supplemented with 0.3 mM 2,4-D, and harvested following 0 or 2.5 h of incubation, during cell adaptation to growth, and at the exponential phase of growth, when the standardized OD600 of 1 ± 0.1 was reached. This corresponded to 10 or 22 h of incubation of the wild type or the Δspi1 mutant, respectively, in the presence of 2,4-D. The harvested cells were washed with distilled water and resuspended in 0.1 mM sodium phosphate buffer (pH 7.5). After the addition of 10 μg of lyticase/ml, cell lysis was monitored by measuring the percent decrease of the initial OD600 of the cell suspensions.
Role of Spi1p in 14C-labeled 2,4-D accumulation in energized adapted cells.
Levels of passive accumulation of 14C-labeled 2,4-D in and its active export from wild-type and Δspi1 cells were compared. Cells were first grown in minimal medium to an OD600 of 0.5 ± 0.05 and then adapted to 2,4-D by cultivation in the same basal growth medium supplemented with 0.3 mM 2,4-D until a culture OD600 of 1 ± 0.1 was reached. These cells were harvested by centrifugation, washed twice with ice-cold water, and resuspended in TM buffer (0.1 M MES [morpholineethanesulfonic acid; Sigma], 41 mM Tris [Sigma]) adjusted with HCl to pH 3.5 to obtain a dense cell suspension (OD600 = 5 ± 0.1; equivalent to approximately 2.2 mg [dry weight] ml−1). After 5 min of incubation in TM buffer at 30°C with orbital agitation (150 rpm), 14C-labeled 2,4-D (Sigma) was added to the cell suspension to a concentration of 8 μM, and incubation proceeded for an additional 12 min, which was found to be enough to reach equilibrium. To monitor intracellular accumulation of labeled 2,4-D in the absence of glucose, 200-μl portions of the cell suspension were taken at adequate time intervals, filtered through prewetted glass microfibers (Whatman; GF/C), and washed with ice-cold TM buffer and the radioactivity was measured in a Beckman LS 5000TD scintillation counter. The active efflux of the labeled 2,4-D that had accumulated beforehand was monitored, after the addition of a pulse of glucose (2% [wt/vol]), by measuring cell radioactivity for an additional period of 20 min. Nonspecific adsorption of 14C-labeled 2,4-D to the filters and to the cells (less than 5% of the total radioactivity) was assessed and taken into consideration. The extracellular concentration of labeled acid was estimated by measuring the radioactivity of 100 μl of the supernatant. To calculate the intracellular concentration of labeled acid, the internal cell volume was considered constant and equivalent to 2.5 μl mg of dry weight−1 (22).
pHi.
The pHi values for populations of S. cerevisiae BY4741 and Δspi1 cells were compared by fluorescence microscopy. Cells were harvested by centrifugation (8,600 × g for 8 min at 4°C) at suitable times during growth in the absence or presence of the herbicide. Cell pellets were washed twice with CF buffer (50 mM glycine, 10 mM NaCl, 5 mM KCl, and 1 mM MgCl2 in 40 mM Tris-100 mM MES [pH 5.8]) and resuspended in 2 ml of CF buffer to an OD600 of 10. 5- and 6-carboxyfluorescein diacetate, succinimidyl-ester [5 (6)-CFDA,SE; 4.0 μl; 45 mM in DMSO; Molecular Probes Europe BV, Leiden, The Netherlands] was added to cell suspension for pHi staining (33). The mixture was vortexed in two bursts of 1 min each, interspersed with 15 min on ice. After being washed twice with cold CF buffer, 5 (6)-CFDA,SE-loaded cells were resuspended in 2 ml of CF buffer and examined immediately with a Axioplan microscope equipped with adequate epifluorescence interference filters (BP450-490 and LP520; Zeiss). Fluorescent emission was collected with a cooled charge-coupled device camera (Cool SNAPFX; Roper Scientific Photometrics). Bright-field images for determinations of pHi were obtained concurrently. Images were recorded at 1-min intervals, and each experiment was finished within 15 min. The images were analyzed with MetaMorph, version 3.5. The fluorescence images were background corrected by using dark-current images. The pHi values were calculated for a minimum of 80 cells/experiment. Individual cells were selected from regions of interest obtained from bright-field images recorded before or after the experiment. The value of fluorescence intensity emitted by each cell was obtained pixel by pixel in the region of interest. To estimate average pHi, an in vivo calibration curve was prepared from cell suspensions grown in the absence of herbicide, which were loaded with 5 (6)-CFDA,SE as described above and incubated at 30°C with 0.5 mM carbonyl cyanide m-chlorophenylhydrazone to dissipate the plasma membrane pH gradient (31), before adjustment of external pH (in the range of 3.0 to 7.0) by the addition of HCl or NaOH at 2 M.
RESULTS
MSN2 and MSN4 genes are required for yeast adaptation and resistance to 2,4-D.
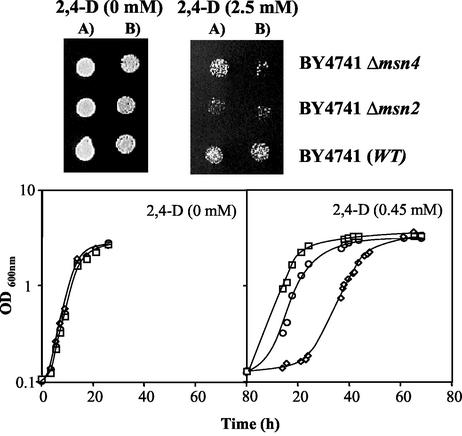
The comparison, by spot assays, of the susceptibilities to the herbicide 2,4-D of the wild-type strain BY4741 and mutants with the MSN2 or MSN4 gene deleted indicated that the two homologous transcription factors Msn2p and Msn4p are determinants of yeast resistance to the herbicide (Fig. 1). Batch cultivation of yeast cell populations that had not been previously adapted to the herbicide revealed that the duration of the adaptation period preceding cell division under 2,4-D stress was longer for the Δmsn4 and the Δmsn2 populations than for the wild-type population (Fig. 1). The effect of MSN2 expression on yeast resistance to 2,4-D, assessed either on solid or liquid medium, was more evident than that of MSN4. Although the expression of the MSN2 or MSN4 gene reduced the duration of the herbicide-induced lag phase, the inhibited growth kinetics of the population adapted to this chemical stress was, apparently, independent of the expression of either gene (Fig. 1).
FIG. 1.
MSN2 and MSN4 genes are required for S. cerevisiae adaptation and growth under 2,4-D stress. Susceptibility assays were based on spot assays (agarized medium at pH 4.5) or on comparison of the growth curves (at pH 3.5), in the same basal medium supplemented or not with 2,4-D (0.45 mM), of wild-type (WT) strain BY4741 (□) and the Δmsn2 (⋄) and Δmsn4 (○) deletion mutants. The growth curves show average values from at least three independent growth experiments. Cells used to prepare the spots or to inoculate the liquid media were grown in the absence of herbicide, until a standardized OD600 of 0.5 ± 0.05 was attained, during exponential phase. The cell suspensions used to prepare the spots in lanes B were 1/4 dilutions of the suspensions (OD600 = 0.05 ± 0.005) in lanes A.
Effects of the expression of Msn2p and Msn4p target genes, in particular of SPI1, on yeast resistance to 2,4-D.
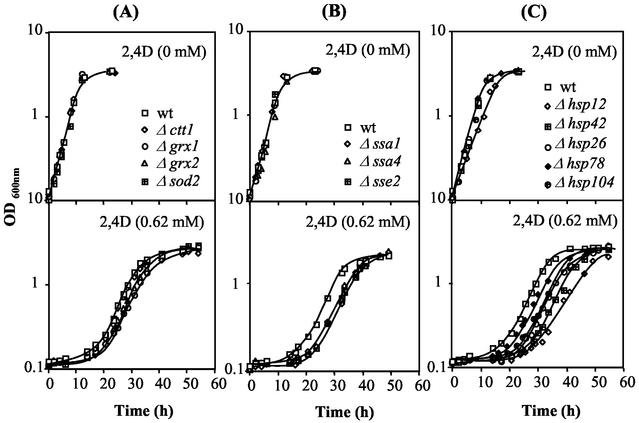
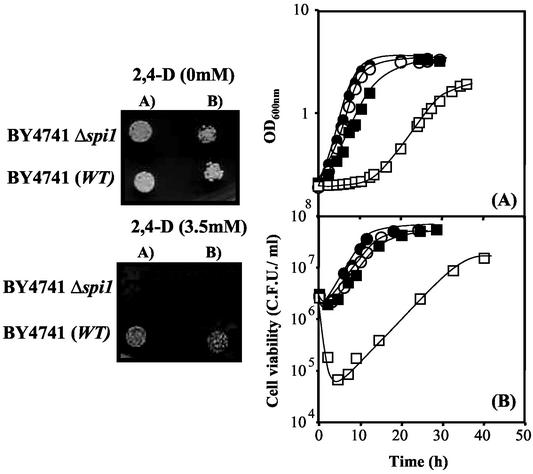
The susceptibilities to 2,4-D of the wild-type strain BY4741 and 13 mutants (Table 1) with known Msn2p or Msn4p target genes deleted were compared in solid and liquid media (Fig. 2 and 3 and results not shown). Spot assays led to the clear identification of the SPI1 and HSP12 genes as 2,4-D resistance determinants in yeast (Fig. 3 and results not shown). The comparison of the growth curves in liquid media supplemented with a highly inhibitory concentration of 2,4-D (0.62 mM, pH 3.5) further indicated that most of the tested deletion mutants are moderately more susceptible to 2,4-D than the wild-type strain, with the exception of the Δsod2 mutant, whose growth curve under herbicide stress was apparently coincident with that of the wild type (Fig. 2). The single deletion of the other genes involved in the oxidative stress response examined had a slight effect (Fig. 2A). More-significant differences, although still small, were observed when any of the genes encoding proteins of the HSP70 family was individually deleted (Fig. 2B). The most significant difference in 2,4-D susceptibility was observed as the result of SPI1 deletion, since no growth of the Δspi1 deletion mutant in medium supplemented with the same inhibitory concentration of 2,4-D was observed (results not shown). The heat shock genes HSP78, HSP26, HSP104, HSP12, and HSP42 also play a role in yeast adaptation to 2,4-D (Fig. 2C). The single deletion of the majority of Table 1 genes led essentially to the reduction of the duration of the lag phase preceding eventual growth resumption under herbicide stress. Indeed, no differences in the specific growth rates of the wild-type and mutant strains were observed, with the exception of those for the Δhsp12 (Fig. 2C) and Δspi1 (Fig. 3) mutants. However, even in the absence of 2,4-D, HSP12 deletion gave rise to a detectable reduction of the specific growth rate compared to that for the wild-type strain, probably due to the HSP12 role on yeast resistance to the low pH of the growth medium. While differences in the specific growth rates of the wild-type and Δhsp12 populations in the presence of 2,4-D at pH 3.5 are closer to those observed at this low pH in the absence of the herbicide, the increase of the specific growth rate under 2,4-D stress due to SPI1 expression appears to be herbicide specific (Fig. 3). Previous adaptation of Δspi1 cells to exponential growth with 2,4-D led to a reduction of the difference in the duration of the lag phase of the unadapted mutant and the wild-type populations following inoculation in 2,4-D-supplemented medium (results not shown). All these results strongly suggest that the SPI1 gene plays an important role during the adaptation period preceding growth resumption under 2,4-D stress and during exponential growth under herbicide stress.
FIG. 2.
Effect of the expression of Msn2p- and Msn4p-regulated genes, encoding antioxidant enzymes and heat shock proteins, on yeast resistance to 2,4D. Growth susceptibilities to 2,4-D for wild-type (wt) strain BY4741 and 12 mutants with the indicated genes deleted were tested by comparing their growth curves in the absence (0 mM) or presence (0.62 mM) of 2,4-D. The growth curves shown are average values from at least three independent growth experiments. Cell suspensions used to prepare the inocula to obtain an initial OD600 of 0.1 ± 0.01 were cultivated in the absence of herbicide until a standardized OD600 of 0.5 ± 0.05 was attained, in mid-exponential phase.
FIG. 3.
The SPI1 gene is required for S. cerevisiae adaptation and growth under 2,4-D stress as indicated by a comparison of susceptibilities by spot assays or growth curves (A) and cell viabilities (B) of wild-type (WT; •, ▪) and Δspi1 (○, □) strains in the absence (•, ○) or presence (▪, □) of 2,4-D (0.45 mM; pH 3.5). Values are representative of the many growth experiments carried out. Cell suspensions used to prepare the spots or to inoculate the liquid media were cultivated in the absence of herbicide until a standardized OD600 of 0.5 ± 0.05 was attained, in mid-exponential phase. The cell suspensions used to prepare the spots in lanes B were 1/4 dilutions of the cell suspensions (OD600 = 0.05 ± 0.005) in lanes A.
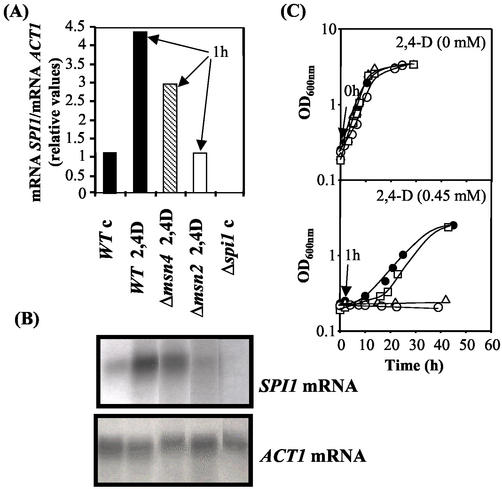
Msn2p- and Msn4p-mediated transcriptional activation of SPI1 following sudden exposure of yeast to 2,4-D.
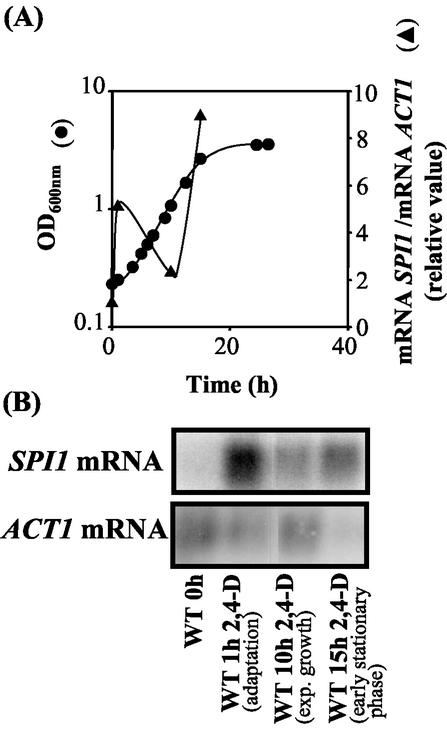
The transcription of the SPI1 gene was activated (fivefold) (Fig. 4) 1 h following sudden exposure of the unadapted wild-type cell population to 2,4-D. However, as soon as the yeast cell population adapted to this chemical stress and started duplication under herbicide stress, the SPI1 mRNA levels were significantly reduced to values that were, however, still above the basal levels detected in unstressed cells (Fig. 4). This rapid transcriptional activation of SPI1 was fully dependent on the presence of Msn2p and partially dependent on the presence of Msn4p (Fig. 5). The longer duration of the adaptation period observed for both the Δmsn2 and the Δspi1 populations, compared with those for the Δmsn4 and wild-type populations (Fig. 5C), may therefore involve the abolishment of the Msn2p-mediated transcriptional activation of SPI1 gene, whose expression is critical to overcome the viability loss during the initial period of adaptation to the herbicide. During transition to the stationary phase of growth in the presence of 2,4-D, SPI1 transcription was dramatically activated (Fig. 4), consistent with results from a previous study reporting the strong stationary-phase-induced activation of SPI1 transcription in the absence of herbicide (21).
FIG. 4.
Results from Northern blot hybridization experiments using SPI1 as probe and the actin-encoding gene, ACT1, as the internal control. Total RNA was extracted from cells of S. cerevisiae BY4741 (WT) harvested during cultivation under 2,4-D stress (0.3 mM) (A). Growth was monitored by measuring culture OD600. Relative SPI1 mRNA/ACT1 mRNA ratios were obtained by densitometry of the autoradiograms (B) and are average values from three experiments. The relative mRNA ratio immediately before cell exposure to the herbicide (0 h) was set as 1.
FIG. 5.
Relative SPI1 mRNA/ACT1 mRNA ratios for the wild type (WT) and Δmsn2 and Δmsn4 mutants 1 h following yeast cell population exposure to 2,4-D (0.3 mM, pH 3.5). Relative SPI1 mRNA/ACT1 mRNA ratios (A) were obtained by densitometry of the autoradiograms (B) and are average values from three experiments. The relative mRNA ratio for the wild type immediately before exposure to the herbicide (control [c]) was set as 1. (C) Corresponding growth curves for the wild-type BY4741 strain (•) and Δspi1 (○), Δmsn2 (▵), and Δmsn4 (□) deletion mutants. Arrows, cell samples harvested for Northern blot assays.
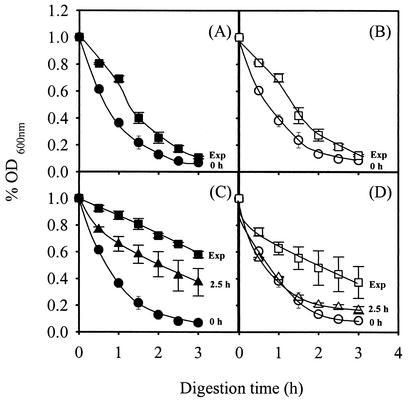
Spi1p is required for lyticase resistance in cells adapted to 2,4-D.
Although the function of Spi1p is not known, the amino acid sequence is 46% identical to that of Sed1p, a cell wall protein whose expression is induced at the stationary phase of growth and that confers resistance to lytic enzymes in stationary-phase cells, presumably because the thicker and less porous wild-type cell wall mannoprotein layer protects the glucan layer from lytic enzymes (7, 14, 24). This prompted us to examine the possible role of Spi1p in the lyticase resistance of yeast cells previously adapted to 2,4-D, in particular those where SPI1 gene transcription was activated. Specifically, the yeast cells used were harvested 2.5 h following cell exposure to 2,4-D and during exponential growth under 2,4-D stress. As reported for SED1, the susceptibilities to lyticase of exponential wild-type and Δspi1 deletion mutant cells grown in the absence of 2,4-D were similar (Fig. 6A and B). However, 2.5 h following cell cultivation in the presence of 2,4-D, the resistance of the wild type to lyticase increased significantly but that of the Δspi1 mutant did not (Fig. 6C and D). Moreover, the lyticase resistance of wild-type cells harvested during exponential growth under 2,4-D stress was higher than the resistance exhibited by this population incubated with 2,4-D for only 2.5 h. Although exponential-phase Δspi1 cells grown under 2,4-D stress were more resistant to lyticase than those grown in its absence, they were still more susceptible to lyticase than exponential-phase wild-type cells adapted to 2,4-D. These results indicate that the higher level of SPI1 expression under 2,4-D stress is one of a number of factors by which yeast cells acquire lyticase resistance, which is presumably also due to other structural cell wall proteins.
FIG. 6.
Lyticase sensitivities of cells of wild type (A and C) and Δspi1 (B and D) strains that were grown in the absence of 2,4-D and then incubated in growth medium unsupplemented (A and B) or supplemented with 0.3 mM 2,4-D (C and D) and harvested during the growth represented by the growth curves shown in Fig. 3, specifically, at time zero, after 2.5 h of incubation, and at the exponential phase when culture OD600 reached 1 (Exp). The different cell populations were washed with water and resuspended in 0.1 M sodium phosphate buffer, pH 7.5. After addition of 10 μg of lyticase (Sigma) per ml, the decrease in the OD600 of the cell suspension was measured periodically. Data are the means ± standard deviations of at least three independent experiments.
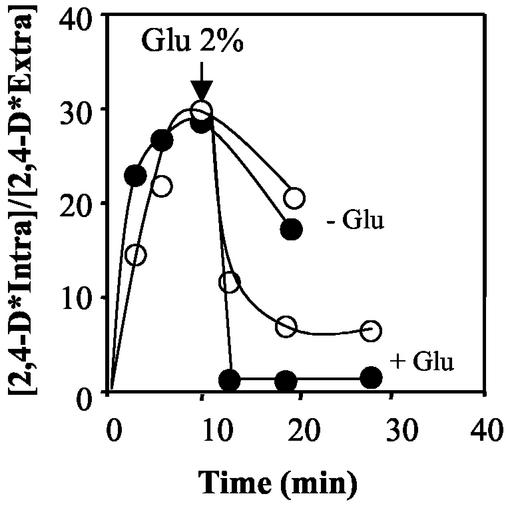
Spi1p is involved in the reduction of the intracellular pool of 2,4-D in adapted cells.
Results from the lyticase susceptibility assays strongly suggested that Spi1p may limit the porosity of the cell wall in 2,4-D-adapted yeast cells. It was therefore hypothesized that, following the cell response to 2,4-D, involving the activation of SPI1 transcription, Spi1p may restrict diffusional entry of the liposoluble acid form of the herbicide across the cell envelope. This is essential to reduce the intracellular pool of 2,4-D through the putative active export of its anionic form mediated by an inducible plasma membrane transporter(s). Results shown in Fig. 7 are consistent with this hypothesis. In the absence of glucose, no significant differences in the amount of labeled 2,4-D that accumulated in cells of the wild-type and Δspi1 strains adapted to growth with 2,4-D were observed. However, following a glucose pulse, both strains became capable of actively reducing the concentration of 2,4-D that had accumulated in the cell beforehand, consistent with the expected induction of a putative transporter(s) for the anionic form of 2,4-D in unadapted cells. At equilibrium, following the glucose pulse, the concentration of labeled 2,4-D that accumulated in the Δspi1 deletion mutant reached a value significantly above the corresponding value for wild-type cells (Fig. 7).
FIG. 7.
Time course variation in the accumulation of 14C-labeled 2,4-D in cells of the wild-type (•) and the Δspi1 mutant (○) strains, in the absence of glucose (− Glu) and its subsequent expulsion after a glucose pulse (+ Glu) (arrow). Cells were previously adapted to the herbicide and were harvested at mid-exponential phase (culture OD600 = 1 ± 0.1) of cultivation in minimal medium supplemented with 0.3 mM 2,4-D at pH 3.5. The corresponding growth curves are shown in Fig. 3. Accumulation values are representative of the several experiments.
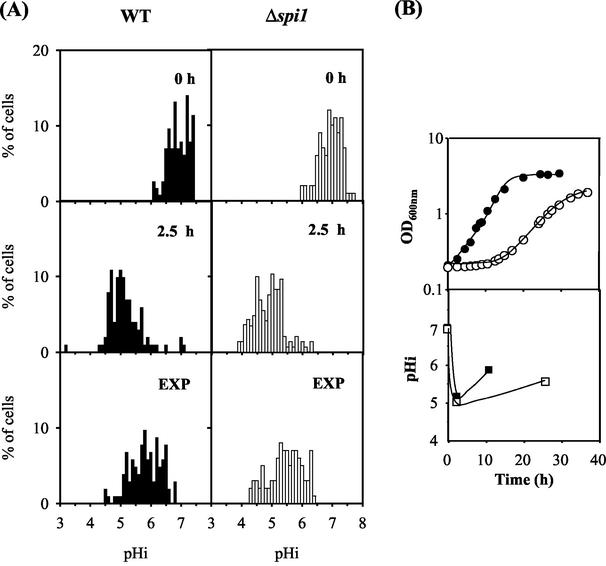
Changes in wild-type and Δspi1 pHi during 2,4-D-stressed cultivation.
The changes in pHi due the exposure of a yeast cell population to 2,4-D at pH 3.5 were monitored by an adaptation of the fluorescence microscopic image processing technique developed by Vindelöv and Arneborg (33). This method, which is different from the techniques based on the distribution of radioactive propionic acid (22), allows a clear-cut picture of the pHi of individual cells and gives information about the distribution of pHi values of the yeast cell population instead of solely an estimation of the average value for the whole population. The wild-type and the Δspi1 populations used to inoculate the growth medium supplemented with 2,4-D (0 h) were heterogeneous as far as pHi values are concerned (Fig. 8). Although both populations showed an average pHi value of 6.9, with 90% of the cells exhibiting pHi values above 6.5, 2.5 h following sudden cell exposure to 2,4-D, the average pHi of the stressed populations decreased significantly; the percentages of the population with pHi below 5.5 reached 76 and 85% for wild-type and Δspi1 strains, respectively (estimated average pHi values were 5.2 and 5.0, respectively). During the exponential growth of yeast cells fully adapted to 2,4-D, pHi values came closer to physiological values (Fig. 8). The percentages of the wild-type and Δspi1 cell populations with pHi values below 5.5 were reduced to 25 and 43%, respectively, while the percentages of cells with pHi above 6 exhibited a corresponding increase. The average pHi values for wild-type and Δspi1 populations in the exponential phase of growth were 5.9 and 5.5, respectively, consistent with the higher accumulation of 2,4-D in Δspi1 cells than in wild-type cells, as suggested by results from active-efflux assays (Fig. 7).
FIG. 8.
(A) Distribution of cells of S. cerevisiae BY4741 (WT) and Δspi1 deletion strains with different pHi values during cultivation under 2,4-D stress. WT, wild type. (B) Average pHi values for wild-type (▪) and Δspi1 (□) 2,4-D-stressed cell populations. Cells were cultivated in the presence of 0.3 mM 2,4-D and harvested at time zero (0 h), after 2.5 h, and at the exponential phase (OD600 = 1 ± 0.1; EXP). Representative growth curves (B, top) for wild-type (•) and Δspi1 (○) strains are shown. Cells were labeled with 5 (6)-CFDA,SE, and pHi was measured by fluorescence imaging. In vivo calibration curves were used to convert measurements of fluorescence to pHi values.
DISCUSSION
The identification of MSN2 and MSN4 genes as determinants of yeast resistance to the highly lipophilic acid herbicide 2,4-D led to the search for Msn2p- and Msn4p-regulated genes required for facilitating yeast cell adaptation to sudden exposure to 2,4-D stress of limited severity. The genes examined are involved in protein folding, defense against reactive oxygen species, and cell wall modification. The great majority belong to a set of genes coinduced in response to a wide range of distinct environmental stresses, attesting to their importance in cellular adaptation to general stress (6, 9). It seems likely that events taking place at the level of the plasma membrane, as the result of the action of a variety of stress factors, presumably including the highly lipophilic acid 2,4-D, may induce a common stress response (18, 32). These events lead to an increase in nonspecific membrane permeability and a consequent decrease in membrane potential and pHi. This study was guided by the large number of indications provided by microarray analysis and other studies concerning the yeast response to acid stress, in particular adaptation to organic acids at low pH (6, 8, 9, 14, 19). The involvement of a number of heat shock proteins and molecular chaperones, believed to be crucial in protein folding, in yeast adaptation to sudden exposure to 2,4-D is consistent with the expected increased damage and aggregation of intracellular proteins under this stress. The anticipated higher levels of expression of several of the HSP genes in yeast cells during adaptation to 2,4-D were recently confirmed by proteomic analysis (M. C. Teixeira, P. M. Santos, and I. Sá-Correia, unpublished results). Although slight, as would be expected in yeast cells in which fermentative metabolism was occurring, the protective role of Msn2p- and -4p-regulated genes encoding oxidative-stress defense mechanisms indicates a pro-oxidant effect of 2,4-D, as reported before for sorbic acid (20).
Among the Msn2p- and Msn4p-regulated genes examined in this work, SPI1 emerged as the major determinant of resistance to 2,4-D. This gene plays a crucial role in facilitating yeast adaptation to a new environment where an herbicide concentration of limited severity is present, providing continuous protection in adapted cells dividing under this chemical stress. Consistently, SPI1 transcription undergoes a rapid and transient fivefold increase following cell exposure to 2,4-D, maintaining levels of transcription slightly above those in unstressed cells during exponential growth in the presence of the herbicide. According to experimental evidence, Spi1p is a GPI-dependent cell wall protein (11). The Spi1p-mediated increased resistance to β-1,3-glucanase of 2,4-D-adapted yeast cells compared with that of unadapted cells indicates that adaptation to herbicide stress involves the modification of cell wall architecture. The molecular organization of the S. cerevisiae cell wall is highly dynamic: the yeast adapts its cell wall composition and structure as a mechanism for protection against cell wall-destabilizing conditions (13, 14). In particular, the molecular organization of the yeast cell wall is strongly affected by decreasing the external pH from 5.5 to 3.5, with the cells becoming increasingly resistant to β-1,3-glucanase lysis (14). Interestingly, DNA microarrays studies revealed the transcriptional activation of four cell wall-related genes at low pH, the SPI1 gene being among them (14). It is possible that this cellular response, attributed by the authors to the decrease of external pH from 5.5 to 3.5, is not caused by the low pH itself but by the presence, in the incubation medium, of succinic acid (5). The antimicrobial effect of weak acids at low pH relies on the predominance of the undissociated liposoluble toxic form, leading to the increase of plasma membrane permeability to different extents depending on the weak acid log P (P is the octanol-water partition coefficient), a value widely used as a measure of the lipophilicity of a chemical. The up-regulation of SPI1 in yeast cells suddenly exposed to the food preservative sorbic acid was also detected by microarrays (8), reinforcing the idea that this gene is induced during weak acid adaptation. Additionally, SPI1 is induced at the stationary phase (21) and is among the yeast genes whose expression is up-regulated under different stress situations, as it is also induced by heat shock, oxidative stress, salt stress, and osmotic stress (6).
Despite the number of indications suggesting that SPI1 is involved in the yeast response to environmental changes, the biological function of the Spi1 protein is not known. In the present work, we have shown that SPI1 is a determinant of resistance to the highly lipophilic weak acid 2,4-D. Adaptation and growth under herbicide stress may involve the rapid alteration of the cell wall mediated by (at least) the increased expression of Spi1p, concomitant with the acquisition of resistance to cell lysis by β-1,3-glucanase digestion. In other words, the external protein layer of the yeast cell wall, which limits the accessibility of the internal glucan layer to lyticase in intact cells, might become more impermeable in cells confronted with the herbicide. It has been proven that Spi1p plays an important role in this protective mechanism, which may also restrict diffusional entry of the undissociated form of 2,4-D across the cell envelope of 2,4-D-adapted cells. The limitation of the diffusional entry of the acid, after sudden exposure of yeast cells to 2,4-D, is crucial to reduce the intracellular pool of the herbicide through the active expulsion of the anion, mediated by 2,4-D-induced plasma membrane transporters, presumably Pdr5p, Tpo1p (1, 26), and, eventually, other still-unidentified exporters, either belonging to the ABC superfamily or to the major facilitator superfamily. Although the global yeast response to 2,4-D shares a number of determinants and mechanisms of resistance with the yeast response to a wide variety of environmental challenges, in particular to weak acids, some determinants appear to be more specific. This is the case for the plasma membrane multidrug resistance (MDR) transporters, which presumably mediate the active efflux of the anion out of the cell. Indeed, although the MDR transporter of the ABC superfamily, Pdr12p, was pointed out as an inducible exporter of benzoate (19), Pdr12p does not confer resistance to 2,4-D (26). Additionally, the putative proton motive force MDR transporters encoded by the AZR1 gene (open reading frame [ORF] YGR224w) (28) and AQR1 gene (ORF YNL065w) (27), required for resistance to short-chain monocarboxylic acids (C2 to C6), could not be implicated in yeast resistance to the highly lipophilic octanoic acid and 2,4-D (27). These observations indicate that the nature of the R groups of the acids (R-COOH) plays a decisive role in the yeast response to their actions, which differ partly because of differences in their hydrophobicities; differences in hydrophobicity lead to differences in the manner in which membrane organization and, consequently, essential membrane functions are disrupted.
Experimental evidence obtained in this work supports the proposed role of Spi1p in avoiding a futile and energetically expensive cycle, in which the acid diffuses into the cells, counteracting the inducible active expulsion of the anion. First, in 2,4-D-adapted cells, the intracellular pool of the acid and the consequent intracellular acidification are higher in cells devoid of the SPI1 gene. Second, a correlation between the rapid and transient patterns of activation of PDR5, TPO1, and SPI1 transcription taking place during 2,4-D adaptation can be established. This stress response is critical to overcome the viability loss during the initial period of adaptation to the herbicide; Spi1p also exerts a protective role in cells dividing under 2,4-D stress. The fall of pHi during the duration of 2,4-D-induced latency is correlated with the loss of cell viability during this period of adaptation, as found before for octanoic acid (31). The heterogeneity concerning the pHi of the exponential-phase yeast cell population that was grown in the absence of 2,4-D and that was used as inoculum strongly suggests that there is a loss of viability in the subpopulation where pHi decreases to nonphysiological values, as registered by the cell-by-cell analysis carried out. According to this concept, the eventual recovery of growth under 2,4-D stress depends on the remaining viable population, consistent with the critical role played by the physiology and number of viable cells in the inoculum during latency under 2,4-D stress (4). For a specific unadapted yeast population suddenly exposed to 2,4-D stress, the role of SPI1 expression in reducing the duration of latency may therefore be the result of a decreased loss of cell viability during this adaptation period, due to its action in reducing the pool of 2,4-D into the cell and the consequent intracellular acidification.
Acknowledgments
This research was supported by the European Union HERBICBIOREM Project (contract QLK3-CT-1999-00041) and by the POCTI program from Fundação para a Ciência e a Tecnologia (FCT), Portugal (contracts POCTI/AGG/38110/2001 and POCTI/BIO/38115/2001) and through Ph.D. grants to T. Simões and M. C. Teixeira and a postdoctoral grant to A. R. Fernandes.
REFERENCES
- 1.Albertsen, M., I. Bellahn, R. Kramer, and S. Waffenschmidt. 2003. Localization and function of the yeast multidrug transporter Tpo1p. J. Biol. Chem. 278:12820-12825. [DOI] [PubMed] [Google Scholar]
- 2.Boy-Marcotte, E., M. Perrot, F. Bussereau, H. Boucherie, and M. Jacquet. 1998. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J. Bacteriol. 180:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradberry, S. M., B. E. Watt, A. T. Proudfoot, and J. A. Vale. 2000. Mechanisms of toxicity, clinical features and management of chlorophenoxy herbicide poisoning: a review. J. Toxicol. Clin. Toxicol. 38:111-122. [DOI] [PubMed] [Google Scholar]
- 4.Cabral, M. G., C. A. Viegas, M. C. Teixeira, and I. Sá-Correia. 2003. Toxicity of chlorinated phenoxyacetic acid herbicides in the experimental eukaryotic model Saccharomyces cerevisiae: role of pH and of growth phase and size of the yeast cell population. Chemosphere 51:47-54. [DOI] [PubMed] [Google Scholar]
- 5.Carmelo, V., H. Santos, and I. Sá-Correia. 1997. Effect of extracellular acidification on the activity of plasma membrane ATPase and on the cytosolic and vacuolar pH of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1325:63-70. [DOI] [PubMed] [Google Scholar]
- 6.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12:323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Nobel, J. G., F. M. Klis, J. Priem, T. Munnik, and H. Van Den Ende. 1990. The glucanase-soluble mannoproteins limit cell wall porosity in Saccharomyces cerevisiae. Yeast 6:491-499. [DOI] [PubMed] [Google Scholar]
- 8.De Nobel, H., L. Lawrie, S. Brul, F. Klis, M. Davis, H. Alloush, and P. Coote. 2001. Parallel and comparative analysis of the proteome and transcriptome of sorbic acid-stressed Saccharomyces cerevisiae. Yeast 18:1413-1428. [DOI] [PubMed] [Google Scholar]
- 9.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Görner, W., E. Durchschlag, M. T. Martínez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schüller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamada, K., S. Fukuchi, M. Arisawa, M. Baba, and K. Kitada. 1998. Screening for glycosylphosphatidylinositol (GPI)-dependent cell wall proteins in Saccharomyces cerevisiae. Mol. Gen. Genet. 258:53-59. [DOI] [PubMed] [Google Scholar]
- 12.Hohmann, S., and W. H. Mager. 1997. Yeast stress responses. Springer R. G. Landes Company, Austin, Tex.
- 13.Kapteyn, J. C., H. Van Den Ende, and F. M. Klis. 1999. The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim. Biophys. Acta 1426:373-383. [DOI] [PubMed] [Google Scholar]
- 14.Kapteyn, J. C., B. Ter Riet, E. Vink, S. Blad, H. De Nobel, H. Van Den Ende, and F. M. Klis. 2001. Low external pH induces HOG1-dependent changes in the organization of the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 39:469-479. [DOI] [PubMed] [Google Scholar]
- 15.Kelly, D. E., D. C. Lamb, and L. Kelly. 2001. Genome-wide generation of yeast gene deletion strains. Comp. Funct. Genom. 2:236-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Pastor, M. T., G. Marchler, C. Schüller, A. Marchler-Bauer, H. Ruis, and F. Estruch. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress-response element (STRE). EMBO J. 15:2227-2235. [PMC free article] [PubMed] [Google Scholar]
- 17.Moskvina, E., C. Schüller, C. T. C. Mauren, W. H. Mager, and H. Ruis. 1998. A search in the genome of Saccharomyces cerevisiae for genes regulated via stress response elements (STREs). Yeast 14:1041-1050. [DOI] [PubMed] [Google Scholar]
- 18.Moskvina, E., E.-M. Imre, and H. Ruis. 1999. Stress factors acting at the level of plasma membrane induce transcription via the stress response element STRE of the yeast Saccharomyces cerevisiae. Mol. Microbiol. 32:1263-1272. [DOI] [PubMed] [Google Scholar]
- 19.Piper, P., C. Ortiz-Calderon, K. Hatzixanthis, and M. Mollapour. 2001. Weak acid adaptation: the stress response that confers yeasts with resistance to organic acid food preservatives. Microbiology 147:2635-2642. [DOI] [PubMed] [Google Scholar]
- 20.Piper, P. W. 1999. Yeast superoxide dismutase mutants reveal a prooxidant action of weak organic acid food preservatives. Free Radic. Biol. Med. 27:1219-1227. [DOI] [PubMed] [Google Scholar]
- 21.Puig, S., and J. E. Pérez-Ortín. 2000. Stress response and expression patterns in wine fermentations of yeast genes induced at the diauxic shift. Yeast 16:139-148. [DOI] [PubMed] [Google Scholar]
- 22.Rosa, M. F., and I. Sá-Correia. 1996. Intracellular acidification does not account for inhibition of Saccharomyces cerevisiae growth in the presence of ethanol. FEMS Microbiol. Lett. 135:271-274. [DOI] [PubMed] [Google Scholar]
- 23.Sá-Correia, I., S. P. Salgueiro, C. A. Viegas, and J. M. Novais. 1989. Leakage induced by ethanol, octanoic and decanoic acids in Saccharomyces cerevisiae. Yeast 5:S123-S127. [Google Scholar]
- 24.Shimoi, H., H. Kitagaki, H. Ohmori, Y. Limura, and K. Ito. 1998. Sed1p is a major cell wall protein of Saccharomyces cerevisiae in the stationary phase and is involved in the lytic resistance. J. Bacteriol. 180:3381-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sicbaldi, F., and A. A. M. Del Re. 1993. Relationship of pesticide octanol/water partition coefficients to their physiochemical properties. Rev. Environ. Contam. Toxicol. 133:59-93. [Google Scholar]
- 26.Teixeira, M. C., and I. Sá-Correia. 2002. Saccharomyces cerevisiae resistance to chlorinated phenoxyacetic acid herbicides involves Pdr1p-mediated transcriptional activation of TPO1 and PDR5 genes. Biochem. Biophys. Res. Commun. 292:530-537. [DOI] [PubMed] [Google Scholar]
- 27.Tenreiro, S., P. A. Nunes, C. A. Viegas, M. S. Neves, M. C. Teixeira, M. G. Cabral, and I. Sá-Correia. 2002. AQR1 gene (ORF YNL065w) encodes a plasma membrane transporter of the major facilitator superfamily that confers resistance to short-chain monocarboxylic acids and quinidine in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 292:741-748. [DOI] [PubMed] [Google Scholar]
- 28.Tenreiro, S., P. C. Rosa, C. A. Viegas, and I. Sá-Correia. 2000. Expression of the AZR1 gene (ORF YGR224w), encoding a plasma membrane transporter of the major facilitator superfamily, is required for adaptation to acetic acid and resistance to azoles in Saccharomyces cerevisiae. Yeast 16:1469-1481. [DOI] [PubMed] [Google Scholar]
- 29.Tomlin, C. D. S. 1997. The pesticide manual, 11th ed. British Crop Protection Council, London, United Kingdom.
- 30.Twonson, J. K. 1997. Herbicide resistance, p. 139-174. In J. D. Hayes and C. R. Wolf (ed.), Molecular genetics of drug resistance. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 31.Viegas, C. A., P. F. Almeida, M. Cavaco, and I. Sá-Correia. 1998. The H+-ATPase in the plasma membrane of Saccharomyces cerevisiae is activated during growth latency in octanoic acid-supplemented medium accompanying the decrease in intracellular pH and cell viability. Appl. Environ. Microbiol. 64:779-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vigh, L., B. Maresca, and J. L. Harwood. 1998. Does the membrane's physical state control the expression of heat shock and other genes? Trends Biochem. Sci. 23:369-374. [DOI] [PubMed] [Google Scholar]
- 33.Vindelöv, J., and N. Arneborg. 2002. Saccharomyces cerevisiae and Zygosaccharomyces mellis exhibit different hyperosmotic shock responses. Yeast 19:429-439. [DOI] [PubMed] [Google Scholar]