Abstract
Salmonella enterica isolates were recovered from swine at a collaborating processing plant over a 2-month period in the spring of 2000. In the present study, molecular subtyping by pulsed-field gel electrophoresis (PFGE) was performed on the 581 confirmed Salmonella isolates from the 84 Salmonella-positive samples obtained from the previous study. A total of 32 different PFGE pulsotypes were observed visually, and a BioNumerics software analysis clustered those pulsotypes into 12 PFGE groups. The B, F, and G groups predominated throughout the sampling period and were isolated from 39, 22, and 13% of the swine, respectively. In addition, multiple isolates were obtained from 67 of the 84 Salmonella-positive samples, and subtyping revealed multiple PFGE profiles in 35 of these 67 (62%) samples. Both carcass and fecal isolates of Salmonella were recovered from 13 swine, resulting in “matched” samples. Molecular typing of the 252 isolates recovered from the matched samples revealed that 7 (54%) of the 13 carcasses were contaminated with Salmonella pulsotypes that were not isolated from the feces of the same animal. Conversely, from 6 of the 13 (46%) matched animals, Salmonella clonal types were isolated from the feces that were not isolated from the carcass of the same animal. These data establish that each lot of swine introduces new contaminants into the plant environment and that swine feces from one animal can contaminate many carcasses. In addition, these results indicate that the examination of multiple Salmonella isolates from positive samples is necessary to determine the variety of potential contaminants of swine carcasses during slaughter and processing.
Salmonella spp. are recognized as major food-borne pathogens in the United States, causing an estimated 1.4 million cases of salmonellosis and over 500 deaths annually (18). Since Salmonella is an enteric pathogen that colonizes the intestinal tracts of a variety of animals, foods that have contributed most to the incidence of salmonellosis are meat and poultry products. Pork products may be particularly risky, since two recent studies detected Salmonella spp. in 10 and 16% of raw retail pork products (10, 26). Contamination of pork products by Salmonella is often the result of feces being spread onto carcasses during the slaughter and processing of swine. Since Salmonella spp. were isolated from 38% of the swine herds sampled in the 1995 National Animal Health Monitoring System survey (10a) and since Salmonella-positive swine are frequently nonsymptomatic, it is critical to prevent or minimize the spread of fecal matter onto carcasses during slaughter and fabrication.
In the last several years, all poultry and meat processors in the United States have been required to adopt the Hazard Analysis and Critical Control Point (HACCP) system. Under the HACCP system, each plant individually identifies critical processing steps and adopts plans to reduce the associated risks of carcass contamination. More recently, the HACCP Implementation Monitoring System (HIMP) was implemented to allow slaughtering facilities to monitor their own HACCP system, rather than relying on USDA Food Safety and Inspection Service (FSIS) inspectors to do so. While a collaborating slaughtering facility was evaluating their HIMP program in the spring of 2000, we were fortunate to be invited to collect carcass and fecal samples from swine over a 10-day period at the postexsanguination and postchiller steps of processing (24). Salmonella strains were subsequently isolated from these samples to address the effectiveness of HIMP and to investigate the prevalence, source, and clonality of Salmonella in swine. Although Salmonella spp. could be initially isolated from swine carcasses at an approximate prevalence of 73%, the processing controls designed to limit carcass contamination were very effective, resulting in only 0.8% of carcasses testing positive for Salmonella after emerging from the chill step (24). These Salmonella prevalence rates were similar to those seen prior to the implementation of the HIMP program, indicating that this particular processing plant was successful at monitoring its own HACCP system. In addition to the carcass testing performed in this study, Salmonella spp. were also isolated from the feces of 33% of the same swine at exsanguination (24).
The confirmed Salmonella recovered during our previous study in 2000 (24) comprised a collection of 581 isolates collected from the 83 positive postexsanguination samples and one positive postchiller sample of swine carcasses. In addition, for 60 of the 100 swine sampled at the postexsanguination step of processing, both carcass and fecal samples were recovered, resulting in “matched” carcass and fecal samples from individual swine. The ability to link certain isolates to specific animals presented a unique opportunity to study Salmonella heterogeneity among swine, the patterns of fecal contamination during slaughter, and the prevalence and persistence of any particular clonal type(s) of Salmonella. To accomplish these tasks, the set of 581 Salmonella isolates were molecularly typed by the technique of pulsed-field gel electrophoresis (PFGE). This highly discriminatory method is a standard technique for the fingerprinting of Salmonella isolates and allowed for clonal types of Salmonella to be tracked by date of isolation, individual animal that yielded the isolate, and source of each isolate (carcass or feces).
MATERIALS AND METHODS
Salmonella isolates.
The Salmonella isolates used in this study were obtained in the course of a previous study in which swine carcasses and feces were sampled (24). In general, 10 carcasses, originating from two to four lots, were sponged on the side of the neck, belly, and ham postexsanguination but prior to dehairing on 10 individual days during April and May 2000. From 60 of the postexsanguination swine sampled by carcass sponging on 6 of the 10 collection days, feces were collected directly from the rectum with a sterile swab, for a total of 60 fecal samples. In addition to the postexsanguination sampling, 122 carcasses from the same lots were sponged about 24 h later at the postchiller step of processing. Carcass samples were tested for the presence of Salmonella spp. by an independent microbiological testing laboratory using selective enrichments and the Vidas method (1). Fecal samples were tested for the presence of Salmonella by a modified version (24) of the selective-enrichment protocol developed by O'Carroll et al. (19). Further testing of 10 to 20 Salmonella isolates from each positive carcass and fecal sample included various biochemical, immunological, and PCR assays (24). The initial tests concluded that 73 of the 100 (73%) carcass samples were positive for Salmonella, 20 of the 60 (33.3%) fecal samples were positive, and 1 of 122 (0.8%) postchiller carcass samples was positive. However, after laboratory passage and further testing, several of the original isolates were either contaminated or determined to be non-Salmonella. Therefore, the final set of 581 Salmonella isolates were recovered from 83 Salmonella-positive postexsanguination samples (64 carcass samples and 19 fecal samples) and one Salmonella-positive postchiller carcass.
PFGE and phylogenetic analysis.
The technique of PFGE was performed in accordance with the standard USDA FSIS protocol (L. V. Cook, Laboratory communication no. 1, The use of pulsed-field electrophoresis for subtyping Salmonella spp. surveillance isolates, FSIS directive, 1998) by using a CHEF mapper XA system (Bio-Rad Laboratories, Hercules, Calif.) in 0.5× Tris-borate-EDTA at 14°C and a 6-V/cm gradient with pulses ramping from 2.16 to 63.8 s over 19 h. Gels were stained with ethidium bromide and photographed by using Multi-Analyst (Bio-Rad) gel documentation software. To perform the phylogenetic analyses of the pulsotypes, TIFF files were analyzed with BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium). Cluster analyses using the Dice correlation for band matching with a 1.0% position tolerance and a hierarchic unweighted pair group method with averaging algorithm were used to generate a dendrogram describing the relationship among Salmonella pulsotypes.
Ribotyping.
Ribotyping studies were conducted with enzyme kits (PvuII) and an automated Riboprinter (Qualicon, Wilmington, Del.) by following the manufacturer's recommendations.
Serotyping.
Isolates from PFGE groups B, F, and G were serotyped by the USDA Animal and Plant Health Inspection Service National Veterinary Services Laboratory, Ames, Iowa.
RESULTS
Heterogeneity of Salmonella pulsotypes.
From the 64 Salmonella-positive swine carcasses sampled postexsanguination, 323 Salmonella isolates were recovered. From the 19 Salmonella-positive fecal samples obtained, 257 Salmonella isolates were recovered. One Salmonella isolate was recovered from a positive postchill carcass. By visual analyses, genomic DNA fingerprinting of these 581 Salmonella isolates by PFGE revealed 32 distinct XbaI restriction patterns, referred to as pulsotypes. However, some of the restriction patterns of the pulsotypes were very similar, differing by three or fewer bands. Since such isolates were likely to be highly related (25), they were considered subtypes of a larger classification of PFGE groups. By these empirical criteria, a total of 12 PFGE groups, designated A to L, were observed. Groups B, F, and G consisted of 12, 4, and 7 pulsotypes, respectively, whereas the remaining nine PFGE groups (A, C, D, E, H, I, J, K, and L) contained only a single pulsotype.
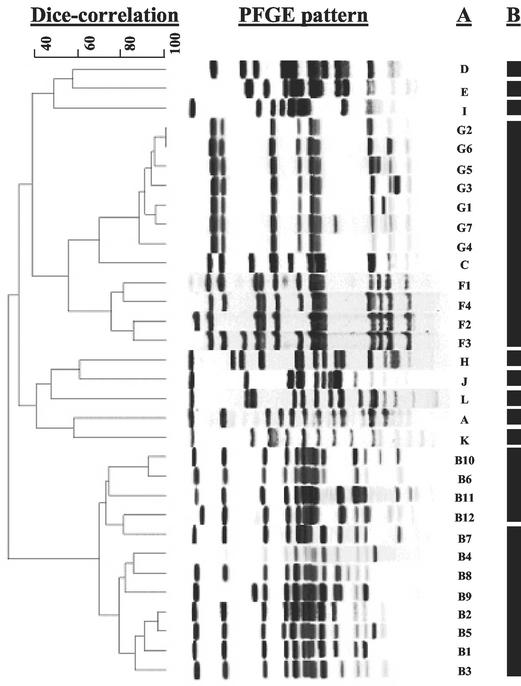
As a method of confirming the visual analyses of the pulsotypes, the relatedness of the 32 pulsotypes was also determined with BioNumerics software (Fig. 1). Analysis by BioNumerics indicated that each of the 12 PFGE groups were from 30 to 69% related to one another as determined by Dice correlation coefficients. In addition, pulsotypes visually identified as subtypes of groups B, F, and G were shown to cluster within their respective groups (68 to 87% related). Analyses by ribotyping revealed 11 “ribogroups” on the basis of indistinguishable patterns generated by the Riboprinter (Fig. 1). Although most of the PFGE groups were identified as distinct ribogroups, PFGE groups C, F, and G were all identified as part of a single ribogroup and two ribogroups were identified within PFGE group B.
FIG. 1.
The relatedness of Salmonella pulsotypes. BioNumerics software was used to analyze the 32 pulsotypes distinguished by visual analyses and determine the phylogenetic relationship among isolates based on the presence or absence of bands. In column A, the 32 pulsotypes are listed adjacent to the corresponding PFGE pattern. In column B, the 11 ribogroups are represented by solid blocks adjacent to the corresponding pulsotypes.
Prevalence of Salmonella pulsotypes and serotyping.
The PFGE profiles of the Salmonella isolates recovered on each of the 10 sampling days were compared (Table 1). On any sampling day, isolates displayed between 1 and 11 different PFGE pulsotype patterns. In addition, different combinations of pulsotypes were isolated on each day. The most prevalent PFGE groups observed were B, F, and G; these three groups comprised 85% of the total number of Salmonella isolates recovered. Specifically, group B isolates comprised 35% (206 of 581) of the total number of isolates recovered and were isolated from 39% (39 of 100) of the swine on 9 of 10 sampling days. Group F isolates comprised 35% (203 of 581) of the total number of isolates and were recovered from 22% (22 of 100) of the swine on 7 of 10 sampling days. Group G isolates comprised 15% (85 of 581) of the isolates and were recovered from 13% (13 of 100) of the swine on 3 of 10 sampling days. Isolates displaying PFGE patterns A, C, D, E, H, I, J, K, and L each comprised less than 4% of the total number of isolates and were recovered from ≤6% of the swine on only 1 or 2 sampling days.
TABLE 1.
Summary of swine Salmonella sources, quantities, and PFGE pulsotypes
| Date | No. of positive samples/ total no. of samples | No. of isolates recovered | PFGE group(s) recovereda |
|---|---|---|---|
| 17 April | 9/10 carcassc | 42 | A (2), B1 (11), B2 (15), D (6), E (8) |
| 22 April | 9/10 carcassc | 38 | B1 (11), B6 (9), B8 (2), B9 (1), F1 (1), G2 (2), H (12) |
| 25 April | 10/10 carcassc | 37 | B10 (2), D (3), F1 (14), J1 (5), L (7), K (6) |
| 27 April | 10/10 carcassc | 72 | B2 (47), B11 (5), B12 (3), F1 (1), G2 (15), G6 (1) |
| 1 May | 8/10 carcasses | 44 | B2 (3), F1 (34), F2 (7) |
| 8/10 fecal | 129 | B3 (1), F1 (126), F2 (1), F3 (1) | |
| 2 May | 9/10 carcass | 54 | B3 (34), B5 (2), G1 (14), G3 (1), G4 (2), G6 (1) |
| 6/10 fecal | 85 | B3 (10), B4 (9), B5 (9), G1 (46), G5 (1), G6 (1), G7 (1), I (8) | |
| 9 May | 1/10 carcass | 1 | F1 (1) |
| 2/10 fecal | 20 | B3 (1), B6 (19) | |
| 11 May | 6/10 carcass | 31 | A (17), B7 (3), F1 (11) |
| 1/10 fecal | 2 | A (2) | |
| 15 May | 1/10 carcass | 5 | B6 (5) |
| 1/10 postchiller carcassb | 1 | F1 (1) | |
| 16 May | 2/10 fecal | 20 | C (10), F1 (9), F4 (1) |
The numbers in parentheses refer to the numbers of isolates displaying each PFGE pulsotype for a given day.
The postchill carcass sampling was performed on 16 May, approximately 24 h after the carcasses were exsanguinated on 15 May.
No fecal samples were collected.
Since PFGE patterns B, F, and G were observed in 85% of the isolates, these Salmonella isolates were chosen for further investigation. Serotyping was performed on representatives of each of the B, F, and G subtypes to determine if each of these PFGE patterns corresponded to a particular serotype. Of the 12 type B isolates that were serotyped, all were identified as S. enterica serotype Derby. In addition, all of the type F (five isolates) and G (seven isolates) isolates that were serotyped were identified as S. enterica serotype Typhimurium subsp. Copenhagen and serotype Typhimurium, respectively.
In addition to the postexsanguination sampling of swine, carcasses were sampled following the chilling step that occurred approximately 24 h after the swine initially entered the plant (24). The 122 postchiller carcasses sampled yielded only one isolate of Salmonella, which was identified as a PFGE type F isolate, a prevalent Salmonella clonal type recovered throughout the study. Although the positive postchill carcass could not be matched to one of the specific carcasses sampled at the postexsanguination step, the positive postchill carcass entered the plant on 15 May, the ninth of 10 postexsanguination sampling days. On this day of postexsanguination sampling, only 1 of 10 carcasses sampled was Salmonella positive (Table 1) and none of the Salmonella isolates were identified as PFGE type F. Instead, all of the Salmonella isolates recovered on 15 May were identified as PFGE type B. These data suggest that the Salmonella-positive postchiller carcass was contaminated by swine that were not sampled or that this carcass was contaminated by an alternate source.
Analysis of multiple Salmonella isolates recovered from individual swine.
Up to 10 carcass and 20 fecal isolates were recovered from each swine sampled, resulting in multiple isolates from individual samples (24). From a total of 84 positive samples (64 carcass samples, 19 fecal samples, and 1 postchill carcass sample), 67 (48 carcass samples and 19 fecal samples) yielded multiple Salmonella isolates. To examine the heterogeneity of Salmonella on the carcass and in the feces of swine at slaughter, the PFGE patterns of the multiple isolates recovered from each sample were compared. Of the 19 fecal samples that yielded multiple isolates of Salmonella, 47% (9 of 19) were positive for two or more Salmonella pulsotypes, indicating that nearly one-half of the Salmonella-positive swine were shedding multiple clonal types of Salmonella. In addition, of the 48 carcass samples that yielded multiple isolates of Salmonella, 54% (26 of 48) were positive for two or more Salmonella pulsotypes, most likely due to fecal spread and the heterogeneity observed among the fecal isolates. Also, these results may indicate the spread of fecal matter from several different animals onto carcasses.
Analysis of matched carcass-fecal isolates.
On sampling days 5 through 10 of the 10-day sampling period, both carcass and fecal samples were obtained from individual swine to compare the Salmonella types contaminating the carcasses with those types isolated from feces. These matched carcass-fecal isolates were useful for determining the source(s) of carcass contamination. Of the 60 swine sampled for both carcass and fecal isolates of Salmonella, both carcass and fecal isolates were recovered from 13 of the 60 swine (Table 2). A comparison of the pulsotypes of the Salmonella isolates obtained from these 13 swine indicated that seven (54%) of the carcasses were contaminated with pulsotypes not isolated from the feces of the same animal, suggesting that feces or carcasses from other animals were the source of about one-half of the carcass contaminants. Likewise, fecal isolates from 6 (46%) of the 13 swine displayed pulsotypes not found on the carcasses of the same animals. These results suggest that approximately one-half of the swine carcasses were contaminated with Salmonella pulsotypes identical to those that they were shedding in their feces.
TABLE 2.
Matched swine carcass-fecal Salmonella isolates and their PFGE profiles
| Piga | Date of sampling | PFGE group(s) inb:
|
|
|---|---|---|---|
| Carcass samples | Fecal samples | ||
| 41 | 1 May | B2 (2), F1 (2) | F1 (12) |
| 44 | 1 May | F1 (6), F2 (2) | F1 (12) |
| 45 | 1 May | F1 (4) | F1 (18) |
| 46 | 1 May | F1 (1) | F1 (19) |
| 47 | 1 May | F1 (4) | F1 (20) |
| 48 | 1 May | B2 (1), F1 (4), F2 (4) | F1 (18), F2 (1), F3 (1) |
| 50 | 1 May | F1 (8) | F1 (10) |
| 51 | 2 May | B3 (10) | G1 (6), G6 (1), G7 (1) |
| 53 | 2 May | B3 (3) | B3 (9), G1 (1) |
| 54 | 2 May | G1 (4) | G1 (10) |
| 57 | 2 May | B3 (2) | B4 (8), B5 (9) |
| 59 | 2 May | B3 (5), B5 (2), G4 (1) | G1 (19), G5 (1) |
| 72 | 11 May | F1 (9) | A (2) |
A total of 100 pigs were sampled at the postexsanguination step of processing and numbered sequentially. Both carcass and fecal samples were taken from pigs 40 to 100, resulting in matched samples. The 13 pigs listed are those swine that yielded both carcass and fecal isolates of Salmonella.
The numbers in parentheses refer to the number of isolates displaying each PFGE pattern for each sample.
DISCUSSION
In the 1995 National Animal Health Monitoring System survey, Salmonella strains were isolated from 38% of pig farms (P. J. Fedorka-Cray, E. Bush, L. A. Thomas, J. T. Gray, J. McKean, and D. L. Harris, Salmonella infection in herds of swine, National Animal Health Monitoring System, 1995, http://www.extension.iastate.edu/Pages/ansci/swinereports/asl-1412.pdf.) and specific geographical areas reported a much higher incidence of Salmonella in their swine populations. Since then, many additional studies have focused on establishing the incidence of Salmonella in swine on the farm and at slaughter (8, 9, 11, 20, 22), as well as on investigating possible links between various preslaughter stresses and increased fecal shedding of Salmonella by swine (5, 13, 14, 17). A natural extension of this research is to investigate the manner in which swine carcasses are contaminated by Salmonella and if certain clonal types of Salmonella pose a comparatively large threat as food contaminants. The present study addressed these issues by using PFGE to determine genetic fingerprints of isolates obtained from swine at a slaughter and fabrication facility, which allowed the carcass contamination to be tracked and the heterogeneity and prevalence of Salmonella clonal types to be investigated.
Of the 581 Salmonella isolates recovered, visual analyses of XbaI-generated PFGE patterns revealed 32 distinct pulsotypes, which were associated into 12 PFGE groups. To confirm that the visual examination was accurate, the 32 pulsotypes were analyzed by BioNumerics software. This software also associated the 32 pulsotypes into 12 PFGE groups, with groups B, F, and G consisting of multiple pulsotypes within a tighter cluster. One difference between the software-based and the visually based analyses is that the software determines the relatedness of isolates by the presence or absence of bands while the visual analysis can also detect differences in the intensities of specific bands. This disparity occurred with pulsotypes G2 and G6, which BioNumerics identified as indistinguishable. Our visual examination detected that the small bands were consistently more intense in the G6 pulsotype; therefore, G2 and G6 were considered empirically distinct pulsotypes. As a second method of confirming our visual analysis, ribotyping was performed on the isolates. This technique associated the 32 pulsotypes into 11 ribogroups, which correlated fairly well with the 12 groups identified by PFGE; however, ribotyping was unable to determine subtle differences that delineated the 32 pulsotypes identified by PFGE. Although these confirmatory methods supported the grouping of isolates as determined by the visual examination of the PFGE patterns, the visual analysis was as effective as the software-based analysis and considerably more discriminatory than ribotyping.
The three most prevalent PFGE groups, patterns B, F, and G, accounted for 85% of the isolates recovered. Although it is difficult to definitively compare PFGE patterns among studies, isolates displaying PFGE patterns similar to our type F pattern have been recovered from environmental, food, clinical, and various animal sources (2, 15, 27). By contrast, we were unable to find published data on Salmonella isolates displaying patterns similar to our PFGE patterns B and G, which is more likely due to the lack of available data than to the uniqueness of the B and G restriction patterns. The lack of available data to compare isolates among studies underscores the importance of researchers being able to disseminate standardized data concerning microbial diversity.
Considerable information on the prevalence of specific Salmonella serotypes has been published; therefore, serotyping was utilized to identify the PFGE group B isolates as serotype Derby, the group F isolates as serotype Typhimurium subsp. Copenhagen, and the group G isolates as serotype Typhimurium. The prevalences of particular serotypes can also be difficult to compare among studies due to regional differences, seasonal changes, and alternate microbiological methodologies; however, several studies performed in North America and Europe reported on the predominance of serotype Derby and/or serotype Typhimurium in swine (4, 7, 9, 13, 16, 20), results similar to those presented here. Of particular interest is that 35% of the Salmonella isolates recovered in this study were PFGE group F isolates, identified as serotype Typhimurium subsp. Copenhagen. The isolation of serotype Typhimurium subsp. Copenhagen from swine at much lower frequencies has been reported in several studies (7, 9, 13, 16, 20). Serovar Typhimurium subsp. Copenhagen is an important pathogen, well characterized for its frequent resistance to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline, the resistance pattern normally associated with DT104 strains. In fact, antibiograms will be established for the entire set of 581 Salmonella isolates, which may allow for correlations between the PFGE profile and the antibiotic resistance phenotype for each isolate.
The heterogeneity of Salmonella isolated on any given sampling day was evident; different combinations of Salmonella clonal types were recovered on each of the 10 sampling days. These data indicate that each group of animals introduces new clonal types of Salmonella into the plant environment and that each lot of animals is, therefore, a potential source for contaminating other carcasses. These results agree with a study conducted in Iowa, where swine sampled from farm to slaughter over a period of 4 months showed a weekly change in the predominant Salmonella serotype isolated at the abattoir but only one serotype was isolated on the farm prior to transport (13). Indeed, there are a few reports of several serotypes of Salmonella being isolated from animals on individual farms (9, 12), but most studies have found that only a small percentage of farms are contaminated with more than two serotypes (5, 13) and that a single serotype is frequently isolated from farms for long periods of time (3, 23). In contrast, there are many reports that a herd sheds several different Salmonella serotypes prior to slaughter, a phenomenon attributed to the shedding of latent organisms or rapid infections as a result of transport, “lairage,” and/or feed withdrawal (5, 13, 21). Since in the present study the 10 swine sampled daily were selected from two to four different herds, the increased diversity of the Salmonella isolates recovered on specific days is likely due to multiple lots being sampled. In addition, we isolated up to 10 carcass and 20 fecal isolates per sample, which was likely to increase the observed heterogeneity of Salmonella isolates recovered. Data regarding transport conditions, as well as the duration of feed withdrawal, were not available for this study. Information on these factors in future studies may help elucidate the influence on Salmonella shedding by swine.
To our knowledge, there have been no studies that examined multiple isolates from a single fecal or carcass sample to determine if individual swine may carry several clonal types of Salmonella. From 67 of the 85 positive carcass and fecal samples obtained in this study, multiple isolates of Salmonella were recovered. Analysis of these multiple isolates by PFGE indicated that at least two different clonal types of Salmonella were isolated from the feces of nearly one-half of the swine (47%), while 54% of swine carried more than one clonal type of Salmonella on their carcasses. Since other data from this study indicated that fecal spread among the swine was common during slaughter, it was not unexpected to find multiple clonal types on individual swine carcasses. However, the isolation of multiple types of Salmonella from fecal samples at slaughter suggests that, although only one Salmonella serotype may be isolated from swine feces on the farm, perhaps preslaughter stress promotes the shedding of multiple serotypes. Since the scope of this study did not include on-farm sampling, there is no evidence that the swine only shed one serotype of Salmonella on the farm. The more significant implication of these results is that multiple isolates must be recovered from a fecal sample to fully characterize the shedding capabilities of animals at slaughter.
Comparing the carcass-feces matched isolates from individual animals revealed that 54% of the Salmonella-positive carcasses were contaminated with Salmonella types not found in the feces of the same animal. These data suggest that approximately one-half of the contaminated carcasses were likely contaminated through carcass-to-carcass or feces-to-carcass contact, possibly in the slaughterhouse holding pens. This cross-contamination indicates that the presence of a pathogen in the feces of only a few swine can contaminate many carcasses during processing.
The objectives of this study were to track the origin of Salmonella contamination of swine carcasses and to determine if any specific Salmonella clonal type was more prevalent than others during swine processing. The results presented here demonstrated that three clonal types of Salmonella predominated in the swine, PFGE groups B, F, and G, and that serotyping identified these as serotypes Derby, Typhimurium subsp. Copenhagen, and Typhimurium, respectively. In addition, the isolation of multiple clonal types of Salmonella from many of the carcass and fecal samples is significant, since standard animal testing usually includes the analysis of a single isolate per sample. Also, these data indicated that approximately one-half of the Salmonella-positive carcasses were likely contaminated by carcass-to-carcass or feces-to-carcass contact, suggesting that fecal spread is common and should be minimized to prevent pathogens from entering the food supply.
Acknowledgments
We extend our appreciation to Alan Oser and Lisa Yoder (Hatfield Quality Meats, Hatfield, Pa.) and to Robert Dudley and Sam Palumbo (USDA/ARS/ERRC, Wyndmoor, Pa.) for helpful discussions and/or technical assistance.
Mention of brand or firm name does not constitute an endorsement by the U.S. Department of Agriculture above others of a similar nature not mentioned.
REFERENCES
- 1.Anonymous. 1998. AOAC official method 996.08, 16th ed. AOAC International, Gaithersburg, Md.
- 2.Baggeson, D. L., D. Sandvang, and F. M. Aarestrup. 2000. Characterization of Salmonella enterica serovar Typhimurium DT104 isolated from Denmark and comparison with isolates from Europe and the United States. J. Clin. Microbiol. 38:1581-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baloda, S. B., L. Christensen, and S. Trajcevska. 2001. Persistence of a Salmonella enterica serovar Typhimurium DT12 clone in a piggery and in agricultural soil amended with Salmonella-contaminated slurry. Appl. Environ. Microbiol. 67:2859-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barber, D. A., P. B. Bahnson, R. Isaacson, C. J. Jones, and R. M. Weigel. 2002. Distribution of Salmonella in swine production ecosystems. J. Food Prot. 65:1861-1868. [DOI] [PubMed] [Google Scholar]
- 5.Berends, B. R., H. A. Urlings, J. M. Snijders, and F. van Knapen. 1996. Identification and quantification of risk factors in animal management and transport regarding Salmonella spp. in pigs. Int. J. Food Microbiol. 30:37-53. [DOI] [PubMed] [Google Scholar]
- 6.Carlson, A. R., and T. Blaha. 1999. Investigations into the infection-contamination-infection cycle of zoonotic Salmonella on swine farms: investigation into the occurrence of Salmonella on 25 selected Minnesota swine farms, p. 113-118. In P. B. Bahnson (ed.), Proceedings of the 3rd International Symposium on the Epidemiology and Control of Salmonella in Pork. Biomedical Communications Center, College of Veterinary Medicine, University of Illinois, Urbana-Champaign.
- 7.Currier, M., M. Singleton, J. Lee, and D. Lee. 1986. Salmonella in swine at slaughter: incidence and serovar distribution at different seasons. J. Food Prot. 49:366-368. [DOI] [PubMed] [Google Scholar]
- 8.Davies, P. R., F. Gerardus, E. M. Bovee, J. A. Funk, W. E. M. Morrow, F. T. Jones, and J. Deen. 1998. Isolation of Salmonella serotypes from feces of pigs raised in a multiple-site production system. J. Am. Vet. Med. Assoc. 212:1925-1929. [PubMed] [Google Scholar]
- 9.Davies, P. R., W. E. M. Morrow, F. T. Jones, J. Deen, P. J. Fedorka-Cray, and I. T. Harris. 1997. Prevalence of salmonella in finishing swine raised in different production systems in North Carolina, USA. Epidemiol. Infect. 119:237-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duffy, E. A., K. E. Belk, J. N. Sofos, G. R. Bellinger, A. Pape, and G. C. Smith. 2001. Extent of microbial contamination in United States pork retail products. J. Food Prot. 64:172-178. [DOI] [PubMed] [Google Scholar]
- 11.Funk, J. A., P. R. Davies, and M. A. Nichols. 2001. Longitudinal study of Salmonella enterica in growing pigs reared in multiple-site swine production systems. Vet. Microbiol. 83:45-60. [DOI] [PubMed] [Google Scholar]
- 12.Funk, J. A., P. R. Davies, and M. A. Nichols. 2000. The effect of fecal sample weight on detection of Salmonella enterica in swine feces. J. Vet. Diagn. Investig. 12:412-418. [DOI] [PubMed] [Google Scholar]
- 13.Hurd, H. S., J. D. McKean, I. V. Wesley, and L. A. Karriker. 2001. The effect of lairage on Salmonella isolation from market swine. J. Food Prot. 64:939-944. [DOI] [PubMed] [Google Scholar]
- 14.Isaacson, R. E., L. D. Firkins, R. M. Weigel, F. A. Zuckermann, and J. A. DiPietro. 1999. Effect of transportation and feed withdrawal on shedding of Salmonella Typhimurium among experimentally infected pigs. Am. J. Vet. Res. 60:1155-1158. [PubMed] [Google Scholar]
- 15.Kotetishvili, M., O. C. Stine, A. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2002. Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J. Clin. Microbiol. 40:1626-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letellier, A., S. Messier, J. Pare, J. Menard, and S. Quessy. 1999. Distribution of Salmonella in swine herds in Quebec. Vet. Microbiol. 67:299-306. [DOI] [PubMed] [Google Scholar]
- 17.McKean, J. D., H. S. Hurd, S. Larsen, M. Rostagno, R. Griffith, and I. Wesley. 2001. Impact of commercial pre-harvest processes on the prevalence of Salmonella enterica in cull sows. Berl. Muench. Tieraerztl. Wochenschr. 114:353-355. [PubMed] [Google Scholar]
- 18.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Chapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Carroll, J. M., P. R. Davies, M. T. Correa, and B. D. Slenning. 1999. Effects of sample storage and delayed secondary enrichment on detection of Salmonella spp. in swine feces. Am. J. Vet. Res. 60:359-362. [PubMed] [Google Scholar]
- 20.Qunke, A.-M., N. Leonard, G. Kelly, J. Egan, P. B. Lynch, and P. J. Quinn. 2001. Prevalence of Salmonella serotypes on pig carcasses from high- and low-risk herds slaughtered in three abattoirs. Berl. Muench. Tieraerztl. Wochenschr. 114:360-362. [PubMed] [Google Scholar]
- 21.Rajkowski, K. T., S. Eblen, and C. Laubauch. 1998. Efficacy of washing and sanitizing trailers used for swine transport in reduction of Salmonella and Escherichia coli. J. Food Prot. 61:31-35. [DOI] [PubMed] [Google Scholar]
- 22.Saide-Albornoz, J. J., C. L. Knipe, E. A. Murano, and G. W. Beran. 1995. Contamination of pork carcasses during slaughter, fabrication, and chilled storage. J. Food Prot. 58:993-997. [DOI] [PubMed] [Google Scholar]
- 23.Sandvang, D., L. B. Jensen, D. L. Baggesen, and S. B. Baloda. 2000. Persistence of a Salmonella enterica serotype Typhimurium clone in Danish pig production units and farmhouse environment studied by pulsed field gel electrophoresis (PFGE). FEMS Microbiol. Lett. 187:21-25. [DOI] [PubMed] [Google Scholar]
- 24.Tamplin, M. L., I. Feder, S. A. Palumbo, A. Oser, L. Yoder, and J. B. Luchansky. 2001. Salmonella spp. and Escherichia coli biotype I on swine carcasses processed under the hazard analysis and critical control point-based inspection models project. J. Food Prot. 64:1305-1308. [DOI] [PubMed] [Google Scholar]
- 25.Tenover, F. C., R. D Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White, D. G., S. Zhao, R. Sudler, S. Ayers, S. Friedman, S. Cheng, P. F. McDermott, S. McDermott, D. D. Wagner, and J. Meng. 2001. The isolation of antibiotic-resistant Salmonella from retail ground meats. N. Engl. J. Med. 345:1147-1154. [DOI] [PubMed] [Google Scholar]
- 27.Zhao, T., M. P. Doyle, P. J. Fedorka-Cray, P. Zhao, and S. Ladley. 2002. Occurrence of Salmonella enterica serotype Typhimurium DT104A in retail ground beef. J. Food Prot. 65:403-407. [DOI] [PubMed] [Google Scholar]