Abstract
Tolerance to digestive stresses is one of the main factors limiting the use of microorganisms as live probiotic agents. Susceptibility to bile salts and tolerance acquisition in the probiotic strain Propionibacterium freudenreichii SI41 were characterized. We showed that pretreatment with a moderate concentration of bile salts (0.2 g/liter) greatly increased its survival during a subsequent lethal challenge (1.0 g/liter, 60 s). Bile salts challenge led to drastic morphological changes, consistent with intracellular material leakage, for nonadapted cells but not for preexposed ones. Moreover, the physiological state of the cells during lethal treatment played an important role in the response to bile salts, as stationary-phase bacteria appeared much less sensitive than exponentially growing cells. Either thermal or detergent pretreatment conferred significantly increased protection toward bile salts challenge. In contrast, some other heterologous pretreatments (hypothermic and hyperosmotic) had no effect on tolerance to bile salts, while acid pretreatment even might have sensitized the cells. Two-dimensional electrophoresis experiments revealed that at least 24 proteins were induced during bile salts adaptation. Identification of these polypeptides suggested that the bile salts stress response involves signal sensing and transduction, a general stress response (also triggered by thermal denaturation, oxidative toxicity, and DNA damage), and an alternative sigma factor. Taken together, our results provide new insights into the tolerance of P. freudenreichii to bile salts, which must be taken into consideration for the use of probiotic strains and the improvement of technological processes.
Microorganisms must constantly sense and react quickly to variations in their environment. Tolerance to harsh conditions can be due to an intrinsic resistance or to an adaptive response (42). The latter may include various modifications, such as morphological changes (22) or induction of proteins (45), appearing after exposure to moderate stresses. The stress response can, at least partly, be specific to bacterial species (7, 39). It is thus difficult to extrapolate a general behavior common to all organisms.
The use of microorganisms as probiotic products is of increasing economic importance. These microorganisms are commonly defined as “live microbial feed supplements which beneficially affect the host animal by improving its intestinal microbial balance” (10). Their main use is the treatment of intestinal disorders (16). They may be subjected to various physical and chemical stresses before ingestion by the human host.
Bile salts are synthesized from cholesterol in the liver, stored in the gallbladder, and released into the duodenum (15). Their maximal concentrations in healthy people reach 12.6 mmol/liter for glycocholate, 6.9 mmol/liter for taurocholate, and 0.7 mmol/liter for unconjugated bile salts (cholate, deoxycholate, chenodeoxycholate, and lithocholate) in the duodenum (25). Their concentrations decrease to 1.0 to 3.0 mmol/liter for glycocholate and 0.4 to 1.2 mmol/liter for taurocholate in the jejunal space and 25 to 50 μmol/liter for cholate in the ileal space (15). These detergents, which are highly toxic for microorganisms, act on their membranes, exposing the bacterial periplasm and cytoplasm (44). Many studies have explored the mechanisms leading to increased tolerance of enteric bacteria to bile salts and other detergents (8, 44). Studies concerning probiotic strains and their tolerance to digestive stresses typically have consisted of screening for acid- and bile salts-tolerant strains (6) or the development of complex media leading to the selective enumeration of probiotic bacteria (43). Only a few of these studies have searched for molecular causes of the observed tolerance.
Traditionally used as cheese starters, dairy propionibacteria, including Propionibacterium freudenreichii, display a number of probiotic effects, such as increased levels of fecal bifidobacteria in humans (4, 29), inhibition of undesirable flora (23), beneficial modification of enzymatic activities within the gut (34), and treatment of lactose intolerance (47). More recently, P. freudenreichii was shown to induce in vitro cell death of human colorectal carcinomas by apoptosis (17). During the cheese-making process, P. freudenreichii resists harsh physical and chemical stresses, including significant heat and salt stresses (28). To exert beneficial effects in the intestine, it also needs to survive digestive stresses. Zarate et al. (47) showed that dairy propionibacteria can survive exposure to artificial gastric and intestinal fluids. Moreover, Bougle et al. (4) demonstrated that some dairy propionibacteria can survive in vivo passage through the digestive tract.
In this probiotic context, the acid susceptibility of P. freudenreichii has already been studied (18, 19). We investigated susceptibility to bile salts and tolerance acquisition in P. freudenreichii and the mechanisms of these responses by using proteomic tools.
MATERIALS AND METHODS
Microorganisms and growth conditions.
The strain of P. freudenreichii subsp. shermanii used in this study, SI41, was kindly provided by Standa Industrie and is part of the probiotic preparation Propiofidus. It was routinely cultured in YEL medium containing 114 mM lactate (24). Growth was carried out at 30°C without shaking and monitored spectrophotometrically at 650 nm (DU 7400 spectrophotometer; Beckman Instruments, Inc., Fullerton, Calif.) as well as by counting of CFU. A chemically defined medium (MDP medium) (18) was used for protein synthesis analysis and for radioactive labeling experiments. This medium was shown to fulfill the nutrient requirements of P. freudenreichii, yielding growth characteristics very similar to those obtained with the rich and complex YEL medium.
Adaptation conditions.
When YEL medium cultures reached a density of 5 × 108 cells per ml (optical density at 650 nm = 0.5), corresponding to the middle of the exponential growth phase, adaptation was performed. For homologous adaptation, an equimolar mixture of cholate and deoxycholate (Sigma Chemical Co., St. Louis, Mo.; referred to as “bile salts” in this report) was used at a final concentration of 0.2 g/liter. Bile salts adaptation was performed for between 5 min and 4 h at 30°C, before cells were subjected to a lethal concentration of bile salts (challenge conditions).
For heterologous pretreatments, experimental conditions were as follows. Various temperatures were applied to log-phase cultures for 1 h: 4, 37, 42, or 55°C. Moderate osmotic stress was induced by adding NaCl to a final concentration of 0.3 M to these cultures for 1 h. Acid pretreatment was carried out by maintaining the cultures at pH 5.0 for 1 h (the pH was adjusted by using HCl). Sodium dodecyl sulfate (SDS) adaptation was carried out by adding 0.06 g of SDS (Sigma)/liter for 4 h. After these pretreatments, cells were harvested by centrifugation (6,000 × g, 30°C, 5 min) and resuspended in YEL medium at pH 7.0 prior to bile salts challenge. Stationary-phase cultures were obtained after 100 h of culture in YEL medium.
Bile salts challenge.
Bile salts (1.0 g/liter) were added to nonadapted or adapted log-phase cells. Viable cell counts (CFU) were determined after 0, 15, 30, and 60 s of bile salts contact at 30°C. Samples (90 μl) were diluted in 9 ml of peptone water (0.1% peptic digest of meat; Biokar Diagnostics, Beauvais, France) adjusted to pH 7.0 and containing 0.9% NaCl, and serial dilutions were poured into YEL medium containing 1.5% agar for maximal recovery. CFU were counted after 5 days of anaerobic incubation at 30°C. Each experiment was repeated at least three times.
Electron microscopy.
Log-phase P. freudenreichii cells were placed by gentle filtration on 0.2-μm-pore-size membranes (Isopore membrane filters; Millipore, Bedford, Mass.). Membranes were fixed for 48 h in 2% (wt/vol) glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 6.8) and rinsed in the same buffer. Samples were dehydrated with ethanol (10, 25, 50, 75, and 95% for 10 min each and finally 100% for 1 h), critical-point dried by the CO2 method, and coated with gold as described previously (18). Cells were examined and photographed with a Philips XL 20 scanning electron microscope operating at 10 kV. Average cell length was calculated from at least 100 measurements for each observation.
Radioactive labeling and whole-cell protein extraction.
Exponential-phase cells grown in MDP medium were labeled essentially as described previously (18). Bacteria were harvested by centrifugation (6,000 × g, 30°C, 5 min) and resuspended in an equal volume of MDP medium devoid of cysteine and methionine, either without (control cells) or with 0.2 g of bile salts/liter or 0.3 M NaCl or at pH 5.0 (adapted cells). One-milliliter subsamples of the bacterial suspension were labeled with 500 μCi of [35S]methionine-cysteine protein labeling mixture (175 Ci/mmol; ICN Pharmaceuticals, Orsay, France) between the following times of bile salts adaptation: 0 to 30 min, 0 to 60 min, 60 to 120 min, and 120 to 240 min. As an alternative, labeling was performed in the absence of bile salts but at 42°C, at pH 5.0, or in the presence of 0.3 M NaCl for 60 min. Incorporation was stopped by rapidly washing the bacteria, and proteins were extracted by sonication in SDS (0.3%) prior to acetone precipitation as described previously (18).
Two-dimensional electrophoresis.
Air-dried protein pellets were solubilized in sample solution, containing 7 M urea, 2 M thiourea, 25 mM dithiothreitol, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), and 2% IPG buffer (Amersham Biosciences, Uppsala, Sweden). Equal amounts of radioactivity (106 dpm) were loaded onto the gel in the first dimension. Isoelectric focusing was carried out by using pH 4 to 7 Immobiline Dry Strips and a Multiphor II electrophoresis system (Amersham). Two-dimensional separation was performed according to a standardized procedure (18). The radioactivity in the dried gels was detected by using a Storm Phosphorimager (Amersham). Image analysis, gel matching, and quantification of the radioactivity in individual spots were performed by using Melanie II software (Bio-Rad). Molecular weights and isoelectric points were calibrated by using comigrating standards (Amersham). Relative rates of synthesis (RRS) were determined by calculating the ratio of radioactivity in a spot to radioactivity in the entire gel. Results are the means of at least three independent experiments. Several control gels (in nonstress conditions) were compared. Because autoradiography is highly proportional compared to nonlinear staining methods, the RRS ratio for a given spot in two similar gels was always below 1.1. In the studies of differences between two autoradiograms (control and adapted), proteins were considered to be significantly induced if the mean RRS for an individual protein was at least 1.7-fold higher than that for the control (induction factor, ≥1.7). However, induction factors of >1.4 were also noted (± in Table 1).
TABLE 1.
BSSPs and induction factors in P. freudenreichii SI41
| BSSP | pIa | Molecular mass (kDa) | Induction during the following period (min)b
|
Maximal induction factorc | |||
|---|---|---|---|---|---|---|---|
| 0-30 | 0-60 | 60-120 | 120-240 | ||||
| 1 | 4.8 | 93 | + | + | UD | UD | 3.29 |
| 2 | 4.4 | 77 | ± | + | − | − | 1.70 |
| 3 | 4.4 | 78 | + | + | − | − | 2.66 |
| 4 | 4.5 | 55 | ± | + | UD | − | 1.77 |
| 5 | 5.2 | 62 | ± | + | − | − | 4.40 |
| 6 | 5.4 | 43 | NS | NS | UD | UD | NS |
| 7 | 5.4 | 40 | − | + | UD | + | 2.36 |
| 8 | 5.3 | 40 | − | − | + | + | 2.42 |
| 9 | 5.2 | 38 | − | + | + | + | 6.12 |
| 10 | 5.0 | 37 | − | − | − | + | 2.29 |
| 11 | 4.7 | 35 | − | ± | UD | − | 1.65 |
| 12 | 5.7 | 35 | − | ± | + | UD | 5.24 |
| 13 | 6.0 | 35 | NS | NS | UD | UD | NS |
| 14 | 5.4 | 32 | − | + | UD | + | 2.80 |
| 15 | 4.6 | 32 | NS | NS | UD | UD | NS |
| 16 | 4.7 | 32 | NS | NS | UD | UD | NS |
| 17 | 5.1 | 26 | − | ± | + | + | 3.04 |
| 18 | 5.3 | 26 | ± | + | + | + | 11.86 |
| 19 | 5.3 | 25 | NS | NS | NS | NS | NS |
| 20 | 5.2 | 22 | − | − | + | + | 3.03 |
| 21 | 4.4 | 20 | + | ± | + | + | 5.60 |
| 22 | 4.5 | 19 | − | ± | + | + | 5.00 |
| 23 | 4.6 | 19 | + | + | + | + | 9.01 |
| 24 | 4.9 | 12 | + | ± | + | + | 8.96 |
Determined from two-dimensional gels calibrated by using molecular mass and isoelectric point calibration kits (Amersham) and Melanie II software.
+, induction factor above 1.7; UD, protein undetectable for this labeling period; ±, induction factor between 1.4 and 1.7; −, induction factor below 1.4; NS, protein newly synthetized in response to bile salts (undetectable on control gels; incalculable induction factor).
Deduced from Melanie II software analysis.
N-terminal amino acid sequence determination.
Cells from a 200-ml culture grown in MDP medium were lysed, and proteins were extracted and electrophoresed as described above. The resulting two-dimensional gels were transferred to polyvinylidene difluoride membranes (Hyperbond; Beckman) by immersion electroblotting in 10 mM 3-[cyclohexylamino]-1-propanesulfonic acid (CAPS) as described by Matsudaira (27). Proteins were visualized by Coomassie blue R-250 staining. Spots were cut from the membranes and applied to an automatic Beckman Porton LF3000 protein sequencer as described by the manufacturer. Sequence homologies were searched for by using the FASTA program (33) from the European Bioinformatics Institute (http://www.ebi.ac.uk).
Whole-genome sequencing of the 2.7-Mb chromosome of P. freudenreichii subsp. shermanii (type strain CIP103027) is currently in progress in our laboratory (Laboratoire de Recherches de Technologie Laitière, INRA, Rennes, France). The actual draft of genomic data contains 513 clusters (average GC content, 67%) corresponding to 92% of the genome (average threefold coverage). In this work, available N-terminal protein sequences were compared against the incomplete (gapped) genome by using the BLAST program (2). The corresponding coding sequences were deposited at the EMBL nucleotide sequence database (http://www.ebi.ac.uk/embl).
Peptide mass fingerprinting by matrix-assisted laser desorption ionization (MALDI)-time-of-flight (TOF) mass spectrometry.
In-gel tryptic digestion of two-dimensional protein spots was performed as follows. Gel pieces were washed twice in 25 mM ammonium bicarbonate buffer (pH 8.0) containing 50% (vol/vol) acetonitrile prior to vacuum drying. Rehydration was done with 50 mM ammonium bicarbonate buffer (pH 8.0) containing 0.02 g of sequencing-grade modified porcine trypsin (Promega, Madison, Wis.)/liter. Trypsin digestion was performed for 18 h with a Thermomixer (Eppendorf, Hamburg, Germany) at 37°C and 500 rpm and stopped by the addition of 0.4% (vol/vol) trifluoroacetic acid. Resulting peptides were recovered in the supernatant and spotted directly onto a MALDI plate. A 0.5-μl aliquot was allowed to dry at room temperature before the addition of a 0.5-μl aliquot of the matrix solution. This dried-droplet sampling method was used with the following matrix solution (5 g/liter), prepared fresh daily: α-cyano-4-hydroxycinnamic acid in 50% (vol/vol) acetonitrile containing 0.1% (vol/vol) trifluororacetic acid. Mass spectra were acquired on a Voyager-DE-STR TOF mass spectrometer (Applied Biosystems, Framingham, Mass.) equipped with a nitrogen laser (Laser Science, Franklin, Mass.; emitting at a wavelength of 337 nm). The accelerating voltage used was 20 kV. All spectra were recorded in positive reflector mode and with a delayed extraction of 130 ns and a 62% grid voltage. The spectra were calibrated by using an external calibration mixture. Peptide masses were queried against entries for eubacteria in the NCBInr database by using the Mascot peptide mass fingerprinting program from Matrix Science (http://www.matrixscience.com). For a match to be considered, a minimum of three matching peptides were required.
Nucleotide sequence accession numbers.
Accession numbers for the sequences reported in this article are given in Table 2.
TABLE 2.
Sequence analysis of P. freudenreichii SI41 BSSPs
| BSSP | N-terminal sequence | P. freudenreichii proteina | EMBL accession no.a | Molecular mass (kDa)b | pIb | Homologous protein (accession no.)c | Molecular mass (kDa)b | pIb | Function | E valued |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | MSRDAVTAAVRLALTKGNP | ClpB1 (partial) | AJ535195 | 66% Identity to ClpB of Mycobacterium tuberculosis (sw O53719)e | 92.6 | 5.22 | ATP-binding chain of ATP-dependent protease | 0e | ||
| 2 | QIPEPISLXXDS | 67% Identity to RecR of Bacillus subtilis (owl P24277)f | 22.0 | 5.44 | Recombinase | 13f | ||||
| 3 | ARSVGIDLGTTX | Hsp70_1 (DnaK1) | AJ535194 | 65.4 | 4.77 | 84% Identity to DnaK of Propioni bacterium acnes (sw Q9L7P1)e | 66.4 | 4.64 | Heat shock protein, Hsp70 family, molecular chaperone | 0e |
| 10 | SAIHDSITELIFNTPIVR | Cys2 | AJ535197 | 33.6 | 5.14 | 47% Identity to CysK of B. subtilis (sw P37887)e | 32.7 | 5.65 | Cysteine synthase (pyridoxal-5′-dependent); EC 4.2.99.8 | 1e-66e |
| 11 | MIIPTTXVXD | ORF0001 | AJ535198 | 26.9 | 4.99 | 73% Identity to putative two-component system response regulator of Streptomyces coelicolor (sptr Q9ADN7)e | 30.2 | 5.54 | Two-component system regulatory protein, signal transducer | 7e-70e |
| 15 | AGPNAPELSTRP | ORF0002 | AJ535199 | 30.5 | 5.02 | 49% Identity to oxidoreductase of S. coelicolor (sptr Q9KYM9)e | 30.6 | 5.52 | NADPH-dependent aldo- or keto-oxidoreductase; EC1.1.1g | 8e-63e |
| 17 | TFYTLPELPYDY | Sod | AJ535193 | 22.7 | 5.06 | 97% Identity to SodA of P. freudenreichii (sw P80293)e | 22.6 | 5.28 | Superoxide dismutase; EC 1.15.1.1 | 1e-107e |
| 21 | QRGDR | 100% Identity to AlgU of Azotobacter vinelandii (sptr Q9RCH9)f | 22.2 | 5.71 | Putative alternative sigma factor | 30f | ||||
| 22 | QRAEADXGD | 78% Identity to HtrII of Halobacterium salinarum (owl P71410)f | 79.2 | 4.02 | Sensory rhodopsin II transducer | 3.6f | ||||
| 23 | IALDPFXE | Hsp20_1 | AJ535200 | 17.0 | 4.64 | 50% Identity to 18-kDa antigen 2 of Mycobacterium avium (sw P46731)e | 15.9 | 5.37 | Heat shock protein, Hsp20 family, molecular chaperone | 2e-21e |
| 24 | MKLKVTV | BCCP | AJ535201 | 12.4 | 5.40 | 100% Identity to BCCP of P. freudenreichii (sw P02904)e | 12.4 | 5.40 | Biotin-containing carboxyl carrier protein (1.3S subunit of methylmalonyl coenzyme A transcarboxylase); EC 2.1.3.1 | 2e-36e |
BSSPs for which the N-terminal sequence allowed identification of the corresponding coding sequence (deposited at the EMBL database) in the P. freudenreichii genome. BCCP, biotin-containing 1.3S carboxyl carrier subunit.
Determined by using ProtParam (http://www.expasy.ch/tools/protparam.html).
sw, Swiss-Prot; owl, Owl nonredundant library; sptr, sptrembl+rem library. Databases searched by using the FASTA program.
Expected values (E values) were obtained from the BLAST or FASTA program for open reading frame or N-terminal protein sequence comparisons, respectively. Sequences with an E value of less than 0.01 were almost always found to be homologous.
Determined from a BLAST comparison of open reading frame sequences.
Determined from a FASTA comparison of N-terminal protein sequences.
Enzyme family-substrate specificity as yet uknown.
RESULTS AND DISCUSSION
The gastrointestinal tract of mammals constitutes an efficient barrier for ingested microorganisms. As a consequence, live probiotic microorganisms, which commonly exert their beneficial effects in the intestine, first have to survive digestive stresses. Dairy propionibacteria display a number of probiotic effects. Their ability to adapt to acid stress was recently investigated (18). The aims of the present study were to characterize the response of P. freudenreichii SI41 to bile salts and to seek an efficient pretreatment.
Bile salts susceptibility and tolerance response of P. freudenreichii SI41.
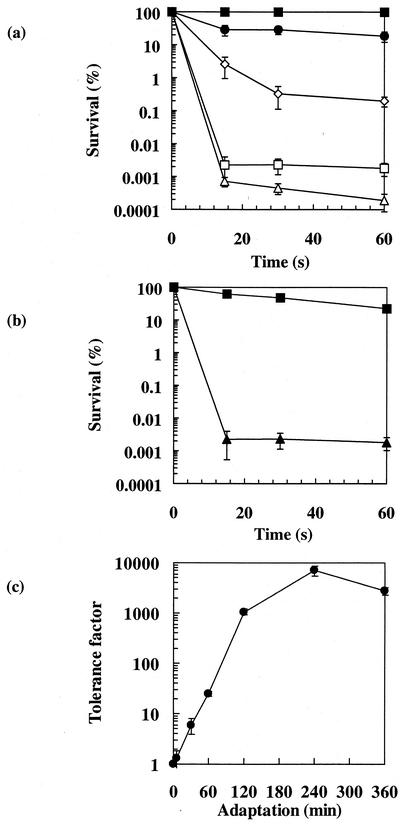
We first investigated the ability of probiotic strain SI41 grown in rich YEL medium to survive increasing concentrations of bile salts. No significant loss of culturability was observed in cultures exposed to 0.2 g of bile salts/liter (Fig. 1a), even after 6 h of exposure (data not shown). In contrast, an extremely rapid loss of culturability was observed when the bile salts concentration reached 1.0 g/liter. We noted a 5-log-unit decrease in survival during the first minute, while no further decrease was observed during the next 30 min. These observations led us to choose for the following experiments 0.2 g/liter as the sublethal bile salts concentration (for adaptation experiments) and 1.0 g/liter for 60 s as the lethal concentration.
FIG. 1.
Bile salts susceptibility and tolerance acquisition in P. freudenreichii SI41. (a) Survival rates. Exponential-phase cells were exposed to bile salts at 0.2 g/liter (▪), 0.5 g/liter (•), 0.8 g/liter (◊), 1.0 g/liter (□), and 2 g/liter (▵). After 15, 30, and 60 s of challenge, surviving bacteria were counted by CFU enumeration. (b) Survival rates of exponential-phase cells, either not treated (▴) or adapted to bile salts at 0.2 g/liter for 120 min (▪), upon exposure to bile salts at 1.0 g/liter for 60 s. (c) Kinetics of bile salts tolerance acquisition. Exponential-phase cells, adapted for various times in the presence of bile salts at 0.2 g/liter, were subjected to a lethal challenge (1.0 g/liter, 60 s). The tolerance factor is plotted as a function of the duration of adaptation (minutes). Error bars indicate standard deviations.
It is known that tolerance to some lethal treatments can be triggered by preexposure to sublethal pretreatments. Therefore, we sought adaptation conditions that could increase survival of an otherwise lethal bile salts challenge. Cells preexposed to 0.2 g of bile salts/liter for 120 min were much less sensitive to the lethal bile salts challenge than nonadapted ones (Fig. 1b). To determine the conditions for optimal adaptation, sublethal pretreatments were carried out in the presence of 0.2 g of bile salts/liter for various time periods. No significant tolerance was observed for a 5-min pretreatment. Pretreatment for 30 min enhanced survival in otherwise lethal conditions (tolerance factor [ratio of survival of adapted cells to survival of control cells] of 5). The optimal duration for adaptation was 4 h, leading to a tolerance factor of 7,000 (Fig. 1c).
We investigated the response of P. freudenreichii SI41 to bile salts in MDP medium, a chemically defined medium that is convenient for the analysis of protein synthesis in this species. The growth characteristics of MDP or YEL medium cultures are similar (18). Furthermore, we showed that lethal and sublethal bile salts concentrations were identical regardless of the culture medium. A bile salts concentration of 1.0 g/liter triggered a 5-log-unit loss of culturability, but 59% of an MDP medium culture survived a lethal challenge preceded by a 120-min adaptation period in the presence of 0.2 g of bile salts/liter (data not shown).
Adapted bacteria thus survived exposure to 1.0 g/liter, corresponding to 2.5 mmol of unconjugated bile salts/liter; the maximum unconjugated bile salts concentration measured in the duodenum of healthy humans was 0.7 mmol/liter (25). Demonstration of bile salts tolerance acquisition was also reported for Enterococcus faecalis (8, 9). For this species, Flahaut et al. (8) demonstrated that a very short exposure (5 s) to 0.8 g of bile salts/liter induced significant homologous tolerance, and the induction of tolerance was nearly maximal after a 1-min adaptation period. Thus, while we also observed bile salts tolerance in P. freudenreichii, the adaptation characteristics appeared to be different from those described for E. faecalis.
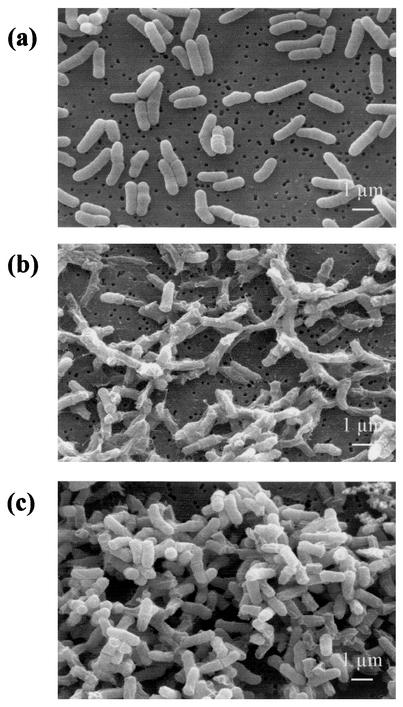
Morphological changes during bile salts stress.
Cells grown under standard conditions exhibited the characteristic pleomorphic rod-shaped morphology of dairy propionibacteria. Their average length was 1.63 ± 0.28 μm (mean and standard deviation) (Fig. 2a). Incubation for 1 h with 1.0 g of bile salts/liter (challenge conditions) caused drastic changes in cell morphology (Fig. 2b). Most bacteria displayed a shrunken and empty appearance, suggesting leakage of intracellular material, which was detected by SDS-polyacrylamide gel electrophoresis analysis of the extracellular fraction (data not shown). Leakage of proteins was previously observed after bile treatment in Lactobacillus acidophilus (32). Bile also caused permeabilization of L. acidophilus and of P. freudenreichii (47). In contrast, adaptation to 0.2 g of bile salts/liter did not cause any notable changes in morphology in this work. However, the cell length seemed to decrease to 1.26 ± 0.23 μm (data not shown). Furthermore, bile salts-adapted bacteria (0.2 g/liter) exposed to challenge conditions (1.0 g/liter) did not show modifications comparable to those shown by nonadapted bacteria. There were only a few morphologically abnormal cells (Fig. 2c), and the extracellular fraction was devoid of proteins.
FIG. 2.
Morphological changes in P. freudenreichii SI41 during bile salts stress. Nontreated cells (a), cells exposed to extreme bile salts challenge (1.0 g/liter, 1 h) (b), and bile salts-adapted cells (0.2 g/liter, 4 h) exposed to extreme bile salts challenge (1.0 g/liter, 1 h) (c) were examined by scanning electron microscopy.
These results illustrate, at the morphological level, the protective effect of bile salts adaptation, which prevents the disruption of cell integrity by higher bile salts concentrations. Similar stress-induced changes were reported previously for P. freudenreichii (18) and for other bacteria (13). Shrinkage of Escherichia coli was shown to result from the induction of the bolA morphogene belonging to the rpoS regulon. A reduction in the intracellular volume has often been linked to environmental stress adaptation (14). Moreover, disruption of the gls-24 gene provoked bile salts sensitivity and drastic morphological changes in E. faecalis (11). In P. freudenreichii, bile salts adaptation triggers a cell response which in turn protects cells from disruption and from cytoplasm leakage. This response was shown here to be accompanied by morphological modifications consistent with potential alterations of cell wall properties.
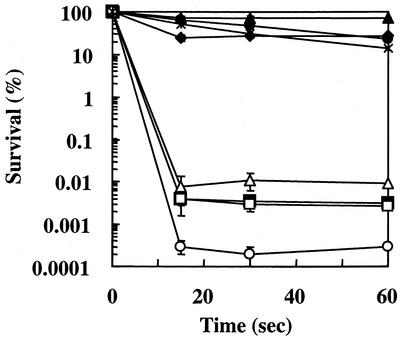
Effects of heterologous pretreatments.
It is generally acknowledged that the response of microorganisms to environmental stresses can be divided into two aspects: the mechanisms that are specifically induced by a given stress (and most probably involved in adaptation) and the mechanisms that are induced by numerous stresses and that should be involved in the development of a multiresistant state. Cheeses have been proposed as potential vectors for the administration of live probiotic strains. We thus sought cross-protection induced by stresses encountered during cheese production processes as well as during passage through the stomach. Figure 3 shows that stationary-phase P. freudenreichii SI41 cells were as tolerant to lethal challenge as bile salts-adapted ones. This effect could be mimicked by withdrawing lactate from growing cultures (data not shown), showing that cross-protection is conferred by carbon starvation. As observed for E. faecalis (8), pretreatment with a sublethal dose of the detergent SDS also conferred significant protection, with a tolerance factor of 4,000. This result suggests a response triggered by the surfactant property of SDS. It is thus tempting to speculate that protection of dairy propionibacteria may be afforded by pretreatment with food-grade surfactants (such as glyceride citrates) in a probiotic food process. Thermal pretreatment was also promising in this respect. Pretreatment at 42°C induced cross-protection against bile salts, with a tolerance factor of 5,000. Pretreatment at 37°C induced cross-protection to a lesser extent, with a tolerance factor of 33, while pretreatment at 55°C did not do so. In contrast, other stresses did not improve significantly the tolerance of P. freudenreichii to bile salts. For cold, osmotic, and acid pretreatments, various doses of stress and various durations of exposure were used but did not trigger any cross-protection. Acid-adapted cells were even less tolerant (approximately 10-fold) to bile salts than nonadapted ones (control).
FIG. 3.
Survival of pretreated P. freudenreichii during bile salts challenge. The effects of different pretreatments on survival rates for exponential-phase cells exposed to bile salts challenge are shown. The pretreatments were as follows: 100 h of stationary phase (•), bile salts pretreatment (0.2 g/liter, 4 h) (⧫), SDS pretreatment (0.06 g/liter, 4 h) (×), heat pretreatment (42°C, 1 h) (▴), cold pretreatment (4°C, 1 h) (▵), osmotic pretreatment (0.3 M NaCl, 1 h) (□), and acid pretreatment (pH 5.0, 1 h) (○). The control received no pretreatment (▪). Error bars indicate standard deviations.
To our knowledge, such cross-protection was previously investigated only for E. faecalis. In this organism, acid pretreatment does not induce protection against bile salts but also does not induce sensitivity, as was observed for P. freudenreichii. Moderate heat or salt stresses do confer enhanced tolerance to bile salts in E. faecalis (9, 37), while salt stress has no protective effect in P. freudenreichii. Such differences between Firmicutes suggest distinct protection systems; comparative analyses would be required in order to better understand the mechanisms involved.
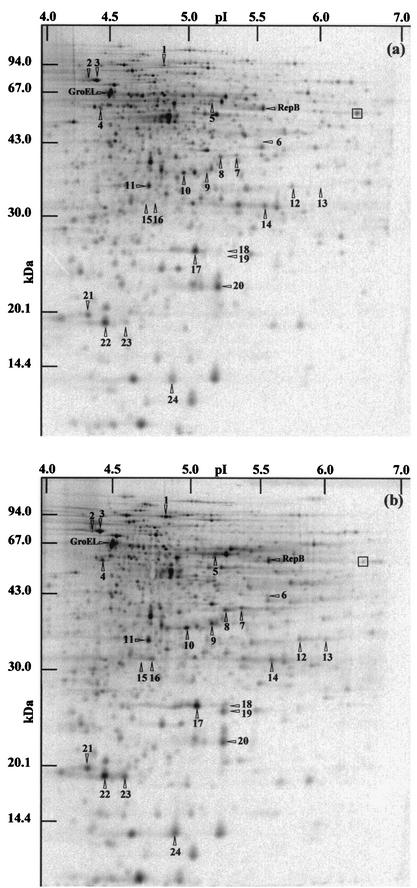
Changes in protein synthesis during bile salts adaptation.
Stress adaptation often involves modifications of gene expression. We investigated protein synthesis in both nonadapted cells (control) of P. freudenreichii SI41 and cells exposed to 0.2 g of bile salts/liter (adaptation) by two-dimensional electrophoresis. Protein synthesis rates were determined at adaptation times ranging from 30 to 240 min. Only proteins with induction factors above 1.7 were regarded as significant in the bile salts response. Figure 4 shows two representative autoradiograms of two-dimensional gels. This experiment led to the selection of the 24 most relevant distinct polypeptides induced during bile salts adaptation; these polypeptides are referred to as bile salts stress proteins (BSSPs). In contrast, most of the cellular proteins whose synthesis was detected under control conditions (Fig. 4a) were down-regulated for adaptation times above 60 min (data not shown). Indeed, of the 733 proteins shown in Fig. 4, only 183 were still detectable after 240 min of adaptation (conferring maximal tolerance). As shown in Table 1, some of the BSSPs were induced throughout the bile salts adaptation period (BSSPs 18, 19, 21, 23, and 24). Early BSSPs were induced only in the first phase of adaptation (BSSPs 1, 2, 3, 4, 5, 6, 13, 15, and 16), while late ones were induced in the last phase (BSSPs 8, 9, 17, 20, and 22). Five BSSPs, in turn, were newly synthesized in response to bile salts and could not be detected on control gels (BSSPs 6, 13, 15, 16, and 19).
FIG.4.
Two-dimensional analysis of protein expression during bile salts adaptation in P. freudenreichii SI41. Protein synthesis was monitored by labeling nontreated cells (a) and bile salts-adapted cells (0.2 g/liter) (b) for 60 min. Whole-cell protein extracts were analyzed by two-dimensional electrophoresis followed by autoradiography. The arrowheads and identifying numbers indicate polypeptides displaying an increased relative rate of synthesis (an induction factor >1.7) during bile salts adaptation compared to the results obtained for nonadapted cells. Maximal induction was evident on this gel (0 to 60 min of adaptation) or on another gel, depending on the induction kinetics (Table 1). A polypeptide displaying a reduced relative rate of synthesis (box) is also shown. GroEL and RepB were previously identified by N-terminal sequencing (18).
Bile salts adaptation has already been correlated with protein synthesis modifications in E. faecalis (9). A set of 44 proteins showed increased expression during bile salts treatment; these included both specific and general stress proteins (39). Moreover, Phan-Thanh and Gormon (36) showed that 0.3 g of deoxycholate/liter induced 13 newly synthesized and 18 up-regulated proteins in Listeria monocytogenes. Of the latter 18 proteins, 2 were also up-regulated during heat adaptation, and none were up-regulated during acid adaptation. The acid susceptibility of P. freudenreichii was studied previously, and an analysis of protein synthesis modifications was performed (18). A comparison of the resulting two-dimensional gels with those used for bile salts stress analysis showed that only six proteins were induced during both acid and bile salts adaptations (BSSPs 1, 2, 6, 20, 22, and 24). In contrast, most of the stress proteins were specifically induced by one of these stresses. Of these differentially induced proteins, those homologous to GroEL (also induced by acid and bile salts in E. faecalis [8]) and RepB (involved in DNA replication in Lactococcus lactis [41]) were induced during acid but not bile salts adaptation in P. freudenreichii.
Identification of BSSPs.
Of the 24 BSSPs, only 15 were detectable on overloaded Coomassie blue-stained two-dimensional gels; they were subjected to identification both by N-terminal sequencing and by peptide mass fingerprinting. Clear N-terminal sequences were obtained for 11 BSSPs. These were used to query the as-yet-incomplete P. freudenreichii genomic database, allowing identification of the coding sequences corresponding to eight BSSPs. Table 2 shows the 11 N-terminal sequences and the eight accession numbers corresponding to the deposited P. freudenreichii coding sequences. Data about the homologies found are also given in Table 2. In parallel, the 15 BSSPs were analyzed by MALDI-TOF mass spectrometry. While all of them produced peptide mass fingerprinting data, most failed to show a match in the general NCBInr database. Indeed, the lack of available P. freudenreichii genomic data in databases is the major limitation for the assignment of putative functions to BSSPs. Matches were found for 10 of them and are shown in Table 3.
TABLE 3.
Peptide mass fingerprinting analysis of P. freudenreichii SI41 bile salts stress
| Spot | Homologous protein | Microorganism | NCBInr accession no.a | No. of peptide mass values matched | Mowse scoreb | % Sequence coverage | Migration
|
|||
|---|---|---|---|---|---|---|---|---|---|---|
| Observedc
|
Theoriticald
|
|||||||||
| pI | Molecu- lar mass (kDa) | pI | Molecu- lar mass (kDa) | |||||||
| 1 | ATP-dependent proteinase ATP-binding chain (ClpB) | Streptomyces coelicolor | T36551 | 19 | 68 | 27 | 4.8 | 93 | 4.9 | 93 |
| 3 | DnaK | Propionibacterium acnes | Q9L7P1 | 4 | 37 | 10 | 4.4 | 78 | 4.6 | 66 |
| 5 | Putative two-component sensor kinase | S. coelicolor | NP_629573 | 6 | 67 | 17 | 5.2 | 62 | 5.5 | 58 |
| 6 | Repressor protein of class I heat shock genes | Mesorhizobium loti | NP_105447 | 12 | 38 | 44 | 5.4 | 43 | 5.8 | 39 |
| 7 | SodA | P. freudenreichii | CAA62838 | 3 | 49 | 29 | 5.4 | 40 | 5.4 | 21 |
| 10 | Unknown protein | Pasteurella multocida | NP_246193 | 6 | 42 | 31 | 5.0 | 37 | 5.1 | 37 |
| 14 | RNA polymerase subunit | Lactococcus lactis | 1908232B | 6 | 65 | 19 | 5.4 | 32 | 5.0 | 39 |
| 17 | SodA | P. freudenreichii | CAA62838 | 5 | 77 | 28 | 5.1 | 26 | 5.4 | 21 |
| 22 | Hypothetical protein | Campylobacter jejuni | NP_282143 | 4 | 44 | 42 | 4.5 | 19 | 5.0 | 19 |
| 24 | BCCPe | P. freudenreichii | P02904 | 8 | 37 | 54 | 4.9 | 12 | 5.4 | 12 |
The NCBInr library was searched by using the Mascot program (http://www.matrixscience.com/cgi/index.pI?page=../home.html).
Probability-based Mowse score; (−10 · log (P), where P is the probability that the observed match is a random event); protein scores of greater than 67 are significant at a P value of <0.05.
Determined from two-dimensional gels calibrated by using molecular mass and isoelectric point calibration kits (Amersham) and Melanie II software.
Determined from the gene sequence by using ProtParam (http://www.expasy.ch/tools/protparam.html).
BCCP, biotin-containing 1.3S carboxyl carrier subunit.
Bile salts induce general stress protein synthesis.
BSSPs were shown to include general stress proteins involved in protein protection or degradation. Indeed, BSSP 3 was identified as the universal chaperone DnaK both by N-terminal sequencing and by peptide mass fingerprinting. Likewise, BSSP 23 was shown to be the highly conserved chaperone Hsp20. Furthermore, BSSP 1 was identified by both identification methods as ClpB, the ATP-binding chain of an ATP-dependent proteinase. Mutation of the clpB gene in E. coli was shown by Rajagopal et al. (38) to be accompanied by hypersensitivity to SDS. They suggested that ClpB is one of the previously described detergent stress proteins in this bacterium. On the other hand, proteins belonging to the Clp family were proposed to play a central role in the Bacillus subtilis signal transduction network. In addition, BSSP 24 showed 100% identity to BCCP, a biotin-containing 1.3S carboxyl carrier subunit of P. freudenreichii methylmalonyl coenzyme A transcarboxylase (EC 2.1.3.1) (31). Jan et al. previously showed this protein also to be induced by acid (18), and it was also shown to be induced by heat and salt stresses (unpublished data). BSSP 2 showed 67% identity to B. subtilis RecR in a 12-amino-acid sequence located just behind the first methionine of the sequence. Confirmation of this homology would point out the requirement for such a protein, involved in DNA repair and genetic recombination in B. subtilis (1), to face DNA damage which is thought to be caused by bile salts. Indeed, Bernstein et al. (3) described for E. coli the bile salts activation of 13 specific stress response genes involved in the cellular responses to membrane perturbation, oxidative stress, and DNA damage. BSSP 17 was shown here to be P. freudenreichii superoxide dismutase SodA (40) by both methods, and BSSP 7, which was much larger, was also matched with this enzyme by peptide mass fingerprinting. Superoxide dismutases are involved in oxidative damage remediation in bacteria. However, nonoxidative stimuli have been shown to enhance the expression of Sod enzymes. SodA was shown to be a σB-dependent general stress protein in B. subtilis (35). This finding is consistent with previous observations showing that, in other bacteria, many stresses induce, via a general sigma factor, a set of general stress proteins. Moreover, BSSP 10 was identified as cysteine synthase, an enzyme also known to participate in oxidative (and cold) stress adaptation in B. subtilis (12), and BSSP 15 was identifed as an NADPH-dependent aldo- or keto-oxidoreductase (EC 1.1.1). This last protein belongs to a superfamily of enzymes metabolizing a wide range of substrates, including monosaccharides and steroids, in both prokariotic and eukaryotic cells (20). It is noteworthy that these enzymes participate in an antioxidant response in human cells (5).
Bile salts response may include signal sensing and gene expression regulation.
BSSP 11 was clearly identified as the response regulator of a two-component signal transduction system from its complete genomic coding sequence. These types of highly conserved proteins, when phosphorylated, bind DNA and cause, in B. subtilis, specific alterations of gene expression, allowing sensing of high salt concentrations, phosphate starvation, or attractants and repellents in the chemotaxis system (30). Moreover, the gene encoding this putative signal transducer (ORF0001; EMBL accession no. AJ535198) slightly overlaps (18 bp) and is followed by a gene encoding a putative two-component system histidine kinase (ORF0003; EMBL accession no. AJ535198). The calculated size and isoelectric point for this protein are 64 kDa and 5.5, consistent with those (62 kDa and 5.2) estimated for BSSP 5. In addition, BSSP 5 was shown, by use of peptide mass fingerprinting, to match a Streptomyces coelicolor histidine kinase that is the sensor of a two-component system. Such a chromosomal colocalization is typical of genes encoding partners of two-component systems and suggests an operon-like structure. It is thus tempting to speculate that the bile salts response in P. freudenreichii involves extracellular stimulus detection and signal transduction.
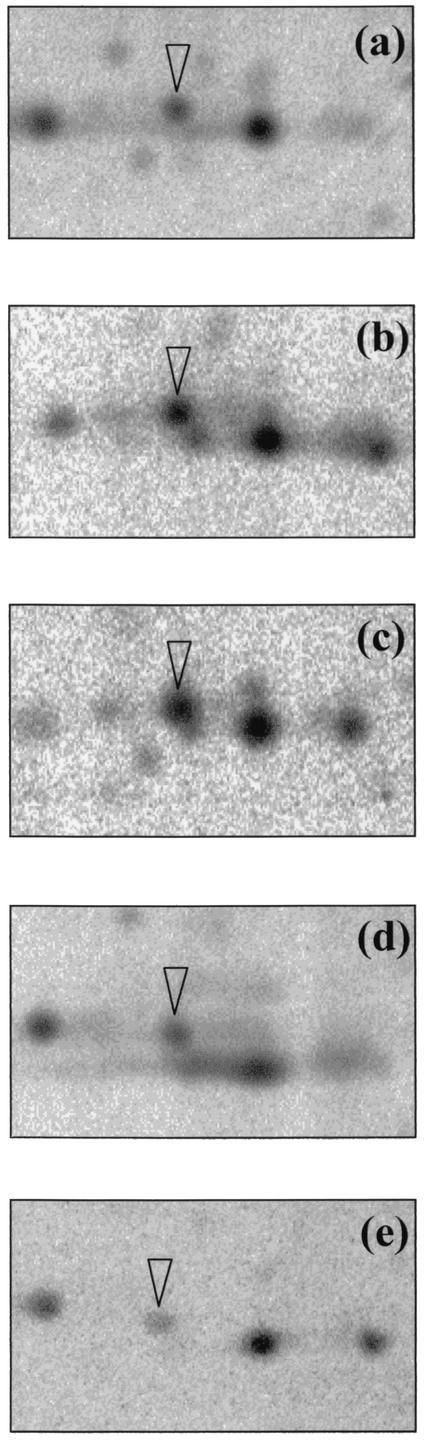
The N terminus of BSSP 21 displayed 100% identity, as well as similar isoelectric point and molecular mass, to the AlgU alternative sigma factor of Azotobacter vinelandii (26). AlgU, homologous to and functionally interchangeable with E. coli RpoE (46), has been shown to be involved in tolerance to multiple environmental stresses in members of the order Pseudomonadales (21). In order to better characterize the involvement of BSSP 21 in the P. freudenreichii stress response, we determined the effects of various pretreatments on its rate of synthesis. As shown in Fig. 5, BSSP 21 was induced to the same degrees by both bile salts and thermal pretreatments. In contrast, salt and acid pretreatments did not induce this protein but led to a decrease in its rate of synthesis. These results are consistent with the observed cross-protection suggesting an overlap between responses to bile salts and heat.
FIG. 5.
Two-dimensional analysis of BSSP 21 expression during stress adaptation in P. freudenreichii SI41. Protein synthesis was monitored by metabolic labeling during 60 min of bile salts adaptation (0.2 g/liter) (b), thermal pretreatment (42°C) (c), saline pretreatment (0.3 M NaCl) (d), or acid pretreatment (pH 5.0) (e). As a control, nontreated cells were labeled for 60 min (a). The arrowhead indicates BSSP 21. The portion of the two-dimensional autoradiograms shown covers molecular masses ranging from 17 kDa (down) to 22 kDa (up) and isoelectric points ranging from 4.1 (left) to 4.7 (right).
In conclusion, the present work constitutes a first insight into the susceptibility to bile salts and the mechanisms leading to effective tolerance in dairy propionibacteria. The elucidation of the mechanisms involved in tolerance acquisition by a proteomic approach is limited mainly by the lack of an available completed Propionibacterium genome. Ongoing study should allow identification of the genes differentially expressed under stress conditions, once this obstacle is overcome. This effort will in turn reveal a series of candidates for gene inactivation or overexpression as well as for structural and functional studies. These investigations will lead to crucial information on stress protection conferred by either homologous or heterologous stimuli and on the cellular and molecular mechanisms triggered in P. freudenreichii. They will also provide molecular markers corresponding to these tolerance states as well as efficient tools for screening of collections of propionibacteria for tolerant strains. This work should allow a rational choice of a strain(s), a pretreatment(s), and a probiotic vector(s) and thus help to optimize probiotic preparations in a pragmatic and experienced manner.
Acknowledgments
We thank A. Rouault for expert technical assistance, J. Berrier for scanning electron microscopy experiments, and C. Henry for MALDI-TOF analysis. J. C. Giard and J. Goodwins are acknowledged for critical reading of the manuscript and for English correction, respectively.
Standa Industrie is gratefully acknowledged for financial support and for constant interest in this work.
REFERENCES
- 1.Alonso, J. C., K. Shirahige, and N. Ogasawara. 1990. Molecular cloning, genetic characterization and DNA sequence analysis of the recM region of Bacillus subtilis. Nucleic Acids Res. 18:6771-6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, C., H. Bernstein, C. M. Payne, S. E. Beard, and J. Schneider. 1999. Bile salt activation of stress response promoters in Escherichia coli. Curr. Microbiol. 39:68-72. [DOI] [PubMed] [Google Scholar]
- 4.Bougle, D., N. Roland, F. Lebeurrier, and P. Arhan. 1999. Effect of propionibacteria supplementation on fecal bifidobacteria and segmental colonic transit time in healthy human subjects. Scand. J. Gastroenterol. 34:144-148. [DOI] [PubMed] [Google Scholar]
- 5.Burczynski, M. E., H. K. Lin, and T. M. Penning. 1999. Isoform-specific induction of a human aldo-keto reductase by polycyclic aromatic hydrocarbons (PAHs), electrophiles, and oxidative stress: implications for the alternative pathway of PAH activation catalyzed by human dihydrodiol dehydrogenase. Cancer Res. 59:607-614. [PubMed] [Google Scholar]
- 6.Chou, L. S., and B. Weimer. 1999. Isolation and characterization of acid- and bile-tolerant isolates from strains of Lactobacillus acidophilus. J. Dairy Sci. 82:23-31. [DOI] [PubMed] [Google Scholar]
- 7.Duwat, P., B. Cesselin, S. Sourice, and A. Gruss. 2000. Lactococcus lactis, a bacterial model for stress responses and survival. Int. J. Food Microbiol. 55:83-86. [DOI] [PubMed] [Google Scholar]
- 8.Flahaut, S., J. Frere, P. Boutibonnes, and Y. Auffray. 1996. Comparison of the bile salts and sodium dodecyl sulfate stress responses in Enterococcus faecalis. Appl. Environ. Microbiol. 62:2416-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flahaut, S., A. Hartke, J. C. Giard, A. Benachour, P. Boutibonnes, and Y. Auffray. 1996. Relationship between stress response toward bile salts, acid and heat treatment in Enterococcus faecalis. FEMS Microbiol. Lett. 138:49-54. [DOI] [PubMed] [Google Scholar]
- 10.Fuller, R. 1989. Probiotics in man and animals. J. Appl. Bacteriol. 66:365-378. [PubMed] [Google Scholar]
- 11.Giard, J. C., A. Rince, H. Capiaux, Y. Auffray, and A. Hartke. 2000. Inactivation of the stress- and starvation-inducible gls-24 operon has a pleiotrophic effect on cell morphology, stress sensitivity, and gene expression in Enterococcus faecalis. J. Bacteriol. 182:4512-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graumann, P., K. Schroder, R. Schmid, and M. A. Marahiel. 1996. Cold shock stress-induced proteins in Bacillus subtilis. J. Bacteriol. 178:4611-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harley, V. S., B. S. Drasar, and G. Tovey. 1997. The ultrastructure of stressed Legionella pneumophila. Microbios 91:73-78. [PubMed] [Google Scholar]
- 14.Hartke, A., J. C. Giard, J. M. Laplace, and Y. Auffray. 1998. Survival of Enterococcus faecalis in an oligotrophic microcosm: changes in morphology, development of general stress resistance, and analysis of protein synthesis. Appl. Environ. Microbiol. 64:4238-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann, A. F., G. Molino, M. Milanese, and G. Belforte. 1983. Description and simulation of a physiological pharmacokinetic model for the metabolism and enterohepatic circulation of bile acids in man. Cholic acid in healthy man. J. Clin. Investig. 71:1003-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holzapfel, W. H., P. Haberer, J. Snel, U. Schillinger, and J. H. J. Huis in't Veld. 1998. Overview of gut flora and probiotics. Int. J. Food Microbiol. 41:85-101. [DOI] [PubMed] [Google Scholar]
- 17.Jan, G., A. S. Belzacq, D. Haouzi, A. Rouault, D. Metivier, G. Kroemer, and C. Brenner. 2002. Propionibacteria induce apoptosis of colorectal carcinoma cells via short-chain fatty acids acting on mitochondria. Cell Death Differ. 9:179-188. [DOI] [PubMed] [Google Scholar]
- 18.Jan, G., P. Leverrier, V. Pichereau, and P. Boyaval. 2001. Changes in protein synthesis and morphology during acid adaptation of Propionibacterium freudenreichii. Appl. Environ. Microbiol. 67:2029-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jan, G., A. Rouault, and J. L. Maubois. 2000. Acid stress susceptibility and acid adaptation of Propionibacterium freudenreichii subsp. shermanii. Lait 80:325-336. [Google Scholar]
- 20.Jez, J. M., T. G. Flynn, and T. M. Penning. 1997. A new nomenclature for the aldo-keto reductase superfamily. Biochem. Pharmacol. 54:639-647. [DOI] [PubMed] [Google Scholar]
- 21.Keith, L. M., and C. L. Bender. 1999. AlgT (sigma22) controls alginate production and tolerance to environmental stress in Pseudomonas syringae. J. Bacteriol. 181:7176-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lange, R., and R. Hengge-Aronis. 1991. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor sigma S. J. Bacteriol. 173:4474-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyon, W. J., J. K. Sethi, and B. A. Glatz. 1993. Inhibition of psychrotrophic organisms by propionicin PLG-1, a bacteriocin produced by Propionibacterium thoenii. J. Dairy Sci. 76:1506-1513. [DOI] [PubMed] [Google Scholar]
- 24.Malik, A. C., G. W. Reinbold, and E. R. Vedamuthu. 1968. An evaluation of the taxonomy of Propionibacterium. Can. J. Microbiol. 14:1185-1191. [DOI] [PubMed] [Google Scholar]
- 25.Mallory, A., F. J. Kern, J. Smith, and D. Savage. 1973. Patterns of bile acids and microflora in the human small intestine. I. Bile acids. Gastroenterology 64:26-33. [PubMed] [Google Scholar]
- 26.Martinez-Salazar, J. M., S. Moreno, R. Najera, J. C. Boucher, G. Espin, G. Soberon-Chavez, and V. Deretic. 1996. Characterization of the genes coding for the putative sigma factor AlgU and its regulators MucA, MucB, MucC, and MucD in Azotobacter vinelandii and evaluation of their roles in alginate biosynthesis. J. Bacteriol. 178:1800-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsudaira, P. 1987. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262:10035-10038. [PubMed] [Google Scholar]
- 28.Mocquot, G. 1979. Rev. of the progress of dairy science: Swiss-type cheese. J. Dairy Res. 46:133-160. [Google Scholar]
- 29.Mori, H., Y. Sato, N. Taketomo, T. Kamiyama, Y. Yoshiyama, S. Meguro, H. Sato, and T. Kaneko. 1997. Isolation and structural identification of bifidogenic growth stimulator produced by Propionibacterium freudenreichii. J. Dairy Sci. 80:1959-1964. [DOI] [PubMed] [Google Scholar]
- 30.Msadek, T. 1999. When the going gets tough: survival strategies and environmental signaling networks in Bacillus subtilis. Trends Microbiol. 7:201-207. [DOI] [PubMed] [Google Scholar]
- 31.Murtif, V. L., C. R. Bahler, and D. Samols. 1985. Cloning and expression of the 1.3S biotin-containing subunit of transcarboxylase. Proc. Natl. Acad. Sci. USA 82:5617-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noh, D. O., and S. E. Gilliland. 1993. Influence of bile on cellular integrity and beta-galactosidase activity of Lactobacillus acidophilus. J. Dairy Sci. 76:1253-1259. [DOI] [PubMed] [Google Scholar]
- 33.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pérez Chaia, A., M. E. Nader de Macias, and G. Oliver. 1995. Propionibacteria in the gut: effect on some metabolic activities of the host. Lait 75:435-445. [Google Scholar]
- 35.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phan-Thanh, L., and T. Gormon. 1997. Stress proteins in Listeria monocytogenes. Electrophoresis 18:1464-1471. [DOI] [PubMed] [Google Scholar]
- 37.Pichereau, V., S. Bourot, S. Flahaut, C. Blanco, Y. Auffray, and T. Bernard. 1999. The osmoprotectant glycine betaine inhibits salt-induced cross-tolerance towards lethal treatment in Enterococcus faecalis. Microbiology 145:427-435. [DOI] [PubMed] [Google Scholar]
- 38.Rajagopal, S., N. Sudarsan, and K. W. Nickerson. 2002. Sodium dodecyl sulfate hypersensitivity of clpP and clpB mutants of Escherichia coli. Appl. Environ. Microbiol. 68:4117-4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rince, A., S. Flahaut, and Y. Auffray. 2000. Identification of general stress genes in Enterococcus faecalis. Int. J. Food Microbiol. 55:87-91. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt, M. 1999. Manipulating the coordination number of the ferric iron within the cambialistic superoxide dismutase of Propionibacterium shermanii by changing the pH-value. A crystallographic analysis. Eur. J. Biochem. 262:117-127. [DOI] [PubMed] [Google Scholar]
- 41.Schouler, C., F. Clier, A. L. Lerayer, S. D. Ehrlich, and M. C. Chopin. 1998. A type IC restriction-modification system in Lactococcus lactis. J. Bacteriol. 180:407-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Segal, G., and E. Z. Ron. 1998. Regulation of heat-shock response in bacteria. Ann. N. Y. Acad. Sci. 851:147-151. [DOI] [PubMed] [Google Scholar]
- 43.Shah, N. P. 2000. Probiotic bacteria: selective enumeration and survival in dairy foods. J. Dairy Sci. 83:894-907. [DOI] [PubMed] [Google Scholar]
- 44.Thanassi, D. G., L. W. Cheng, and H. Nikaido. 1997. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179:2512-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volker, U., H. Mach, R. Schmid, and M. Hecker. 1992. Stress proteins and cross-protection by heat shock and salt stress in Bacillus subtilis. J. Gen. Microbiol. 138:2125-2135. [DOI] [PubMed] [Google Scholar]
- 46.Yu, H., M. J. Schurr, and V. Deretic. 1995. Functional equivalence of Escherichia coli sigma E and Pseudomonas aeruginosa AlgU: E. coli rpoE restores mucoidy and reduces sensitivity to reactive oxygen intermediates in algU mutants of P. aeruginosa. J. Bacteriol. 177:3259-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zarate, G., A. P. Chaia, S. Gonzalez, and G. Oliver. 2000. Viability and beta-galactosidase activity of dairy propionibacteria subjected to digestion by artificial gastric and intestinal fluids. J. Food Prot. 63:1214-1221. [DOI] [PubMed] [Google Scholar]