Abstract
The formation of the important flavor compound 4-hydroxy-2,5-dimethyl-3[2H]-furanone (HDMF; Furaneol) from d-fructose-1,6-bisphosphate by the yeast Zygosaccharomyces rouxii was studied with regard to the identification of intermediates present in the culture medium. Addition of o-phenylenediamine, a trapping reagent for α-dicarbonyls, to the culture medium and subsequent analysis by high-pressure liquid chromatography with diode array detection revealed the formation of three quinoxaline derivatives derived from d-fructose-1,6-bisphosphate under the applied growth conditions (30°C; pH 4 to 5). Isolation and characterization of these compounds by tandem mass spectrometry and nuclear magnetic resonance spectroscopy led to the identification of phosphoric acid mono-(2,3,4-trihydroxy-4-quinoxaline-2-yl-butyl) ester (Q1), phosphoric acid mono-[2,3-dihydroxy-3-(3-methyl-quinoxaline-2-yl)-propyl] ester (Q2), and phosphoric acid mono-[2-hydroxy-3-(3-methyl-quinoxaline-2-yl)-propyl] ester (Q3). Q1 and Q2 were formed independently of Z. rouxii cells, whereas Q3 was detected only in incubation systems containing the yeast. Identification of Q2 demonstrated for the first time the chemical formation of 1-deoxy-2,3-hexodiulose-6-phosphate in the culture medium, a generally expected but never identified intermediate in the formation pathway of HDMF. Since HDMF was detected only in the presence of Z. rouxii cells, additional enzymatic steps were presumed. Incubation of periplasmic and cytosolic protein extracts obtained from yeast cells with d-fructose-1,6-bisphosphate led to the formation of HDMF, implying the presence of the required enzymes in both extracts.
4-Hydroxy-3[2H]-furanones are exceptional aroma compounds due to their attractive flavor and low odor thresholds (34). They are biosynthesized by plants, microorganisms, and insects but are also formed during the thermal treatment of food in the so-called Maillard reaction. In the 1970s 4-hydroxy-3[2H]-furanones were disclosed as secondary metabolites of the yeast Zygosaccharomyces rouxii. Nunomura et al. (23) identified for the first time 2 (or 5)-ethyl-5 (or 2)-methyl-4-hydroxy-3[2H]-furanone (homofuraneol) as the flavor impact compound in soy sauce (shoyu). In further studies 5-methyl-4-hydroxy-3[2H]-furanone (norfuraneol) and 2,5-dimethyl-4-hydroxy-3[2H]-furanone (HDMF; Furaneol) were found as flavor compounds in soy sauce (24, 25) as well. The authors assumed the formation of the furanones during preparation of the soy sauce due to a Maillard reaction. Sugawara (39) as well as Hayashida et al. (12) investigated the flavor of miso, a fermentation product similar to soy sauce but made from rice. They showed that the furanones are formed during fermentation, indicating an enzymatically catalyzed bioformation performed by Z. rouxii. Few attempts were undertaken to identify the precursors of 4-hydroxy-3[2H]-furanones in soy sauce. d-Xylulose-5-phosphate, d-ribulose-5-phosphate, and d-sedoheptulose-7-phosphate were determined as potential precursors of homofuraneol and norfuraneol (31). Hecquet et al. (13) demonstrated that HDMF is produced by Z. rouxii from d-fructose-1,6-bisphosphate (FBP), and results from Roscher et al. (30) confirmed FBP as the most effective precursor in ripe strawberry fruits. Z. rouxii produced approximately 80 ppm of HDMF after 11 days of incubation in nutrient solutions containing 10% FBP and 5% d-glucose (carbon source). Addition of [1-13C]FBP to the nutrient medium revealed that only exogenously supplied FBP was transformed to HDMF (6). It was assumed that at least one metabolic step takes place in the cell wall or membrane of the yeast. However, the biosynthesis in plants and microorganisms is still unclear since no enzyme or intermediate involved in HDMF formation has been identified so far. Recently, the chemical formation of HDMF from FBP in the presence of NAD(P)H was reported (11). The authors proposed the dicarbonyl compound 1-deoxy-2,3-hexodiulose-6-phosphate as a possible intermediate which is subsequently reduced by NAD(P)H to the final product, HDMF. A similar biosynthetic pathway is anticipated in plants and microorganisms.
Prior to the usage of Z. rouxii for the biotechnological production of the economically important flavor compound HDMF, it is of utmost significance to know the biosynthetic pathway of HDMF. The objective of this study was to obtain detailed information about HDMF formation from FBP by the yeast Z. rouxii. Here, we report on potential intermediates, such as α-dicarbonyls in the culture medium. Employment of the α-dicarbonyl-trapping reagent o-phenylenediamine revealed three highly reactive α-dicarbonyl structures derived from FBP in the nutrient medium. Structure elucidation of the corresponding quinoxaline derivatives demonstrated the chemical formation of the often postulated but never identified intermediate 1-deoxy-2,3-hexodiulose-6-phosphate, and incubation of periplasmic and cytosolic protein extracts obtained from Z. rouxii confirmed the existence of oxidoreductases involved in the bioformation of HDMF.
MATERIALS AND METHODS
General methods.
High-pressure liquid chromatography (HPLC) analysis with UV detection of the quinoxalines was performed using an HPLC system equipped with a Gilson Abimed (Abimed, Langenfeld, Germany) model 231 sample injector connected to two Gilson Abimed pumps, model 303, and a Knauer variable wavelength monitor (Knauer, Berlin, Germany). Gilson 712 HPLC controller version 1.02 software was used for data acquisition and evaluation. HDMF quantifications were performed on an HPLC system equipped with a Spark Holland Basic marathon autosampler (Spark Holland, Emmen, The Netherlands) connected to a Knauer Maxistar pump and a Knauer variable wavelength monitor (285 nm). Knauer Eurochrom 2000 software was used for data acquisition and evaluation. The HDMF yields were quantified using a standard curve of commercial HDMF. For HPLC analysis with diode array detection (DAD) of the quinoxalines and HDMF, a Hewlett-Packard (Waldbronn, Germany) 1100 HPLC gradient pump and a Hewlett-Packard 1100 photodiode array detector were used including Hewlett-Packard Chemstation software for data acquisition and evaluation. A Eurospher 100-C18 column (250-mm length, 4-mm inside diameter [i.d.], 5-μm particle size) (Knauer) was employed for the quinoxalines and HDMF analysis. A binary gradient starting from 95% (A, 0.05% [vol/vol] formic acid in water) and 5% (B, acetonitrile) to 80% A within 10 min and then to 0% A in 30 min was used at a flow rate of 1 ml min−1. The injection volume was 20 μl. An anion-exchange HPLC column (Nucleosil 100-10SB; Macherey-Nagel, Düren, Germany) (length, 240 mm; diameter, 4 mm) was employed at a flow rate of 1 ml min−1 for the separation of phosphorylated carbohydrates. A binary gradient starting from 95% A (water) and 5% B (1 M NH4HCOO) to 100% B within 20 min was used. HPLC radiodetection was performed with a Canberra-Packard (Dreieich, Germany) A100 on-line radiodetector applying a solid scintillation cell. Thin-layer chromatography radiodetection was conducted with a Berthold (Bad Wildbad, Germany) linear analyzer with Ar:CH4 (90:10) as the counting gas at 1,381 V. HPLC-tandem mass spectrometry (HPLC-MS/MS) was performed with a TSQ 7000 tandem mass spectrometer system equipped with an electrospray ionization (ESI) interface (Finnigan MAT, Bremen, Germany) and an Applied Biosystems (BAI, Bensheim, Germany) 140b pump. Data acquisition and evaluation were conducted on a DEC 5000/33 (Digital Equipment, Unterföhring, Germany) computer with Finnigan MAT ICIS 8.1 software. HPLC separation with MS detection of the quinoxalines and HDMF was carried out on a Knauer Eurospher-100 C18 column (100-mm length, 2-mm i.d., 5-μm particle size) with a binary gradient. Solvent A was 0.05% (vol/vol) trifluoroacetic acid (TFA) in water, and solvent B was acetonitrile. HPLC was programmed as described above except for the use of a flow rate of 200 μl min−1. The injection volume was 5 μl, and mass spectra were acquired in the positive ion mode. For pneumatically assisted ESI, the spray voltage was set to 3.5 kV, and the temperature of the heated capillary was 210°C. Nitrogen served both as sheath (70 lb/in2) and as auxiliary gas (10 U). Product ion scanning was performed at a collision gas pressure of 2.0 mtorr and a collision energy of 25 eV with a total scan duration of 1.0 s for a single spectrum. The UV maxima were determined by means of HPLC-DAD analysis. Gas chromatography-MS (GC-MS) analysis was performed with a Fisons Instruments (Fisons, Engelsbach, Germany) GC 8000 Series chromatograph coupled to a Fisons Instruments MD800 quadrupole mass detector fitted with a split injector (1:20) at 280°C. A J & W DB-5 fused silica capillary column (30 m by 0.25 mm [i.d.]; film thickness, 0.25 μm) (J & W, Folsom, Calif.), which was programmed from 60 to 300°C (for 10 min) with a temperature increase of 6°C min−1, was employed with a 2-ml · min−1 flow rate of helium gas. The software Xcalibur for Windows was used for data acquisition. Significant MS operating parameters were as follows: ionization voltage, 70 eV (electron impact ionization); ion source temperature, 250°C; and interface temperature, 280°C. Constituents were identified by comparison of their mass spectra and retention indices with those of authentic reference compounds. Nuclear magnetic resonance (NMR) spectra were acquired with a Bruker (Rheinstetten, Germany) DMX 600 spectrometer, calibrating the chemical shifts with the help of the solvent signal (CD3OD: 3.31 ppm for 1H NMR and 49.0 ppm for 13C NMR) as reference.
Reagents.
Chemicals, salts, and solvents of high purity were obtained from Fluka (Deisenhofen, Germany), Sigma (Deisenhofen, Germany), and Aldrich (Deisenhofen, Germany). Yeast lytic enzyme (from Achromobacter sp.) was purchased from ICN (Eschwege, Germany). Solvents were redistilled prior to use. Water of HPLC-gradient grade was from Merck (Darmstadt, Germany), and acetonitrile of HPLC-gradient grade was from Fisher (Loughborough, United Kingdom).
Strain, growth media, and culture conditions.
The yeast strain used in this investigation was Z. rouxii ATCC 13356. The YPD medium (yeast extract-peptone-dextrose) (pH 4.6) consisted of 5 g of yeast extract liter−1, 5 g of peptone liter−1, 4 g of KH2PO4 liter−1, 5 g of MgSO4 · 7H2O liter−1, 10 g of d-glucose liter−1, and 170 g of NaCl liter−1. The medium was sterilized by being autoclaved at 120°C and 200 kPa for 16 min. After the medium was cooled to room temperature, 50 g of d-fructose-1,6-diphosphate liter−1 was added. The d-fructose-1,6-diphosphate stock solution (200 g liter−1) was filtered (0.2-μm-pore-size filter) prior to use. One-hundred-milliliter aliquots of the medium were inoculated with 0.2 ml of a Z. rouxii culture (approximately 2 × 107 cells) and incubated at 30°C in a GFL 3033 shaker (GFL, Burgwedel, Germany) at 150 rpm. A control medium without yeast cells and a second control medium without yeast cells and without d-fructose-1,6-diphosphate were treated similarly. For the isolation of Q1, Q2, and Q3, 1 liter of YPD medium supplemented with 5% FBP was incubated with yeast cells (approximately 2 × 107 cells) and cells were allowed to grow over a period of 6 days.
Incubation with o-phenylenediamine.
After 6 days the HDMF concentration in the growth medium was determined by RP18-HPLC with UV detection at 285 nm as described previously (6). An o-phenylenediamine solution (100 mM) was added (14 mM final concentration), and the mixture was incubated overnight at 30°C.
Extraction.
The yeast cells were withdrawn by centrifugation at 5,000 × g for 10 min, and the supernatant was subjected to solid-phase extraction on an XAD-2 (30- by 2.5-cm) column. After the stationary phase was washed with 100 ml of water, nonpolar compounds were eluted by 200 ml of diethyl ether and phosphorylated compounds were obtained by elution with 200 ml of methanol. For the isolation of Q1 to Q3 10-fold volumes were applied. The methanol extract was concentrated by rotary evaporation at 40°C to a final volume of approximately 20 ml and then by a stream of nitrogen to a final volume of approximately 6 ml. Subsequently 15 ml of water was added, and the formed precipitate was removed by centrifugation at 5,000 × g for 10 min.
Separation and isolation.
The aqueous extract was applied to a Lichroprep C18 column (33 by 3.3 cm) (40- to 63-μm particle size; Merck) equilibrated with water-formic acid (100:0.05, vol/vol). The column was eluted with 2.5 liters of water-acetonitrile-formic acid (95:5:0.05, vol/vol/vol) and then with 1 liter of water-acetonitrile-formic acid (90:10:0.05, vol/vol/vol). Fractions of approximately 10 ml were collected, measured by UV spectroscopy at 318 nm against water, and analyzed by RP18-HPLC with UV detection at 318 nm. Fractions containing Q1, Q2, and Q3 were pooled respectively, and the solvents were removed by rotary evaporation at 40°C and lyophilization. The three samples were separately applied onto a second glass column (12 by 2.5 cm) filled with Lichroprep RP18 material equilibrated with water-TFA (100/0.05, vol/vol). Separation was carried out by employing a gradient composed of mobile phase A (water-TFA [100:0.05, vol/vol]) and B (acetonitrile). The portion of solvent B was increased from 0 to 24% B in 4% steps with 50-ml fractions. Fractions (approximately 10 ml) were collected and analyzed by HPLC with UV detection at 318 nm. Fractions containing the target compounds were combined and freeze-dried after rotary evaporation at 40°C. The described procedures yielded the following compounds as pale yellow amorphous solids: Q1, ESI-MS, [M + H]+ m/z 331; ESI-MS/MS (precursor ion m/z 331, 25 eV) m/z 331, 233, 215; UV λmax (H2O-TFA-acetonitrile, 84:0.05:16, vol/vol/vol): 318 nm, 238 nm; 1H NMR (600 MHz, CD3OD) δ 9.14 (s, 1H, H-3′), 8.08 (d, 2H, H-6′ and H-7′), 7.82 (m, 2H, H-5′ and H-8′), 5.34 (s, 1H, H-4), 4.29 (m, 1H, H-1), 4.16 (m, 1H, H-1), 4.06 (m, 1H, H-2), 3.97 (d, 1H, H-3); 13C NMR (150 MHz, CD3OD) δ 159.8 (C-2′), 145.8 (C-3′), 142.7 (C-8a′), 142.5 (C-4a′), 131.5 (C-5′), 130.9 (C-8′), 129.8 (C-7′), 129.7 (C-6′), 74.8 (C-3), 73.5 (C-4), 71.5 (C-2), 69.6 (C-1); 2JC-1,P = 5.2 Hz, 3JC-2,P = 7.5 Hz; Q2, ESI-MS, [M + H]+ m/z 315; ESI-MS/MS (precursor ion m/z 315, 25 eV) m/z 315, 217; UV λmax (H2O-TFA-acetonitrile, 84:0.05:16, vol/vol/vol): 320 nm, 238 nm; 1H NMR (600 MHz, CD3OD) δ 8.1 (m, 1H, H-6′), 7.98 (m, 1H, H-7′), 7.77 (m, 2H, H-5′ and H-8′), 5.14 (d, 1H, H-3), 4.38 (m, 1H, H-2), 4.28 to 4.35 (m, 2H, H-1), 2.87 (s, 3H, H-3a′); 13C NMR (150 MHz, CD3OD) δ 157.6 (C-2′), 155.3 (C-3′), 142.1 (C-8a′), 141.9 (C-4a′), 131.2 (C-5′), 130.4 (C-8′), 129.8 (C-7′), 128.7 (C-6′), 74.8 (C-2), 71.2 (C-3), 69.2 (C-1), 22.5 (C-3a′); 2JC-1,P = 5.5 Hz, 3JC-2,P = 7.5 Hz; Q3, ESI-MS, [M + H]+ m/z 299; ESI-MS/MS (precursor ion m/z 299, 25 eV) m/z 299, 201, 183; UV λmax (H2O-TFA-acetonitrile, 80:0.05:20, vol/vol/vol): 320 nm, 240 nm; 1H NMR (600 MHz, CD3OD) δ 8.02 (m, 1H, H-6′), 7.97 (m, 1H, H-7′), 7.74 (m, 2H, H-5′ and H-8′), 4.51 (m, 1H, H-2), 4.09 (m, 2H, H-1), 3.25 (m, 2H, H-3), 2.81 (s, 3H, H-3a′); 13C NMR (150 MHz, CD3OD) δ 156.1 (C-2′), 155.7 (C-3′), 142 (C-8a′), 141.6 (C-4a′), 130.7 (C-5′), 130.4 (C-8′), 129.3 (C-7′), 128.7 (C-6′), 70.8 (C-2), 70.7 (C-1), 39 (C-3), 22.5 (C-3a′); 2JC-1,P = 5.7 Hz, 3JC-2,P = 8.3 Hz.
Preparation of Z. rouxii cytosolic protein extract.
Z. rouxii cells were grown at 30°C in YPD medium (240 ml). During the mid-logarithmic phase the yeast cells were harvested by centrifugation at 5,000 × g for 10 min, washed twice in cooled (4°C) buffer (20 mM Tris-Cl at pH 7.5, 17% NaCl), and resuspended in 4 ml of cooled (4°C) lysis buffer (50 mM Tris-Cl at pH 7.5; 150 mM NaCl; 30 μg each of leupeptin, pepstatin, and antipain per ml). The cell suspension was mixed with an equal volume of cold glass beads (diameter, 0.5 mm; Roth, Karlsruhe, Germany), and cells were broken by vortexing the mixture six times for 1 min each with 1-min cooling intervals on ice as described previously (19, 44). Unbroken cells and cell wall debris were removed by centrifugation at 1,000 × g, and the supernatant was centrifuged at 27,000 × g for 1 h at 4°C. The supernatant (soluble cytosolic proteins) was removed and subjected to dialysis against 0.1 M potassium phosphate buffer (pH 7.0) at 4°C.
Preparation of the periplasmic fraction.
Z. rouxii cells were grown to mid-logarithmic phase (4 days) in YPD medium and harvested by centrifugation for 15 min at 5,000 × g and 4°C. Cells were washed three times with 25 ml of 20 mM Tris-Cl buffer (pH 7.5) containing 17% NaCl. After incubation with 20 ml of 20 mM Tris-Cl buffer (pH 7.5) containing 17% NaCl, 1 mM dithiothreitol, and 10 U of zymolyase (yeast lytic enzyme, obtained from ICN) ml−1 at 30°C for 2.5 h, the released periplasmic fraction was separated from the spheroplasts by centrifugation at 8,420 × g and 4°C for 10 min. The supernatant (containing the periplasmic proteins) was removed and subjected to dialysis against 0.1 M potassium phosphate buffer (pH 7.0) at 4°C. For the time course experiment 2-ml aliquots were withdrawn 20, 40, 60, 90, 120, 150, and 180 min after the addition of the zymolyase-containing solution. Samples were immediately centrifuged and dialyzed as described above. The digestion of the cell wall was monitored by measuring the absorbance at 800 nm. After 4- to 10-min intervals 25 μl of the cell solution was withdrawn, 2,975 μl of a 5% sodium dodecyl sulfate solution was added, and the mixture was measured against a control sample containing 25 μl of the digestion buffer and 2,975 μl of the 5% sodium dodecyl sulfate solution.
General procedures.
Two-milliliter aliquots of the protein extract were supplemented with 50 mg of substrate (FBP) and 1 mg of NADPH. The solutions were kept with gentle agitation at 30°C for 24 h. In the case of the periplasmic extracts NADPH was omitted.
HDMF analysis.
The samples were subjected to solid-phase extraction with RP18 cartridges (Supelco, 500 mg/3 ml), preconditioned with 6 ml of methanol and 6 ml of water. After application the cartridges were rinsed with 1 ml of water and eluted with 3 ml of diethyl ether. The water remaining in the diethyl ether extract was removed by freezing (−18°C). Water (200 μl) was added to the organic phase, and the diethyl ether was removed by a stream of nitrogen. The aqueous phase was analyzed by HPLC with UV detection at 285 nm.
Proof of aldolase activity.
A periplasmic extract was incubated with d-[U-14C]fructose-1,6-diphosphate (106 dpm, 13.3 MBq mg−1; Sigma) under gentle agitation at 28°C overnight. The supernatant was removed by centrifugation and directly analyzed by HPLC, by applying an anion-exchange column with on-line scintillation detection and by thin-layer chromatography with radiodetection. Periplasmic extracts were also incubated with unlabeled d-fructose-1,6-diphosphate at 28°C overnight. GC-MS analysis was carry out on the alditol acetates formed from the transformation products according to the method of Higgins et al. (14). The aldolase assay was performed according to the method of Schwab et al. (33).
RESULTS
Detection of α-dicarbonyl compounds derived from FBP.
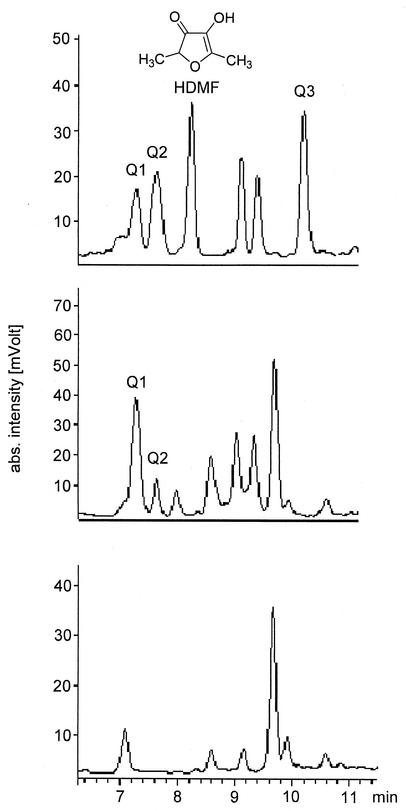
Three nutrient solutions were prepared according to the method of Hecquet et al. (13). Two media were supplemented with FBP, and one of the phosphohexose-containing media was inoculated with Z. rouxii cells. After 7 days of incubation each of the three solutions was supplemented with o-phenylenediamine, a widely used trapping reagent for highly reactive α-dicarbonyl compounds (10). After overnight incubation yeast cells were removed by centrifugation, and the supernatant as well as the incubations without yeast cells was analyzed by HPLC with DAD. In the sample containing yeast cells and FBP three compounds (Q1, Q2, and Q3) exhibiting the characteristic quinoxaline UV spectrum (35) with two absorption maxima at 238 and 318 nm were detected (Fig. 1) in addition to HDMF. Q1 and Q2 were also present in the sample supplemented with FBP but devoid of yeast cells (Fig. 1). Since none of these compounds was detected in the nutrient solution without FBP and yeast cells (Fig. 1), it was deduced that Q1 and Q2 are formed by the reaction of o-phenylenediamine with compounds formed nonenzymatically from FBP under moderate conditions. However, formation of Q3 and HDMF was observed only in the presence of FBP and yeast cells, implying a decisive participation of the yeast cells. Since the other compounds shown in Fig. 1 did not show quinoxaline or furanone characteristic UV spectra, they were not further examined. Solid-phase extraction on XAD-2 according to the method of Beuerle et al. (2) and subsequent analysis of the diethyl ether extracts as well as methanol extracts by reversed-phase HPLC with UV detection at 318 nm revealed that Q1, Q2, and Q3 are exclusively eluted with methanol while HDMF was detected only in the diethyl ether extract. After further purification on RP18 material, pseudomolecular ions at m/z 331 [M + H]+ for Q1, m/z 315 [M + H]+ for Q2, and m/z 299 [M + H]+ for Q3 were determined by HPLC-ESI-MS analysis. The product ion spectra of all compounds under study as obtained by low-energy collision-induced dissociation were dominated by the loss of 98 AMU presumably due to the abstraction of phosphoric acid. Thus, the combined UV, MS, and MS/MS data indicated the formation of phosphorylated quinoxalines by the reaction of o-phenylenediamine and phosphorylated α-dicarbonyl compounds derived from FBP under the applied conditions. Due to their different molecular masses it appeared that the three compounds differed only in their hydroxylation status.
FIG. 1.
HPLC-DAD analysis of an extract obtained by incubation of YPD medium supplemented with FBP and Z. rouxii cells with o-phenylenediamine overnight (top), of YPD medium supplemented with FBP with o-phenylenediamine overnight (middle), and of YPD medium with o-phenylenediamine overnight (bottom). Column effluent was monitored at 318 nm. Unlabeled peaks did not show absorption maxima at 238 and 318 nm.
Identification of Q1, Q2, and Q3 by NMR spectroscopy.
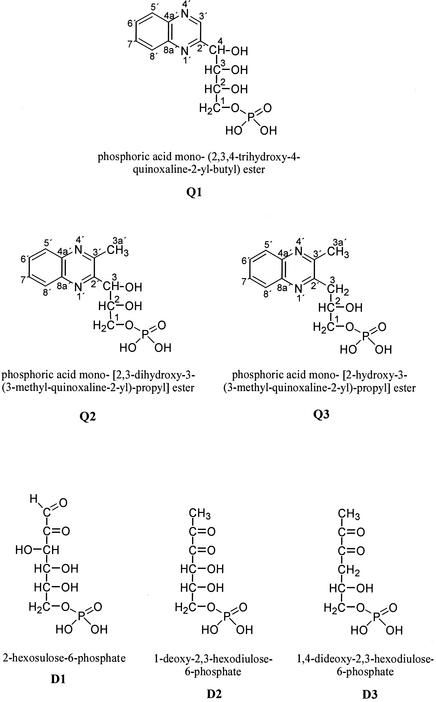
Q1, Q2, and Q3 were isolated from a culture medium containing FBP and Z. rouxii cells for structure elucidation. Identification was achieved by NMR spectroscopy including 1H, 13C, distortionless enhancement by polarization transfer, homonuclear correlation spectroscopy, heteronuclear multiple-bond connectivity, and 1H-detected heteronuclear multiple-quantum coherence experiments. All three substances showed similar signals in the 1H and 13C NMR spectra arising from the protons H-5′ and H-8′ and from the carbons C-4a′, C-5′, C-6′, C-7′, C-8′, and C-8a′ of the quinoxaline skeletal structure (9, 16, 20, 42). In contrast, Q1 to Q3 displayed clear differences in the carbohydrate backbone at C-2′ and C-3′. The NMR data for Q1 exhibited a proton bound to C-3′ of the quinoxaline skeleton due to a singlet signal in the lower field at 9.14 ppm and a 1H-detected heteronuclear multiple-quantum coherence correlation between H-3′ and C-3′. The NMR data for Q2 and Q3 revealed the presence of a methyl group at C-3′ due to the chemical shift of C-3a′ (δC = 22.5 ppm for Q2 and Q3) and the heteronuclear multiple-bond connectivity correlation between H-3a′ and C-2′. For Q1 the NMR data showed the presence of a 1-phospho-2,3,4-trihydroxybutyl moiety, and for Q2 the data showed the presence of a 1-phospho-2,3-dihydroxypropyl moiety at C-2′. The linkage of the phosphate group to C-1 was proved by the split signals in the 13C spectra due to the phosphorus-carbon coupling and the corresponding coupling constants (2JC-1,P = 5.2 Hz and 3JC-2,P = 7.5 Hz for Q1, 2JC-1,P = 5.5 Hz and 3JC-2,P = 7.5 Hz for Q2) (36). The presence of phosphorus was additionally confirmed by 31P NMR (data not shown). Thus, Q1 and Q2 were unequivocally identified as phosphoric acid mono-(2,3,4-trihydroxy-4-quinoxaline-2-yl-butyl) ester and phosphoric acid mono-[2,3-dihydroxy-3-(3-methyl-quinoxaline-2-yl)-propyl] ester, respectively (Fig. 2). For Q3 the NMR spectra confirmed the attachment of a 1-phospho-2-hydroxypropyl moiety to C-2′ of the quinoxaline skeleton. The position of the methylene group at C-3 was evidenced by the chemical shift (δC-3 = 39 ppm) and the negative distortionless enhancement by polarization transfer signal. Analogously to Q1 and Q2 the presence of the phosphate group was confirmed by the split signals in the 13C spectra (2JC-1,P = 5.7 Hz and 3JC-2,P = 8.3 Hz), and therewith the formation of phosphoric acid mono-[2-hydroxy-3-(3-methyl-quinoxaline-2-yl)-propyl] ester (Fig. 2) was unambiguously demonstrated. Consequently, the α-dicarbonyl structures D1, D2, and D3 are formed in the nutrient solutions as can be deduced from their quinoxaline derivatives Q1, Q2, and Q3, respectively (Fig. 2).
FIG. 2.
FBP-derived quinoxaline derivatives and deduced FBP-derived α-dicarbonyls.
Incubation with protein extracts obtained from Z. rouxii.
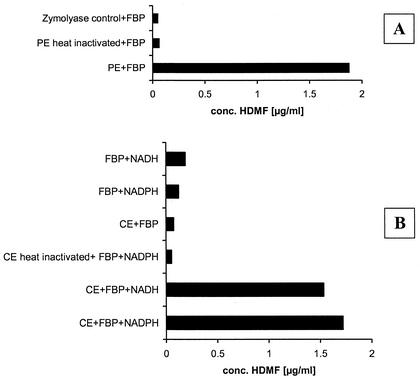
HDMF was formed only in media containing FBP and yeast cells, but the predicted precursor 1-deoxy-2,3-hexodiulose-6-phosphate (D2) was formed independently of the yeast cells. As a reduction is required to transform the intermediate into HDMF, we concluded that there was an additional enzymatic reduction catalyzed by a nonspecific yeast ketoreductase. In order to localize this reductase activity, periplasmic protein extracts were prepared by zymolyase (yeast lytic enzyme, lyticase) digestion of the cell wall according to the method of Yoda et al. (45), and cytosolic protein extracts were prepared by vortexing Z. rouxii cells with glass beads followed by high-speed centrifugation (19, 44). Both extracts were dialyzed to remove low-molecular-mass components (<12 kDa) and were subsequently incubated at 30°C for 24 h with an excess of FBP to ensure the in situ formation of the required precursor. After solid-phase extraction on RP18 cartridges samples were analyzed by HPLC with UV detection at 285 nm. In Fig. 3A the HDMF amounts obtained from incubations of an active periplasmic extract and a heat-inactivated periplasmic extract and of a control incubation including the zymolyase solution are displayed. HDMF was exclusively formed in the active sample. Addition of reducing agents such as NADH or NADPH was not required, and they did not increase the yield of HDMF. For further characterization of HDMF formation in the periplasmic extract, a time course experiment was performed revealing the liberation of the HDMF-forming enzymatic activity during cell wall digestion. Aliquots were withdrawn periodically from a zymolyase digestion and treated as described above. The HDMF-forming enzymatic activity was gradually liberated from the cells and reached a maximum level. In parallel, the efficiency of the cell wall digestion was monitored by measurement of the absorbance at 800 nm.
FIG. 3.
Formation of HDMF by periplasmic extracts (A) and cytosolic extracts (B) obtained from Z. rouxii. (PE, periplasmic extract; CE, cytosolic extract). Formation of HDMF in periplasmic extracts is independent of NAD(P)H addition. Two-milliliter aliquots of the protein extracts were supplemented with 50 mg of substrate (FBP) and 1 mg of NAD(P)H. The solutions were kept with gentle agitation at 30°C for 24 h. In the case of the periplasmic extracts NAD(P)H was omitted
In the case of the cytosolic extracts significant amounts of HDMF were detected only after the addition of NADPH or NADH and FBP (Fig. 3B). When the enzyme extract was inactivated by heat treatment prior to the incubation or when the cofactor was omitted, HDMF formation was not observed. Due to the recently noted chemical formation of HDMF from FBP in the presence of NADs (11), control experimental mixtures including the appropriate buffer instead of the extract were treated similarly. The HDMF formation in the control incubations was well below the formation in the active extracts (Fig. 3B).
Regeneration of NAD(P)H.
It was assumed that the bound NAD(P)H accounts for only a single turnover, which does not account for the amount of reduced product that was formed in the incubation. Thus, an NAD(P)H regeneration system was proposed for the periplasmic extract. Such a system could consist of FBP aldolase and glyceraldehyde-3-phosphate dehydrogenase. Enzymatically active glyceraldehyde-3-phosphate dehydrogenase has already been described as a constituent of the cell wall of Candida albicans, Streptococcus pyogenes, and Schistosoma mansoni (3, 28, 37). Aldolase activity in the periplasmic extract was demonstrated by radiochemical analysis of the products formed after the incubation with d-[U-14C]fructose-1,6-diphosphate as well as GC-MS analysis of the alditol acetate derivatives of the products generated after the incubation with unlabeled FBP. The experiments clearly proved the formation of dihydroxyacetone phosphate and glyceraldehyde-3-phosphate, products formed by aldolase activity. Final evidence was provided by an aldolase enzyme assay. Addition of EDTA inhibited the enzymatic reaction as expected for a yeast aldolase. Thus, the combined action of glyceraldehyde-3-phosphate dehydrogenase and FBP aldolase on the very high concentration of FBP would account for the formation of NADH, which could serve as the reductant, allowing multiple turnovers. We assume that aldolase is present in the yeast cell wall like other glycolytic enzymes described previously (1, 8), but we cannot rule out that it is released from the cytoplasm during the extraction procedure (3).
DISCUSSION
We demonstrated for the first time the formation of highly reactive α-dicarbonyl compounds from FBP. This transformation occurred under moderate growth conditions in the Z. rouxii culture medium (30°C; pH 4 to 5). The structures of 2-hexosulose-6-phosphate (D1), 1-deoxy-2,3-hexodiulose-6-phosphate (D2), and 1,4-dideoxy-2,3-hexodiulose-6-phosphate (D3) were deduced from the quinoxalines Q1, Q2, and Q3. The formation of α-dicarbonyls from carbohydrates has been intensively studied by the use of trapping reagents, especially o-phenylenediamine derivatives and aminoguanidines, but carbohydrate phosphates have rarely been used as precursors (10, 15, 16, 17, 21, 22). Since α-dicarbonyls represent a major group of reactive intermediates formed during the so-called Maillard reaction, most of the researches studied their formation in heated mixtures of pentoses or hexoses with amino acids (10, 17, 21) and determined their in vivo generation during the advanced Maillard reaction (26). Only a few examples are available of investigations of the formation of α-dicarbonyls derived from carbohydrate phosphates in consequence of a phosphate elimination (27). Larimer et al. (18) trapped for the first time a phosphorylated α-dicarbonyl derived from d-ribulose-1,5-diphosphate with o-phenylenediamine. However, Q1 to Q3 have not been detected until now. This is the first report on the formation of α-dicarbonyls derived from a hexose phosphate under moderate conditions.
We assume that D1 is formed by oxidation of d-fructose-6-phosphate generated from FBP by hydrolysis due to the slightly acid pH of the nutrient medium (pH 4 to 5). This assumption was substantiated by the detection of Q1 by means of HPLC-ESI-MS/MS analysis in a phosphate buffer containing d-fructose-6-phosphate and o-phenylenediamine (data not shown). According to the report of Glomb and Tschirnich (10) formation of dicarbonyls is effected by the trapping reagent itself due to its oxidative potency. Incubation of the Amadori product of Nα-tert-butoxycarbonyl-lysine and glucose in the presence of o-phenylenediamine led to a significant formation of glucosone, the nonphosphorylated analog of the observed 2-hexosulose-6-phosphate.
1-Deoxy-2,3-hexodiulose-6-phosphate (D2) is formed by β-elimination of the phosphate group at C-1 in consequence of an initial 2,3-enolization. The formation of Q2 confirms results obtained by incubation of d-ribulose-1,5-diphosphate with o-phenylenediamine for 15 h in a phosphate buffer leading to the quantitative conversion of the trapping reagent to a quinoxaline derivative. ESI-MS analysis revealed a molecular ion at m/z 285[M + H]+, in accordance with the structure proposed by Larimer et al. (18).
The formation of D3 can be explained by dehydration of D2 followed by a reduction step. The exclusive generation in the presence of Z. rouxii cells indicates that the reduction is obviously performed by the yeast cells.
The α-dicarbonyl compounds formed either chemically or by yeast biocatalysis represent possible HDMF precursors. Recently, the chemical formation of HDMF in solutions composed of FBP and reducing agents was demonstrated, and D2 was postulated as a precursor of HDMF formed from FBP (11). Since a very large amount of FBP is required in the nutrient medium of Z. rouxii (up to 8%) to obtain a reasonable concentration of HDMF (6, 13), we expected a chemical transformation rather than a biotransformation as the initial step of HDMF formation. Generally, a phosphate elimination via 2,3-enolization was assumed, but this mechanism of formation of D2 (11, 32) was never confirmed. However, in a similar reaction sequence dihydroxyacetone phosphate (29) is transformed to methylglyoxal and d-ribulose-1,5-diphosphate forms the corresponding phosphorylated 1-desoxypentosone (27).
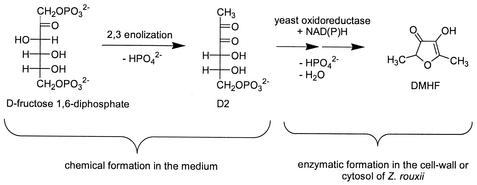
We trapped and characterized D2 for the first time as a quinoxaline derivative. Our results indicate that the first step of HDMF formation by the yeast Z. rouxii is the chemical formation of D2 in the culture medium, since only exogenously supplied FBP is transformed to HDMF, as shown by Dahlen et al. (6), and D2 is formed in the culture medium independently of the yeasts. The formation of D2 might be the limiting step in the bioformation of HDMF because very large amounts of the sugar phosphate are necessary (up to 8% in the culture medium) to yield reasonable amounts of the target molecule. We conclude that a combination of chemical reactions leading to the elimination of at least one phosphate group and an enzymatic reduction mediated by an oxidoreductase of the yeast cells is responsible for HDMF formation in the culture media (Fig. 4). Indeed, we detected the generation of HDMF in cell periplasmic and cytosolic protein extracts. Thus, we assume the existence of nonspecific ketoreductases in the yeast cells participating in HDMF formation. Yeast-mediated reductions have been intensively investigated (5, 7, 38). Tadashi et al. (40, 41) purified a carbonyl reductase from baker's yeast, exhibiting high enantioselectivity and broad substrate specificity. Costello et al. (4) isolated and characterized a ketoreductase from cytosolic protein extracts obtained from Z. rouxii, able to reduce methylketones, α-ketolactones, and diketones effectively and enantioselectively.
FIG. 4.
Proposed bioformation pathway of HDMF by Z. rouxii.
Since the periplasmic extract shows activity without the addition of a cofactor, the presence of enzyme-bound NAD(P)H is anticipated. The well-characterized periplasmic NADP-containing glucose-fructose oxidoreductase detected in the gram-negative bacterium Zymomonas mobilis represents such a candidate (43).
Acknowledgments
We thank M. Grüne, E. Ruckdeschel, and C. Schollmayer for assistance with NMR spectroscopy.
Financial support from Firmenich SA is gratefully acknowledged.
REFERENCES
- 1.Alloush, H. M., J. L. López-Ribot, B. J. Masten, and W. L. Chaffin. 1997. 3-Phosphoglycerate kinase: a glycolytic enzyme protein present in the cell wall of Candida albicans. Microbiology 143:321-330. [DOI] [PubMed] [Google Scholar]
- 2.Beuerle, T., P. Schreier, and W. Schwab. 1997. (R)-3-Hydroxy-5(Z)-octenyl-β-d-glucopyranoside from Malus sylvestris fruits. Nat. Prod. Lett. 10:119-124. [Google Scholar]
- 3.Chaffin, W. L., J. L. López-Ribot, M. Casanova, D. Gozalbo, and J. P. Martínez. 1998. Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol. Mol. Biol. Rev. 62:130-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costello, C. A., R. A. Payson, M. A. Menke, J. L. Larson, K. A. Brown, J. E. Tanner, R. E. Kaiser, C. L. Hershberger, and M. J. Zmijewski. 2000. Purification, characterization, cDNA cloning and expression of a novel ketoreductase from Zygosaccharomyces rouxii. Eur. J. Biochem. 267:5493-5501. [DOI] [PubMed] [Google Scholar]
- 5.Csuk, R., and B. I. Glaenzer. 1991. Baker's yeast mediated transformations in organic chemistry. Chem. Rev. 91:49-97. [Google Scholar]
- 6.Dahlen, T., T. Hauck, M. Wein, and W. Schwab. 2001. 2,5-Dimethyl-4-hydroxy-3(2H)-furanone as a secondary metabolite from d-fructose 1,6-diphosphate metabolism by Zygosaccharomyces rouxii. J. Biosci. Bioeng. 91:352-358. [DOI] [PubMed] [Google Scholar]
- 7.D'Arrigo, P. G., G. Pedrocchi-Fantoni, and S. Servi. 2000. Stereoselective synthesis of chiral compounds using whole-cell biocatalysts. Stereoselective Biocatalysis 1:365-396. [Google Scholar]
- 8.Edwards, S. R., R. Braley, and W. L. Chaffin. 1999. Enolase is present in the cell wall of Saccharomyces cerevisiae. FEMS Microbiol. Lett. 177:211-216. [DOI] [PubMed] [Google Scholar]
- 9.Glomb, M. A., and C. Pfahler. 2000. Synthesis of 1-deoxy-d-erythro-hexo-2,3-diulose, a major hexose Maillard intermediate. Carbohydr. Res. 329:515-523. [DOI] [PubMed] [Google Scholar]
- 10.Glomb, M. A., and R. Tschirnich. 2001. Detection of α-dicarbonyl compounds in Maillard reaction systems and in vivo. J. Agric. Food Chem. 49:5543-5550. [DOI] [PubMed] [Google Scholar]
- 11.Hauck, T., C. Landmann, T. Raab, F. Bruhlmann, and W. Schwab. 2002. Chemical formation of 4-hydroxy-2,5-dimethyl-3[2H]-furanone from d-fructose 1,6-diphosphate. Carbohydr. Res. 337:1185-1191. [DOI] [PubMed] [Google Scholar]
- 12.Hayashida, Y., K. Nishimura, and J. C. Slaughter. 1998. The importance of the furanones HDMF and HEMF in the flavour profile of Japanese barley miso and their production during fermentation. J. Sci. Food Agric. 78:88-94. [Google Scholar]
- 13.Hecquet, L., M. Sancelme, J. Bolte, and C. Demuynck. 1996. Biosynthesis of 4-hydroxy-2,5-dimethyl-3[2H]-furanone by Zygosaccharomyces rouxii. J. Agric. Food Chem. 44:1357-1360. [Google Scholar]
- 14.Higgins, M. K., R. S. Bly, and S. L. Morgan. 1994. Differentiation of isomeric alditol hexaacetates and identification of aldohexoses by electron impact mass spectrometry. Anal. Chem. 66:2656-2668. [Google Scholar]
- 15.Hirsch, J., V. V. Mossine, and M. S. Feather. 1995. The detection of some dicarbonyl intermediates arising from the degradation of Amadori compounds (the Maillard reaction). Carbohydr. Res. 273:171-177. [Google Scholar]
- 16.Hofmann, T. 1998. Characterization of precursors and elucidation of the reaction pathway leading to a novel colored 2H,7H,8aH-pyrano[2,3-b]pyran-3-one from pentoses by quantitative studies and application of 13C-labeling experiments. Carbohydr. Res. 313:215-224. [Google Scholar]
- 17.Hofmann, T. 1999. Quantitative studies on the role of browning precursors in the Maillard reaction of pentoses and hexoses with l-alanine. Eur. Food Res. Technol. 209:113-121. [Google Scholar]
- 18.Larimer, F. W., M. R. Harpel, and F. C. Hartman. 1994. β-Elimination of phosphate from reaction intermediates by site-directed mutants of ribulose-bisphosphate carboxylase/oxygenase. J. Biol. Chem. 269:11114-11120. [PubMed] [Google Scholar]
- 19.Lu, C., J. Kurjan, and P. A. Lipke. 1994. A pathway for cell wall anchorage of Saccharomyces cerevisiae α-agglutinin. Mol. Cell. Biol. 14:4825-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNab, H. 1982. Carbon-13 nuclear magnetic resonance spectra of quinoxaline derivatives. J. Chem. Soc. Perkin Trans. I 2:357-363. [Google Scholar]
- 21.Naofumi, M., D. Yoshiyuki, and T. Masanosuke. 1984. Quinoxalines derived from pentoses with o-phenylenediamine under acidic refluxed conditions. Agric. Biol. Chem. 48:3161-3163. [Google Scholar]
- 22.Nevidek, W., F. Ledl, and W. Fischer. 1992. Detection of 5-hydroxymethyl-2-methyl-3(2H)-furanone and α-dicarbonyl compounds in reaction mixtures of hexoses and pentoses with different amines. Z. Lebensm. Unters. Forsch. 194:222-228. [Google Scholar]
- 23.Nunomura, N., M. Sasaki, Y. Asao, and T. Yokotsuka. 1976. Isolation and identification of 4-hydroxy-2(or 5)-ethyl-5(or 2)-methyl-3[2H]-furanone. Agric. Biol. Chem. 40:491-495. [Google Scholar]
- 24.Nunomura, N., M. Sasaki, and T. Yokotsuka. 1979. Isolation of 4-hydroxy-5-methyl-3[2H]-furanone: a flavor component in shoyu (soy sauce). Agric. Biol. Chem. 43:1361-1363. [Google Scholar]
- 25.Nunomura, N., M. Sasaki, and T. Yokotsuka. 1980. Shoyu (soy sauce) flavor components: acidic fractions and characteristic flavor components. Agric. Biol. Chem. 44:339-351. [Google Scholar]
- 26.Odani, H., T. Shinzato, Y. Matsumoto, J. Usami, and K. Maeda. 1999. Increase in three α,β-dicarbonyl compound levels in human uremic plasma: specific in vivo determination of intermediates in advanced Maillard reaction. Biochem. Biophys. Res. Commun. 256:89-93. [DOI] [PubMed] [Google Scholar]
- 27.Paech, C., J. Pierce, S. D. McCurry, and N. E. Tolbert. 1978. Inhibition of ribulose-1,5-bisphosphate carboxylase/oxygenase by ribulose-1,5-bisphosphate epimerization and degradation products. Biochem. Biophys. Res. Commun. 83:1084-1092. [DOI] [PubMed] [Google Scholar]
- 28.Pancholi, V., and V. A. Fischetti. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate dehydrogenase with multiple binding activity. J. Exp. Med. 176:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips, S. A., and P. J. Thornalley. 1993. The formation of methylglyoxal from triose phosphates. Investigation using a specific assay for methylglyoxal. Eur. J. Biochem. 212:101-105. [DOI] [PubMed] [Google Scholar]
- 30.Roscher, R., G. Bringmann, P. Schreier, and W. Schwab. 1998. Radiotracer studies on the formation of 2,5-dimethyl-4-hydroxy-3[2H]-furanone in detached ripening strawberry fruits. J. Agric. Food Chem. 46:1488-1493. [Google Scholar]
- 31.Sasaki, M., N. Nunomura, and T. Matsudo. 1991. Biosynthesis of 4-hydroxy-2(or 5)-ethyl-5(or 2)-methyl-3[2H]-furanone by yeast. J. Agric. Food Chem. 39:934-938. [Google Scholar]
- 32.Schieberle, P. 1992. Formation of furaneol in heat-processed foods. ACS Symp. Ser. 490:164-174. [Google Scholar]
- 33.Schwab, W., A. Aharoni, T. Raab, A. G. Perez, and C. Sanz. 2001. Cytosolic aldolase is a ripening related enzyme in strawberry fruits (Fragaria × ananassa). Phytochemistry 56:407-415. [DOI] [PubMed] [Google Scholar]
- 34.Schwab, W., and R. Roscher. 1997. 4-Hydroxy-3(2H)-furanones: natural and Maillard products. Recent Res. Dev. Phytochem. 1:643-673. [Google Scholar]
- 35.Seok, Y.-J., K.-S. Yang, S.-T. Kim, W.-K. Huh, and S.-O. Kang. 1996. Characterization of quinoxaline derivatives of dehydro-d-erythroascorbic acid. J. Carbohydr. Chem. 15:1085-1095. [Google Scholar]
- 36.Serianni, A. S., J. Pierce, and R. Barker. 1979. Carbon-13-enriched carbohydrates: preparation of triose, tetrose, and pentose phosphates. Biochemistry 18:1192-1199. [DOI] [PubMed] [Google Scholar]
- 37.Shoemaker, C. A., A. Gross, A. Gebremichael, and D. Harn. 1992. cDNA cloning and functional expression of the Schistosoma mansoni protective antigen triose-phosphate isomerase. Proc. Natl. Acad. Sci. USA 89:1842-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart, J. D. 2000. Organic transformations catalyzed by engineered yeast cells and related systems. Curr. Opin. Biotechnol. 11:363-368. [DOI] [PubMed] [Google Scholar]
- 39.Sugawara, E. 1991. Identification of 4-hydroxy-2(or 5)-ethyl-5(or 2)-methyl-3[2H]-furanone as a flavor component in miso. Nippon Shokuhin Gakkaishi 38:491-493. [Google Scholar]
- 40.Tadashi, E., M. Hiroyuki, K. Toru, I. Tomomasa, M. Kentaro, U. Masanori, and S. Takashi. 2001. High enantioselectivity and broad substrate specificity of a carbonyl reductase: toward a versatile biocatalyst. J. Org. Chem. 66:8682-8684. [DOI] [PubMed] [Google Scholar]
- 41.Tadashi, E., S. Yasushi, F. Minoru, M. Hiroyuki, C. Jing-Nan, S. Takashi, and U. Masanori. 1998. Highly enantioselective reduction of carbonyl compounds using a reductase purified from bakers' yeast. J. Org. Chem. 63:4996-5000. [Google Scholar]
- 42.Vermeersch, G., J. Marko, N. Febvay-Garot, S. Caplain, and A. Lablache-Combier. 1984. Low-field photochemically induced dynamic nuclear polarization (photo-CIDNP) of diazanaphthalenes. J. Chem. Soc. Perkin Trans. II 12:2027-2030. [Google Scholar]
- 43.Wiegert, T., H. Sahm, and G. A. Sprenger. 1997. The substitution of a single amino acid residue (Ser-116 → Asp) alters NADP-containing glucose-fructose oxidoreductase of Zymomonas mobilis into a glucose dehydrogenase with dual coenzyme specificity. J. Biol. Chem. 272:13126-13133. [DOI] [PubMed] [Google Scholar]
- 44.Wojciechowicz, D., C. Lu, J. Kurjan, and P. Lipke. 1993. Cell surface anchorage and ligand-binding domains of the Saccharomyces cerevisiae cell adhesion protein α-agglutinin, a member of the immunoglobulin superfamily. Mol. Cell. Biol. 13:2554-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoda, K., J.-H. Ko, T. Nagamatsu, Y. Lin, C. Kaibara, T. Kawada, N. Tomishige, H. Hashimoto, Y. Noda, and M. Yamasaki. 2000. Molecular characterization of a novel yeast cell-wall acid phosphatase cloned from Kluyveromyces marxianus. Biosci. Biotechnol. Biochem. 64:142-148. [DOI] [PubMed] [Google Scholar]