Abstract
Metabolic engineering is a powerful method to improve, redirect, or generate new metabolic reactions or whole pathways in microorganisms. Here we describe the engineering of a Saccharomyces cerevisiae strain able to utilize the pentose sugar l-arabinose for growth and to ferment it to ethanol. Expanding the substrate fermentation range of S. cerevisiae to include pentoses is important for the utilization of this yeast in economically feasible biomass-to-ethanol fermentation processes. After overexpression of a bacterial l-arabinose utilization pathway consisting of Bacillus subtilis AraA and Escherichia coli AraB and AraD and simultaneous overexpression of the l-arabinose-transporting yeast galactose permease, we were able to select an l-arabinose-utilizing yeast strain by sequential transfer in l-arabinose media. Molecular analysis of this strain, including DNA microarrays, revealed that the crucial prerequisite for efficient utilization of l-arabinose is a lowered activity of l-ribulokinase. Moreover, high l-arabinose uptake rates and enhanced transaldolase activities favor utilization of l-arabinose. With a doubling time of about 7.9 h in a medium with l-arabinose as the sole carbon source, an ethanol production rate of 0.06 to 0.08 g of ethanol per g (dry weight) · h−1 under oxygen-limiting conditions, and high ethanol yields, this yeast strain should be useful for efficient fermentation of hexoses and pentoses in cellulosic biomass hydrolysates.
Bioethanol is an excellent alternative to fossil transportation fuels, either as a pure fuel with high efficiency and performance or as a gasoline additive. An attractive feedstock that is available in large amounts is cellulosic biomass; examples are herbaceous and woody plants, agricultural and forestry residues, and a large portion of municipal solid waste and industrial waste streams (29, 37). Cellulosic biomass is a complex mixture of carbohydrate polymers. Therefore, hydrolysates will contain hexoses and pentoses, including glucose, galactose, mannose, d-xylose, and l-arabinose (13). Because the feedstock represents a significant portion of all process costs, an economical fermentation process will require rapid and efficient conversion of all sugars present (30).
A lack of microorganisms that will efficiently convert hexoses and pentoses to ethanol is a major constraint to the economical conversion of biomass. Escherichia coli, Klebsiella oxytoca, and Zymononas mobilis have been genetically engineered to produce ethanol efficiently from all hexose and pentose sugars present in the polymers of hemicellulose (7, 23, 39). Most industrial ethanol fermentations use the yeast Saccharomyces cerevisiae, as it exhibits fast sugar consumption, high yields, and ethanol tolerance. But even though S. cerevisiae is able to ferment hexoses rapidly and efficiently, it is unable to ferment pentose sugars and to use these sugars for growth (1).
d-Xylose is the most abundant hemicellulosic sugar. Nevertheless, some of the cellulosic biomass, such as corn fiber and many herbaceous crops, contain significant amounts of l-arabinose (15, 22). Genetic engineering has been used to establish a d-xylose-utilizing pathway in S. cerevisiae by inserting heterologous genes encoding d-xylose reductase and xylitol dehydrogenase and by increasing the expression of its endogenous xylulokinase (10, 16, 18, 27). This process has resulted in yeast strains able to utilize the pentose d-xylose and to ferment it to ethanol. However, efforts to establish an l-arabinose-fermenting S. cerevisiae strain have failed so far (24, 26).
Many yeasts and fungi can aerobically assimilate l-arabinose, but most are unable to ferment it to ethanol or they exhibit only very low ethanol production rates and yields (8, 22). The conversion of l-arabinose to d-xylulose-5-phosphate (d-xylulose-5-P), an intermediate of the pentose phosphate pathway, proceeds via a complex pathway consisting of two reducing and two oxidizing steps and a kinase (5, 36). The genes for this pathway have recently been expressed in S. cerevisiae (24). However, although the recombinant strain was able to grow on l-arabinose medium very slowly (doubling time, ∼5 days), it did not produce any ethanol from l-arabinose. This lack of ethanol production was attributed to the predicted, very low l-arabinose uptake rate in S. cerevisiae and a cofactor imbalance which results from the utilization of NADPH in the two reductions but results in the production of NADH in the oxidation reactions.
The bacterial pathway for l-arabinose utilization does not use redox reactions but consists of l-arabinose isomerase (AraA), l-ribulokinase (AraB), and l-ribulose-5-P 4-epimerase (AraD) converting l-arabinose to l-ribulose, l-ribulose-5-P, and d-xylulose-5-P, respectively (20). However, the expression of the E. coli pathway in S. cerevisiae did not result in either growth on l-arabinose or production of ethanol from l-arabinose (26). It was suggested that the main problem was the low activity of E. coli l-arabinose isomerase in yeast.
When we tried to establish an l-arabinose utilization pathway in S. cerevisiae, we observed that the E. coli araA gene did not produce any l-arabinose isomerase activity in our yeast strains. Nevertheless, the corresponding enzymes from B. subtilis and Mycobacterium smegmatis were active in yeast but did not promote growth on l-arabinose. By using sequential transfer of yeast transformants in media containing l-arabinose as a breeding strategy, a strain that exhibits fast growth on l-arabinose and a high fermentative performance with l-arabinose was selected. Molecular analysis of this strain revealed that the crucial parameter for efficient utilization of l-arabinose is a specifically balanced stoichiometry of the l-arabinose-utilizing enzymes with high l-arabinose uptake and high l-arabinose isomerase, l-ribulose-5-P 4-epimerase, and transaldolase activities but with low l-ribulokinase activity. Our yeast strain and the knowledge derived from this work should greatly enhance the development of an efficient biomass-to-ethanol fermentation process.
MATERIALS AND METHODS
Yeast strains.
All S. cerevisiae strains used in this study (Table 1) were generated from the CEN.PK2-1C wild-type strain (K. D. Entian and P. Koetter, Frankfurt, Germany).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Plasmids |
|---|---|---|
| CEN.PK2-1C (=JBY24) | MATaleu2-3, 112 ura3-52 trp1-289 his3-Δ1 MAL2-8cSUC2 | |
| JBY24-4V | Same as for CEN.PK2-1C | p423H7, p424H7, p425H7, p426H7 |
| JBY24-4P | Same as for CEN.PK2-1C | YEparaA, YEparaB, YEparaD, YEpGAL2 |
| JBY24-4M | Same as for CEN.PK2-2C | YEparaA,a YEparaB,b YEparaD,a YEpGAL2a |
| JBY24-3T | Same as for CEN.PK2-1C | YEparaA,a YEparaB,b YEparaD,a YCpTAL1 |
| JBY25 | MATaleu2-3, 112 ura3-52 trp1-289 his3-Δ1 MAL2-8cSUC2 Unknown mutation(s) promoting growth on l-arabinose | |
| JBY25-4V | Same as for JBY25 | p423H7, p424H7, p425H7, p426H7 |
| JBY25-4M | Same as for JBY25 | YEparaA,a YEparaB,b YEparaD,a YEpGAL2a |
| JBY25-4P | Same as for JBY25 | YEparaA,a YEparaB, YEparaD,a YEpGAL2a |
| JBY25-4C | Same as for JBY25 | YEparaA,a YCparaB, YEparaD,a p424H7 |
| JBY25-3M | Same as for JBY25 | YEparaA,a YEparaB,b YEparaD,a p426H7 |
Reisolated plasmid from strain JBY25-4M.
Contains the mutant araBG361A allele.
Media and cultivation conditions.
The yeast strains were grown on solid or in liquid synthetic media consisting of 0.67% Difco yeast nitrogen base (without amino acids) supplemented with amino acids and adenine and with 2% glucose, 2% maltose, or 2% l-arabinose as the carbon source. The plasmid-selectable markers uracil, tryptophan, histidine, and leucine were omitted (selective medium). For the sequential transfer medium, 1 g of yeast extract/liter and 2 g of peptone/liter were added. Rich medium consisted of 1% yeast extract and 2% Bacto Peptone. Cells were grown aerobically at 30°C on a rotary shaker or on agar plates. Oxygen-limiting conditions were obtained by incubating 50-ml yeast cultures with high cell densities in loosely closed 50-ml bottles with gentle stirring.
Overexpression of genes.
The 0.4-kb SacI/SpeI MET25 promoter fragments of multicopy vectors p423MET25 (HIS3 as the selective marker), p424MET25 (TRP1), p425MET25 (LEU2), and p426MET25 (URA3) were replaced by 0.4-kb DNA fragments containing the very strong and constitutive HXT7 promoter fragment from −392 bp to −1 bp followed by ATG and six-histidine codons as previously described (14), resulting in plasmids p423H7 to p426H7. The coding regions of the E. coli araA, araB, and araD genes; the B. subtilis araA gene; and the M. smegmatis araA gene were amplified by whole-cell PCR from strains JM101 (New England BioLabs), DSM 1092, and DSM 43078 (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH), respectively, and cloned into these vectors by recombination-cloning by using the methods described by Wieczorke et al. (35) and omitting the six-histidine codons. araA genes were cloned into p423H7 (HIS3), araB genes were cloned into p424H7 (TRP1), and araD genes were cloned into p425H7 (LEU2). The GAL2 overexpression plasmid pHL125 (21) was a kind gift of R. Gaber. E. coli araB and mutant araB together with the HXT7 promoter fragment were cut out of p424H7 with SacI and BamHI and cloned into the centromeric vector YCplac33 (URA3) (11). Furthermore, E. coli araB and mutant araB were cloned by recombination-cloning into the vector p424H7 and fusing six-histidine codons at their N-terminal ends. The TAL1 gene with 736-bp promoter and 434-bp terminator sequences was cloned into YCplac33 by recombination-cloning. Molecular biology techniques were performed by using published procedures (25). Yeast transformations and reisolation of plasmid DNA from yeast cells were carried out as described previously (3, 12).
The genomic DNA library from strain JBY25 in plasmid YCplac33 (11) was constructed essentially as described previously (34), yielding about 50,000 E. coli transformants. Yeast transformants were selected on a selective medium, with maltose as a permissive carbon source. More than 300,000 transformant colonies grew up within 3 days at 30°C and were replica plated onto synthetic medium with 2% l-arabinose. Five of about 160 transformants that were able to grow on l-arabinose were further analyzed by plasmid isolation, restriction enzyme analysis, retransformation, and sequencing.
Microarray analysis.
Pan yeast arrays (MWG Biotech) were used for genome-wide transcriptional analysis. Two analyses (one set) were performed with yeast cells incubated overnight in synthetic medium with 2% l-arabinose; two further analyses (second set) were performed with yeast cells grown in synthetic medium with 2% galactose. The cells were harvested at an optical density at 600 nm (OD600) of about 1. mRNA and cDNA preparation, hybridization, array scanning, and data analysis were performed according to the pan yeast array manual (MWG Biotech). Transcript levels were compared after normalization.
Enzyme assays and l-arabinose uptake.
S. cerevisiae strains were grown into the exponential growth phase in synthetic medium with 2% maltose. Crude extracts were prepared by using glass beads for breaking the cells as described by Ciriacy and Breitenbach (6). Assays were performed in imidazole buffer (50 mM imidazole, 10 mM MgCl2, 100 mM KCl, 0.1 mM EDTA, pH 7). Protein levels were determined as described by Zamenhoff (38) by using bovine serum albumin as a standard. l-Arabinose isomerase activity was assayed in two steps. Crude cell extract was mixed in 100 mM l-arabinose in imidazole buffer in a total volume of 500 μl. l-Arabinose isomerase in the crude extract was allowed to convert l-arabinose to l-ribulose for 5 to 50 min at 30°C. The reaction was stopped with 150 μl of 50% trichloric acid and subsequently neutralized with 195 μl of 2 M Na2CO3. In the second step, the decrease of NADH was measured photometrically at an OD340 when l-ribulose was converted to arabinitol by sorbitol dehydrogenase in a mixture of neutralized sample, 0.2 mM NADH, and 0.1 mg of sorbitol dehydrogenase at 30°C in a volume of 1 ml. As sorbitol dehydrogenase has a very low affinity for l-ribulose, the measured amounts of l-ribulose and, therefore, l-arabinose isomerase activity were underestimated. l-Ribulokinase activity was assayed in imidazole buffer with 1 mM ATP and 5 mM l-ribulose at 30°C. For determination of the kinetic parameters, 0.1 to 10 mM l-ribulose was used. The consumption of ATP was determined continuously by the addition of 0.23 mM NADH, 1 mM phosphoenolpyruvate, 1 U of l-lactate dehydrogenase, and 1 U of pyruvate kinase plus 1 mM of its activator fructose-1,6-bisphosphate. The decrease of NADH was measured photometrically. l-Ribulose-5-P 4-epimerase activity was determined as follows. Crude extracts from yeast transformants expressing E. coli AraB were used for the production of l-ribulose-5-P from 50 mM l-ribulose and 2 mM ATP for 30 min at 30°C in 1 ml of imidazole buffer. l-Ribulose-5-P 4-epimerase activity was determined photometrically by measuring the decrease of NADH in a reaction mixture of crude extract, 100 μl of l-ribulose-5-P solution, 1 mM d-ribose-5-P, 0.23 mM NADH, 0.5 U of transketolase, 4.5 U of triosephosphate isomerase, and 1.55 U of glycerol-3-P dehydrogenase in 1 ml of imidazole buffer at 30°C. Transaldolase activity measurements were done as previously described (4). The enzymes and substrates were obtained from Roche, Aldrich, and Sigma. l-Arabinose uptake measurements were based upon the method of Bisson and Fraenkel (2) but with radioactively labeled l-[1-14C]arabinose (American Radiolabeled Chemicals Inc.). Uptake was determined in 100 mM potassium phosphate buffer, pH 6.5, at 30°C with 10 mM l-arabinose with a specific radioactivity of 55 mCi/mmol. Uptake was measured for up to 1 min, as appropriate experiments showed that it was linear for at least 1 min.
Purification of six-His-tagged ribulokinases was done after yeast transformants were harvested from 1-liter cultures, washed with 100 ml of cold lysis buffer (20 mM HEPES [pH 8], 300 mM NaCl, 10% glycerol), and resuspended in 4 ml of resuspension buffer (20 mM HEPES [pH 8], 300 mM NaCl, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitors [Complete; Roche]). Cells were broken with glass beads, and cell debris was removed by centrifugation. Four milliliters of the supernatants was mixed with 1 ml of Ni-nitrilotriacetic acid agarose suspension (Qiagen) and incubated for 1 h at 4°C. The mixture was loaded in columns and washed four times with washing buffer (lysis buffer plus 10 mM imidazole), and six-His-ribulokinases were eluted four times with 500 μl of elution buffer (lysis buffer plus 300 mM imidazole). The various fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, Coomassie blue staining, and Western blot analysis with anti-six-His antibodies (Roche) for l-ribulokinase protein that was detected in the crude extract and elution fractions as a band of 61 kDa. The elution fractions were collected and used for enzyme assays.
Analysis of growth and ethanol.
Growth of the yeast cells was monitored by measuring the OD600 or by comparing the colony diameters on agar plates. Ethanol concentrations in yeast culture supernatants were determined by using an enzymatic test kit (Roche). Ethanol yields are based on the total amount of available sugar.
RESULTS
Heterologous expression of a bacterial l-arabinose-utilizing pathway in S. cerevisiae.
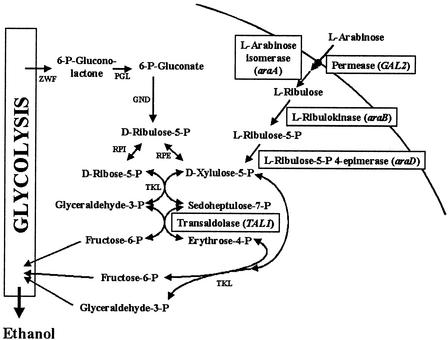
To establish an l-arabinose-utilizing pathway in S. cerevisiae (Fig. 1), the E. coli genes araA (l-arabinose isomerase), araB (l-ribulokinase), and araD (l-ribulose-5-P 4-epimerase) were overexpressed in strain CEN.PK2-1C behind a strong HXT7 promoter fragment on multicopy vectors. As it had been shown that the yeast galactose permease (Gal2) is able to transport l-arabinose (19), the GAL2 gene was simultaneously overexpressed from the strong ADH1 promoter. Determination of enzyme activities in crude extracts of the yeast transformants revealed that E. coli araA did not produce any measurable l-arabinose isomerase activity, araB produced up to 0.7 U of l-ribulokinase activity per mg of protein, and araD produced up to 0.13 U of l-ribulose-5-P 4-epimerase activity per mg of protein. Uptake activity for l-arabinose (at 10 mM) was increased from 0.01 (empty plasmid) to 0.32 (GAL2 plasmid) nmol per min and mg (dry weight). In contrast to the E. coli araA gene, overexpression of the B. subtilis or M. smegmatis araA in yeast resulted in an active protein that produced more than 2.5 mU of l-arabinose isomerase activity per mg of protein. However, all of the transformants were not able to grow with l-arabinose as the sole carbon source.
FIG. 1.
Schematic presentation of the l-arabinose utilization pathway engineered in S. cerevisiae. l-Arabinose is transported into the cell by the galactose permease Gal2 and subsequently converted to d-xylulose-5-P by three enzymatic steps. d-Xylulose-5-P is an intermediate of the pentose phosphate pathway and is directed to glycolysis by transketolase (TKL) and transaldolase (TAL). ZWF, glucose-6-P dehydrogenase; PGL, phosphogluconolactonase; GND, gluconate-6-P dehydrogenase; RPI, ribose-5-P isomerase; RPE, ribulose-5-P 3-epimerase.
Selection for growth on l-arabinose medium.
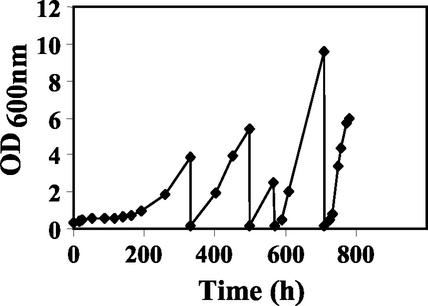
Transformants overexpressing the B. subtilis araA gene together with the E. coli araB and araD genes and the yeast GAL2 gene (strain JBY24-4P) were incubated in synthetic liquid medium with 20 g of l-arabinose/liter, 1 g of yeast extract/liter, and 2 g of peptone/liter. Due to the utilization of components of yeast extract and peptone as carbon and energy sources, slow but limited growth was possible. After 4 to 5 days of incubation with gentle shaking at 30°C, the growth rate of the transformants increased progressively (Fig. 2), in contrast to that of a strain containing only four empty vectors or E. coli araA instead of B. subtilis araA (data not shown). Whenever the cells reached an OD600 of up to 10, they were inoculated in fresh medium at an OD600 of 0.3 and grown further. After about 750 h, the doubling time reached up to 8.5 h. To distinguish between the occurrence of spontaneous suppressor mutations or an adaptation process, the transformants were grown for seven generations on selective medium with glucose as the sole carbon source and then shifted again onto l-arabinose medium. They started to grow on l-arabinose medium with only a short lag phase, excluding an adaptation process. Single colonies were isolated on l-arabinose agar plates, and one of the colonies was taken for further analysis. Plasmids were reisolated from the cells. Prolonged growth of the cells on a rich medium with glucose as the carbon source resulted in a loss of plasmids (strain JBY25) (Table 1). Plasmids were retransformed into this strain in various combinations.
FIG. 2.
Selection of yeast transformants for growth on l-arabinose medium by sequential transfer. Yeast transformants (JBY24-4P) were pregrown in synthetic liquid medium with 2% glucose, washed twice, and inoculated in synthetic medium with 2% l-arabinose, 0.1% yeast extract, and 0.2% peptone at an initial OD600 of about 0.2. Cells were grown aerobically at 30°C in shaker flasks on a rotary shaker. Growth was monitored by measuring the OD600s of the cultures. Whenever the transformants reached an OD600 of up to 10, they were inoculated in fresh medium at an OD600 of about 0.3 and grown further.
Determination of enzyme activities and kinetics.
The activities of all three heterologous enzymes were measured in crude extracts of the mutant transformants (JBY25-4M) and compared to those of the wild-type transformants (JBY24-4P). Whereas the activities of l-ribulose-5-P 4-epimerase and l-arabinose isomerase were similar in both strains, the l-ribulokinase activity was hardly detectable in the mutant transformants (less than 0.01 U of protein per mg compared to 0.7 U of protein per mg in the wild-type transformants). In contrast, l-arabinose uptake activity (at 10 mM) was increased (0.63 nmol per min and mg [dry weight]). Sequencing of the reisolated plasmids revealed no mutations in the araA, araD, and GAL2 genes, whereas the reisolated araB coding sequence exhibited one mutation (G361 → A), which led to an exchange of aspartate for an asparagine at position 121. Structural modeling by using the National Center for Biotechnology Information protein structure database and its Cn3D application (http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?form=6&db=t&Dopt=s&uid = 12196) with E. coli glycerol kinase as the related sequence revealed that this amino acid residue lies in the conserved sugar kinase domain of l-ribulokinase, close to the substrate binding site.
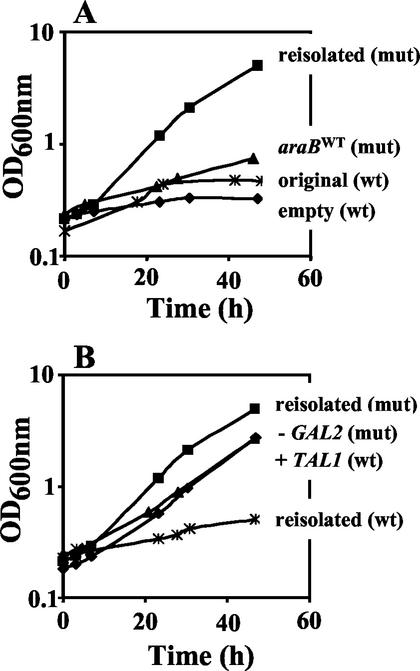
Replacing the reisolated araB (l-ribulokinase) plasmid with the corresponding original plasmid in strain JBY25-4P resulted in strongly reduced growth on l-arabinose (Fig. 3). However, when wild-type araB was expressed on a centromeric plasmid in strain JBY25-4C, the growth rate on l-arabinose increased (data not shown). In contrast, mutant araB expressed on a centromeric plasmid no longer promoted growth on l-arabinose. These results indicate that reduced activity of l-ribulokinase is important for growth on l-arabinose, although a certain level of enzyme activity is required. Six-histidine codons were fused to the N termini of wild-type and mutant araB genes and used to purify the enzymes from crude extracts via Ni-nitrilotriacetic acid-agarose columns. Both constructs behaved similarly to the original enzymes in l-arabinose growth experiments, indicating that the histidine residues did not significantly change their properties. Determination of l-ribulokinase kinetics revealed that the Km value for l-ribulose was increased for the mutant kinase (1.5 versus 0.3 mM for wild-type kinase) and the Vmax was decreased (8 versus 20 μmol per min and mg of protein).
FIG. 3.
Growth of different yeast transformants in l-arabinose medium. Yeast transformants were pregrown in synthetic medium with 2% glucose, washed twice, and inoculated in synthetic medium with 2% l-arabinose at an initial OD600 of about 0.2. Cells were grown aerobically at 30°C in shaker flasks on a rotary shaker. Growth was monitored by measuring the OD600s of the cultures. All growth tests were performed two to four times, with similar results. Representative results are shown here. The strains are JBY25-4M (▪) (A and B); JBY24-4V (♦), JBY24-4P (∗), and JBY25-4P (▴) (A); and JBY25-3M (♦), JBY24-3T (▴), and JBY24-4M (∗) (B). The doubling time of JBY25-4M in the exponential growth phase was 7.9 h. The various plasmid compositions of the transformants are indicated on the right side of the growth curves. wt, wild-type background (JBY24); mut, mutant background (JBY25).
Growth on l-arabinose depends on the complete bacterial pathway.
To find out whether all four plasmids carrying the B. subtilis araA gene, the E. coli araB and araD genes, and the yeast GAL2 genes are necessary for growth on l-arabinose, the mutant strain JBY25 (Table 1) was transformed with different combinations of reisolated and empty plasmids. Transformants lacking the l-arabinose isomerase, the l-ribulokinase, or the l-ribulose-5-P 4-epimerase but transformed with the other three reisolated plasmids did not show any growth on l-arabinose (data not shown), indicating that these genes are absolutely necessary for the utilization of l-arabinose. Transformants lacking the overexpressed galactose permease (JBY25-3M) were able to grow on l-arabinose medium but with slightly decreased growth rates (Fig. 3), indicating that overexpression of a transporter is not necessary for growth on l-arabinose but can improve it.
DNA microarrays and genetic analyses.
The four reisolated plasmids were transformed into a CEN.PK2-1C wild-type strain (JBY24-4M). In contrast to strain JBY25-4M, they did not promote growth on l-arabinose (Fig. 3), indicating that an additional genomic mutation(s) had occurred in JBY25, enabling it to grow efficiently on l-arabinose. A genetic analysis showed that dominant as well as recessive genomic mutations were responsible for the improved growth on l-arabinose (data not shown). To clone the dominant mutant alleles, a genomic DNA library was constructed from the JBY25 strain in the centromeric plasmid YCplac33 and transformed into the CEN.PK2-1C strain together with the reisolated plasmids YEparaA and YEparaD and mutant YEparaB. Five different plasmids enabling growth on l-arabinose medium were isolated and characterized. Sequencing revealed that they all contained the TAL1 gene encoding yeast transaldolase (Fig. 1). However, no mutations were found within its coding or the flanking sequences. Genome-wide transcription patterns were compared between JBY24-4M and JBY25-4M by using MWG pan yeast array gene chips, which represent a DNA array encompassing the entire S. cerevisiae genome. The majority of the yeast genes showed similar transcript levels in the two strains. Only 10 genes showed >1.5-fold-higher transcript levels in strain JBY25-4M on all four chips (Table 2). Interestingly, they included the TAL1 gene. Moreover, all the genes were located on the right arm of chromosome 12 and induction was about twofold, indicating a duplication of this part of the yeast genome including TAL1. No gene was found that showed repression (<75% transcript level on all four chips) in the mutant strain.
TABLE 2.
Genes induced >1.5-fold in the l-arabinose-utilizing yeast mutant strain
| Systematic gene name | Standard name (function)a | Induction (fold) (mean value from results of four experiments) |
|---|---|---|
| YLR342w | FKS1 (1,3-beta-glucan synthase) | 1.99 |
| YLR348c | DIC1 (dicarboxylic acid transporter) | 2.58 |
| YLR351c | NIT3 (unknown) | 2.21 |
| YLR354c | TAL1 (transaldolase) | 2.28 |
| YLR359w | ADE13 (adenylosuccinate lyase) | 3.76 |
| YLR370c | ARC18 (structural constituent of cytoskeleton) | 1.88 |
| YLR387c | — (unknown) | 2.14 |
| YLR396c | VPS33 (intracellular transport) | 2.07 |
| YLR420w | URA4 (dihydroorotase) | 2.03 |
| YLR424w | — (unknown) | 2.15 |
—, not yet named.
Enhanced transaldolase activities improve growth on l-arabinose.
Determination of transaldolase enzyme activities revealed that they were significantly increased in strain JBY25-4M (70 mU per mg of protein) compared to those in the wild-type JBY24-4M strain (40 mU per mg of protein). To demonstrate that the increased transaldolase activities are responsible for enabling JBY25-4M to grow on l-arabinose, the TAL1 gene from the CEN.PK2-1C strain was cloned into a centromeric vector and transformed together with the l-arabinose utilization plasmids into the CEN.PK2-1C wild-type strain (JBY24-3T). Indeed, growth rates on l-arabinose were comparable to those of the JBY25-3M strain (Fig. 3), indicating that enhanced expression of TAL1 plays a major role in the ability of JBY25-4M to utilize l-arabinose.
Ethanol production.
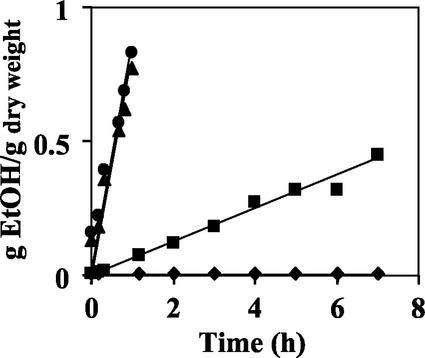
Ethanol production was determined with the JBY25-4M mutant strain, which was incubated in a synthetic growth medium with 2% l-arabinose (Fig. 4). Under oxygen-limiting conditions at a culture OD600 of 15 to 20, ethanol production rates reached 0.06 to 0.08 g of ethanol per g (dry weight) and h. In contrast, the wild-type strain JBY24-4V did not produce any ethanol under the same conditions. On 2% glucose medium, JBY25-4M produced ethanol as rapidly and as efficiently as JBY24-4V, with ethanol production rates of about 0.8 g of ethanol per g (dry weight) and h (Fig. 4). If l-arabinose is metabolized exclusively to ethanol as the sole fermentation product through a combination of pentose phosphate and glycolytic pathways (Fig. 1), the maximum theoretical yield from 20 g of l-arabinose/liter (133 mmol/liter) would be 10.2 g of ethanol/liter (222 mmol/liter). In the overall fermentation reaction, 3 mol of l-arabinose is converted to 5 mol of ethanol with the following stoichiometry: 3 mol of l-arabinose plus 5 mol of ADP plus 5 mol of Pi yields 5 mol of ethanol plus 5 mol of CO2 plus 5 mol of ATP. With an initial l-arabinose concentration of 20 g/liter, strain JBY25-4M achieved about 6 g of ethanol/liter, which is 60% of the maximum theoretical ethanol yield based on the amount of available sugar.
FIG. 4.
Ethanol production from l-arabinose and glucose. The yeast transformants JBY25-4M and JBY24-4V were pregrown in synthetic medium with 2% glucose or 2% l-arabinose until they reached an OD600 of about 2, washed twice, and incubated at 30°C in synthetic medium with 2% glucose or 2% l-arabinose under oxygen-limiting conditions at an OD600 of about 15 to 20. Ethanol concentrations in the culture supernatants were determined at various time points. Representative results of at least two independent experiments for each strain are shown. •, JBY24-4V with glucose; ♦, JBY24-4V with l-arabinose; ▴, JBY25-4M with glucose; ▪, JBY25-4M with l-arabinose. EtOH, ethanol.
DISCUSSION
Genetic engineering together with directed evolution strategies have allowed us to select an efficient l-arabinose-fermenting S. cerevisiae strain. This has been achieved by expression of a bacterial pathway for catabolism of l-arabinose, consisting of l-arabinose isomerase, l-ribulokinase, and l-ribulose-5-P 4-epimerase. In this pathway, l-arabinose is converted to d-xylose-5-P, an intermediate of the pentose phosphate pathway. The same strategy had already been attempted before (26) but without much success.
We found that essentially four problems had to be resolved. The first problem was that the E. coli l-arabinose isomerase AraA was not functional in S. cerevisiae or at least that the activity was too low to be detected in the enzyme assay. Moreover, using E. coli araA, we were not able to select an l-arabinose-utilizing yeast strain. These findings are consistent with those of Sedlak and Ho (26), who found that the expression of E. coli l-arabinose isomerase in yeast reached only 10% of the activity in wild-type E. coli. As similar problems had been observed before with the expression of heterologous xylose isomerases during the construction of xylose-fermenting yeast strains (reviewed in reference 13), it might be assumed that structural features of bacterial isomerases might pose problems to the yeast cells. Improper protein folding, posttranslational modifications, disulfide bridge formation, etc., have been suggested to be responsible. Nevertheless, there are no structural similarities between l-arabinose isomerase and xylose isomerase. Moreover, other bacterial isomerases have been expressed successfully in S. cerevisiae, e.g., xylose isomerase from the thermophilic bacterium Thermus thermophilus (33) or a phosphoglucose isomerase from E. coli (3). Therefore, there seem to be no general obstacles for expression of heterologous isomerases in S. cerevisiae.
Consistent with this assumption, we found that expression of l-arabinose isomerases from B. subtilis and M. smegmatis produced significant enzyme activities in yeast, although the actual values appeared to be very low. However, in our enzyme assay we used a coupled reaction with sorbitol dehydrogenase to detect the l-ribulose originating from l-arabinose. As sorbitol dehydrogenase proved to have only a very low affinity for l-ribulose, the measured amounts of l-ribulose and, therefore, of l-arabinose isomerase activity in our study were clearly underestimated.
Nevertheless, after overexpression of the complete and functional bacterial pathway, the yeast cells did not grow with l-arabinose as the only carbon source. Evolutive screening and molecular analysis of the resulting strain revealed that two further prerequisites were essential for the utilization of l-arabinose by the recombinant yeast cells: a specifically reduced l-ribulokinase activity and enhanced transaldolase activities.
The importance of a reduced l-ribulokinase activity for efficient utilization of l-arabinose is consistent with the recent demonstration that catabolic pathways beginning with an ATP-requiring activation step but finally producing a surplus of ATP need tightly controlled activities of their initial phosphorylating enzymes (28). Therefore, in contrast to what is generally believed, maximizing the enzymatic activities in a metabolic pathway may not always be an optimal strategy. The evolved cells had acquired a mutation in l-ribulokinase that reduced its affinity for l-ribulose and its activity. Moreover, reducing activity of wild-type ribulokinase by expression from a centromeric plasmid instead of a multicopy plasmid had a similar effect on growth on l-arabinose. Nevertheless, the activity must not drop below a certain level, as the expression of the mutant enzyme from a centromeric plasmid no longer promoted growth. However, l-ribulokinase is not the first enzyme of the pathway but is preceded by l-arabinose isomerase, which seems to have very low activity in yeast. Therefore, this enzyme should already limit the delivery of substrate to l-ribulokinase. However, as already discussed above, l-arabinose isomerase activity is clearly underestimated with our enzyme assay and the actual activity might be much higher.
The third problem that obviously hampered growth of the yeast cells on l-arabinose was too-low endogenous activity of transaldolase. Also, in the case of d-xylose utilization by recombinant S. cerevisiae, it had been shown that a d-xylose reductase- and xylitol dehydrogenase-containing strain overexpressing TAL1 exhibited considerably enhanced growth on d-xylose compared with that of a strain containing only d-xylose reductase and xylitol dehydrogenase (32). It was concluded that the transaldolase level in S. cerevisiae is insufficient for the efficient utilization of pentose phosphate pathway metabolites. Using DNA microarray analysis, we found that the TAL1 gene, as well as other genes located in close proximity to it, showed about a twofold induction in the selected strain. This points to a duplication of part of yeast chromosome XII, including TAL1, and is consistent with recent observations that genome rearrangements, especially amplifications and deletions, can generally be observed as responses to sustained application of the same strong selective pressure in yeast populations (9). An amplification can also explain the dominant phenotype of wild-type TAL1 in strain JBY25, although no mutation could be found.
The expression of a fungal l-arabinose utilization pathway led to only very slow growth of the yeast cells on l-arabinose (24). It was speculated that one reason for this is that l-arabinose uptake into the cell is very slow. Therefore, we overexpressed the yeast galactose permease and confirmed that it is able to transport l-arabinose. Moreover, we observed even more enhanced l-arabinose uptake activities in the selected strain, although we did not further investigate the molecular reasons. On the other hand, we found that even transformants lacking the overexpressed galactose permease were able to grow on l-arabinose medium, although with slightly decreased growth rates. These results indicate that overexpression of a transporter is not necessary for growth on l-arabinose but can improve it.
Not only did the selected strain grow with l-arabinose as the only carbon source on agar plates or in liquid shaken-flask cultures (aerobic conditions) but under oxygen-limited conditions it produced up to 0.08 g of ethanol per g (dry weight) and hour (Fig. 4). This rate is comparable to the highest d-xylose fermentation rates ever obtained with recombinant d-xylose-fermenting S. cerevisiae strains (17, 31). Moreover, based on the amount of available sugar, the ethanol yield was about 60% of the maximum theoretical ethanol yield. It must be said that the true ethanol yield should clearly be higher, since under our fermentation conditions ethanol is lost due to its evaporation. On the other hand, the difference in the theoretical yields may be explained by residual unfermented l-arabinose, by-product formation, and partial consumption of the produced ethanol under the not strictly anaerobic conditions.
Our results demonstrate that sustained application of a strong selective pressure is a powerful method to improve metabolic fluxes. It was advantageous in the first rounds of selection that slow growth of the yeast cells was made possible by the addition of small amounts of yeast extract and peptone, which served as poor carbon and energy sources. Similar strategies should expand opportunities to improve or redirect other new or already existing metabolic pathways in microorganisms. The strategies described and the knowledge derived from our work should provide a substantial basis for the development of an industrial l-arabinose fermentation process. For this, of course, further improvements are necessary. For example, it would be desirable to integrate all heterologous genes stably into the yeast genome. Nevertheless, the powerful selection strategy should then force the strain to use l-arabinose as an efficient carbon source.
Acknowledgments
This work was supported by a grant from the European Commission (grant QLK3-CT-1999-00080) to E.B.
We thank all members of the BIO-HUG EC project for fruitful and stimulating discussions.
REFERENCES
- 1.Barnett, J. A. 1976. The utilization of sugars by yeasts. Adv. Carbohydr. Chem. Biochem. 32:125-234. [DOI] [PubMed] [Google Scholar]
- 2.Bisson, L. F., and D. G. Fraenkel. 1983. Involvement of kinases in glucose and fructose uptake by Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 80:1730-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boles, E., and F. K. Zimmermann. 1993. Saccharomyces cerevisiae phosphoglucose isomerase and fructose bisphosphate aldolase can be functionally replaced by the corresponding enzymes of Escherichia coli and Drosophila melanogaster. Curr. Genet. 23:187-191. [DOI] [PubMed] [Google Scholar]
- 4.Brand, K. 1974. Transaldolase, p. 752-756. In H. U. Bergmeyer (ed.), Methoden der enzymatischen analyse 1. Verlag Chemie, Weinheim, Germany.
- 5.Chiang, C., and S. G. Knight. 1961. l-Arabinose metabolism by cell free extracts of Penicillium chrysogenum. Biochim. Biophys. Acta 46:271-278. [DOI] [PubMed] [Google Scholar]
- 6.Ciriacy, M., and I. Breitenbach. 1979. Physiological effects of seven different blocks in glycolysis in Saccharomyces cerevisiae. J. Bacteriol. 139:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deanda, K., M. Zhang, C. Eddy, and S. Picataggio. 1996. Development of an arabinose-fermenting Zymomonas mobilis strain by metabolic pathway engineering. Appl. Environ. Microbiol. 62:4465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dien, B. S., C. P. Kurtzman, B. C. Saha, and R. J. Bothast. 1996. Screening for l-arabinose fermenting yeasts. Appl. Biochem. Biotechnol. 57-58:233-242. [PubMed] [Google Scholar]
- 9.Dunham, M. J., H. Badrane, T. Ferea, J. Adams, P. O. Brown, F. Rosenzweig, and D. Botstein. 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99:16144-16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eliasson, A., C. Christensson, C. F. Wahlbom, and B. Hahn-Hägerdal. 2000. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl. Environ. Microbiol. 66:3381-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 12.Gietz, R. D., and R. A. Woods. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87-96. [DOI] [PubMed] [Google Scholar]
- 13.Hahn-Hägerdal, B., C. F. Wahlbom, M. Gardonyi, W. H. van Zyl, R. R. Cordero Otero, and L. J. Jönsson. 2001. Metabolic engineering of Saccharomyces cerevisiae for xylose utilization. Adv. Biochem. Eng. Biotechnol. 73:53-84. [DOI] [PubMed] [Google Scholar]
- 14.Hamacher, T., J. Becker, M. Gárdonyi, B. Hahn-Hägerdal, and E. Boles. 2002. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology 148:2783-2788. [DOI] [PubMed] [Google Scholar]
- 15.Hespell, R. B. 1998. Extraction and characterization of hemicellulose from the corn fiber produced by corn wet-milling processes. J. Agric. Food Chem. 46:2615-2619. [Google Scholar]
- 16.Ho, N. W. Y., Z. Chen, and A. P. Brainard. 1998. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl. Environ. Microbiol. 64:1852-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kötter, P., R. Amore, C. P. Hollenberg, and M. Ciriacy. 1990. Isolation and characterization of the Pichia stipitis xylitol dehydrogenase gene, XYL2, and construction of a xylose-utilizing Saccharomyces cerevisiae transformant. Curr. Genet. 18:493-500. [DOI] [PubMed] [Google Scholar]
- 18.Kötter, P., and M. Ciriacy. 1993. Xylose fermentation by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 38:776-783. [Google Scholar]
- 19.Kou, S.-C., M. S. Christensen, and V. P. Cirillo. 1970. Galactose transport in Saccharomyces cerevisiae. II. Characteristics of galactose uptake and exchange in galactokinaseless cells. J. Bacteriol. 103:671-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, N., W. Gielow, R. Martin, E. Hamilton, and A. Fowler. 1986. The organization of the araBAD operon of Escherichia coli. Gene 47:231-244. [DOI] [PubMed] [Google Scholar]
- 21.Liang, H., and R. F. Gaber. 1996. A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol. Biol. Cell 7:1953-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMillan, J. D., and B. L. Boynton. 1994. Arabinose utilization by xylose-fermenting yeasts and fungi. Appl. Biochem. Biotechnol. 45-46:569-584. [DOI] [PubMed] [Google Scholar]
- 23.Ohta, K., D. S. Beall, J. P. Mejia, K. T. Shanmugam, and L. O. Ingram. 1991. Genetic improvement of Escherichia coli for ethanol production: chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl. Environ. Microbiol. 57:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richard, P., M. Putkonen, R. Vaananen, J. Londesborough, and M. Penttilä. 2002. The missing link in the fungal l-arabinose catabolic pathway, identification of the l-xylulose reductase gene. Biochemistry 41:6432-6437. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. M. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Sedlak, M., and N. W. Ho. 2001. Expression of E. coli araBAD operon encoding enzymes for metabolizing l-arabinose in Saccharomyces cerevisiae. Enzyme Microb. Technol. 28:16-24. [DOI] [PubMed] [Google Scholar]
- 27.Tantirungkij, M., N. Nakashima, T. Seki, and T. Yoshida. 1993. Construction of xylose-assimilating Saccharomyces cerevisiae. J. Ferment. Bioeng. 75:83-88. [Google Scholar]
- 28.Teusink, B., M. C. Walsh, K. van Dam, and H. V. Westerhoff. 1998. The danger of metabolic pathways with turbo design. Trends Biochem. Sci. 23:162-169. [DOI] [PubMed] [Google Scholar]
- 29.Van Wyk, J. P. H. 2001. Bio/technology and the utilization of biowaste as a resource for bioproduct development. Trends Biotechnol. 19:172-177. [DOI] [PubMed] [Google Scholar]
- 30.von Sivers, M., and G. Zacchi. 1995. A techno-economical comparison of three processes for the production of ethanol from pine. Bioresour. Technol. 51:43-52. [Google Scholar]
- 31.Wahlbom, C. F., and B. Hahn-Hägerdal. 2002. Furfural, 5-hydroxymethyl furfural, and acetoin act as external electron acceptors during anaerobic fermentation of xylose in recombinant Saccharomyces cerevisiae. Biotechnol. Bioeng. 78:172-178. [DOI] [PubMed] [Google Scholar]
- 32.Walfridsson, M., J. Hallborn, M. Penttilä, S. Keränen, and B. Hahn-Hägerdal. 1995. Xylose-metabolizing Saccharomyces cerevisiae strains overexpressing the TKL1 and TAL1 genes encoding the pentose phosphate pathway enzymes transketolase and transaldolase. Appl. Environ. Microbiol. 61:4184-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walfridsson, M., X. Bao, M. Anderlund, G. Lilius, L. Bülow, and B. Hahn-Hägerdal. 1996. Ethanolic fermentation of xylose with Saccharomyces cerevisiae harboring the Thermus thermophilus xylA gene, which expresses an active xylose (glucose) isomerase. Appl. Environ. Microbiol. 62:4648-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weierstall, T., C. P. Hollenberg, and E. Boles. 1999. Cloning and characterization of three genes (SUT1-3) encoding glucose transporters of the yeast Pichia stipitis. Mol. Microbiol. 31:871-883. [DOI] [PubMed] [Google Scholar]
- 35.Wieczorke, R., S. Krampe, T. Weierstall, K. Freidel, C. P. Hollenberg, and E. Boles. 1999. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 464:123-128. [DOI] [PubMed] [Google Scholar]
- 36.Witteveen, C. F., R. Busink, P. J. I. Van de Vondervoort, C. Dijkema, K. Swart, and J. Visser. 1989. l-Arabinose and d-xylose catabolism in Aspergillus niger. J. Gen. Microbiol 135:2163-2171.
- 37.Wyman, C. E. 2001. Twenty years of trials, tribulations, and research progress in bioethanol technology: selected key events along the way. Appl. Biochem. Biotechnol. 91-93:5-21. [DOI] [PubMed] [Google Scholar]
- 38.Zamenhoff, S. 1957. Preparation and assay of desoxyribonucleic acids from animals tissue. Methods Enzymol. 3:696-704. [Google Scholar]
- 39.Zhang, M., C. Eddy, K. Deanda, M. Finkelstein, and S. Picataggio. 1995. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science 267:240-243. [DOI] [PubMed] [Google Scholar]