Abstract
Bacterial proliferations have recurrently been observed for the past 15 years in fermentor cultures of the ectomycorrhizal fungus Laccaria bicolor S238N, suggesting the presence of cryptic bacteria in the collection culture of this fungus. In this study, intracellular bacteria were detected by fluorescence in situ hybridization in combination with confocal laser scanning microscopy in several collection subcultures of L. bicolor S238N. They were small (0.5 μm in diameter), rare, and heterogeneously distributed in the mycelium and were identified as Paenibacillus spp. by using a 16S rRNA-directed oligonucleotide probe initially designed for bacteria isolated from a fermentor culture of L. bicolor S238N.
Bacterial endosymbiosis is characterized by the intracellular localization of the bacteria, regardless of their parasitic or mutualistic behavior (25). Endosymbiotic bacteria are known to colonize numerous eukaryotic organisms, e.g., aphids, Schizaphis graminum (7); bivalves, Bankia setacea (14); and plants, Gunnera spp. (30).
Concerning fungi, only a few cases of bacterial endosymbiosis have been reported as yet, mostly for Glomeromycota species (35). Moreover, except for the cyanobacteria colonizing Geosiphon pyriforme (34), known intrafungal bacteria are nonculturable. Since the 1970s, electron microscopy has allowed the detection of small organelles (less than 1 μm wide) with a bacterial type of cell wall (20, 21, 33). However, the true bacterial nature of these bacterium-like organisms has only been confirmed recently thanks to molecular methods. For example, with the help of PCR amplification and sequencing, Bianciotto et al. (8) identified the endobacteria belonging to several species of endomycorrhizal fungi (Gigaspora spp. and Scutellospora spp.) as Burkholderia spp. (9). More recently, Barbieri et al. (5) discovered, by fluorescence in situ hybridization (FISH), bacteria belonging to the Cytophaga-(Flavobacterium)-Bacteroides phylogroup embedded in the cell wall of the hyphae of Tuber borchii, an ectomycorrhizal fungus (5, 6).
Laccaria bicolor (Maire) P. D. Orton S238N (formerly Laccaria laccata S238N), an ectomycorrhizal fungal strain, has been selected in France for the controlled mycorrhization of Douglas fir plantations. It is commercially used to inoculate seedlings of Douglas fir in French nurseries in order to obtain high-performance planting material for reforestation (19). Since its transfer from Oregon in 1980, it has been subcultured on diluted brewery wort solid medium every year in our laboratory without ever displaying bacterial contamination. In contrast, recurrent bacterial proliferations have been observed sporadically for 15 years in the derived axenic fermentor cultures which produce the commercial inoculum for nurseries. This is why we suspected the fungus to harbor cryptic bacteria, which would be revealed by the particular culture conditions of the fermentor. Therefore we investigated the presence of fungus-associated bacteria, whether intra- or extracellular, in pure cultures of L. bicolor S238N, by using FISH and confocal laser scanning microscopy (CLSM). Further identification of these bacteria was provided by a specific probe, originally designed to target the 16S rRNA of bacteria isolated from a fermentor culture of L. bicolor S238N.
MATERIALS AND METHODS
Fungal strain.
Laccaria bicolor (Maire) P. D. Orton is a member of the ectomycorrhizal family Tricholomataceae. The original strain, S238O, was isolated in 1976 in Oregon by Trappe and Molina from a fruit body collected under Tsuga mertensiana (Bong.) Carr. The subculture transferred to the 4°C fungal collection (Institute National de la Recherche Agronomique, Nancy, France) in 1980 was named S238N (13). In this study, agar plugs from the strain S238N 4°C collection cultures were grown in the dark at 20°C on modified P5 (5 mM NH4+) and P25 (25 mM NH4+) Pachlewski media (28), liquid and solid. For solid cultures, hyphae were grown over a sheet of cellophane to avoid sampling agar fragments, which are autofluorescent.
Reference bacterial strain.
The bacterial strain EJP73 was kindly provided by G. Bending (Horticulture International, Wellesbourne, United Kingdom). It had originally been isolated from Lactarius rufus-Pinus sylvestris macerated ectomycorrhizal roots and identified as Paenibacillus sp. (EMBL accession number AJ302333) (29).
Isolation of bacterial strains from an L. bicolor culture produced in a fermentor.
The fermentor (Setric 50 L) was filled with 35 liters of Opti medium (i.e., optimized for the growth of L. bicolor S238N in fermentor; Durand, Institut National de la Recherche Agronomique, Dijon, France) and heat sterilized. The parameters of fermentation were as follows: temperature, 25°C; shaking, 90 rpm; pH 6.6; and high oxygenation rate. Inoculation under sterile conditions was done 5 days later in order to ensure that no sporulating bacteria had escaped sterilization. The fungal inoculant consisted of 1 liter of a 2-week-old culture of L. bicolor S238N on modified P5 (5 mM NH4+) Pachlewski liquid medium (28), subcultured from the 4°C collection. It was fragmented axenically in a mixer previous to inoculation to ensure homogeneous distribution of the fungus in the fermentor.
Platings on tryptic soy agar (TSA) (10%) medium of the fungal inoculant and of the fermentor broth, before and after inoculation, revealed no bacterial contaminant. Bacterial proliferations were observed in the fermentor L. bicolor S238N culture 7 days after inoculation. Bacteria were isolated from the fermentor broth by cultivation on TSA (10%) medium plates for 2 days at 25°C and then purified by three successive subcultures on the same medium. Two bacterial isolates were finally maintained at −80°C in NB-glycerol (nutrient broth [Difco], 8 g; 150 ml of glycerol per liter). A comparison with noncontaminated batches showed that the growth of the fungus in the fermentor was not hampered by the development of these bacterial proliferations.
Bacterial DNA extraction, PCR, and sequencing.
Bacterial suspensions (optical density at 600 nm, 0.5) were boiled for 5 min and subsequently diluted 100-fold. Almost full-length 16S ribosomal DNA (rDNA) PCR products were obtained by PCR with the eubacterial primers 27f and 1492r (38). The amplifications were performed in Perkin-Elmer thermocyclers 9700 and 2400. The reaction mixture (25 μl) consisted of 1× incubation buffer with MgCl2 (Qbiogene), 0.2 μM deoxynucleoside triphosphate mix, 0.5 μM each primer, 0.6 U of Taq DNA polymerase (Qbiogene), and 2.5 μl of bacterial DNA. The PCR cycling conditions were as follows: one cycle of 94°C for 3 min, 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min, and a final extension step at 72°C for 15 min. The 16S rDNA gene products were purified with a Geneclean Turbo kit (Bio 101) and were fully sequenced by an automated sequencer (Beckman Coulter, Fullerton, Calif.), according to the manufacturer's instructions with the eubacterial primers 27f and 1492r (38) and 341f and 926r (17). Sequences were assembled with Sequencher 3.1.1 software (Gene Codes Corporation) and analyzed with the ARB software package (http://www.mikro.biologie.tu-muenchen.de).
Fixation of bacteria and mycelium prior to FISH.
Bacteria were cultured overnight at 25°C in Luria-Bertani (LB) liquid medium (32), harvested in the logarithmic growth phase (optical density at 600 nm, 0.2 to 0.6), and washed in 1× PBS (130 mM NaCl, 7 mM Na2HPO4, 3 mM NaH2PO4, pH 7.3). One- to six-month-old fungal colonies were collected under aseptic conditions and detached from the sheet of cellophane prior to fixation. Mycelium was aseptically dipped in 1× PBS during sampling for a maximum of 30 min. Bacteria and fungus were fixed by adding 3 volumes of 3% paraformaldehyde to one volume of 1× PBS (2) and incubated at 4°C for 1.5 h to 20.5 h for the fungus and 3 h for the bacteria. Then they were washed three times in 1× PBS and finally resuspended in a 1:1 mixture of 1× PBS and 96% ethanol. Fixed samples were stored at −20°C.
rRNA-targeted oligonucleotide probes.
Oligonucleotide probes were purchased from Thermo Hybaid Division Interactiva GmbH, Ulm, Germany, 5′-end labeled with indocarbocyanine (Cy3), indodicarbocyanine (Cy5), or fluorescein isothiocyanate (FITC). Intracellular bacteria were detected with EUB338mix (an equimolar mixture of EUB338I, EUB338II, and EUB338III [1, 12]), with LGC354mix (an equimolar mixture of LGC354A, LGC354B, and LGC354C [24]), specific for the Firmicutes, and with BIF216 (5′-GCCCATCCCCGAGTAACA-3′), a 16S rRNA-directed probe specific for bacterial isolates from the L. bicolor S238N culture in a fermentor, designed in this study. The probes ALF968 (27), BET42a (22), GAM42a (22), CF319 (23), and HGC69a (31), specific for α-, β-, and γ-proteobacteria, Cytophaga-Flavobacterium, and high-GC-content gram-positive bacteria, respectively, and non-EUB338 (16), the reverse complement of EUB338I, were used as negative controls to detect nonspecific binding.
In situ hybridization.
Fixed fungal samples were dilacerated with pins in order to obtain a thin mycelial layer, facilitating microscopic observation. Hyphae were subsequently immobilized on hydrophobic Teflon-coated slides in 8-mm hybridization wells (Roth). Alternatively, washed (TDF4 detergent; Franklab), gelatin-coated [0.075% gelatin-0.01 CrK(SO4)2] slides were used to improve the attachment of the samples. Each well analyzed contained roughly the equivalent of one square millimeter of fungal colony. The slides were dehydrated in 50, 80, and 96% ethanol for 3 min each. Prior to hybridization, some samples were treated with lysozyme (150,000 U/mg; Serva) to enhance the permeabilization of gram-positive bacterial cell walls (32), with 10 μl per well of a 10-mg/ml solution for 20 min, at room temperature, which yielded optimal results. Lysozyme was then removed with a pipette, and the slides were dehydrated again as previously. Bacteria were treated in the same way after depositing 2 μl of fixed bacterial suspension per well. Once the slides were perfectly dry, hybridization was performed according to Manz et al. (23). Before observation, the slides were mounted in AF1 antifading reagent (Citifluor Ltd.).
Design and evaluation of oligonucleotide probe specific for bacterial isolates from L. bicolor S238N culture produced in a fermentor.
The oligonucleotide probe BIF216, specific for bacterial isolates F2001-L and F2001-O, was designed with the ARB program package (http://www.arb-home.de). Optimal stringency for in situ hybridization was determined by whole-cell hybridization with the Cy3-labeled probe on the reference bacterial strain Paenibacillus sp. strain EJP73, as the nontarget organism, and on F2001-L as the target organism. The stringency was gradually increased by the addition of deionized formamide in 5 to 10% increments in the hybridization buffer and by lowering the salt concentration of the wash buffer accordingly (18). The probe-conferred fluorescence of the bacteria was evaluated semiqualitatively by eye, according to Neef et al. (26). As a reference, formamide gradients were realized with the eubacterial probe EUBmix-Cy3 and to define the background range, without probe and with the antisense probe non-EUB338-Cy3. A discriminant formamide stringency was achieved when the target organism yielded bright signals whereas the nontarget organism stayed in the background range.
Observations.
The fungal samples were observed with a confocal laser scanning microscope (LSM 510 Axiovert 100 M; Zeiss) equipped with an argon laser emitting at 488 nm and two helium-neon lasers emitting at 543 and 633 nm, suitable to excite FITC, Cy3, and Cy5, respectively. Plan-Neofluar 100×/1.3 oil and Apochromat 63×/1.2 water immersion lenses were used for both observation and image recording. Systematically, monochrome images were taken sequentially at each wavelength. Artificial colors were assigned to the images taken for each excitation wavelength: green for 488 nm, red for 543 nm, and blue for 633 nm. Superimpositions were processed with the Zeiss software package LSM 510, version 3.0.
The bacterial pure cultures were observed with an epifluorecent microscope (Nikon), equipped with a filter for Cy3 (excitation, 535 to 550 nm; emission, 610 to 675 nm) and a 100×/1.3 oil immersion lens.
RESULTS
Isolation and characterization of bacteria growing in fermentor culture of L. bicolor S238N.
Whereas all the control platings remained negative, bacterial proliferations appeared in the L. bicolor S238N fermentor culture 7 days after inoculation. Two morphotypes of bacterial colonies, L (limpid) and O (opaque), were identified after plating on TSA (10%) medium and subsequently purified. Each morphotype was conserved as a distinct isolate, named, respectively, F2001-L and F2001-O. A few colonies of the O morphotype were observed to revert to the L morphotype and vice versa.
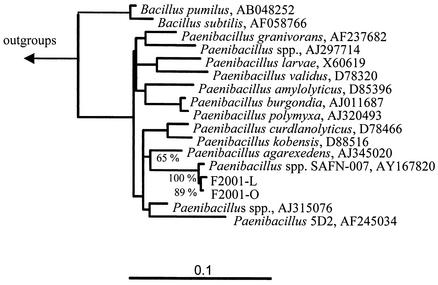
A 1,394-base fragment was sequenced for each of the two bacterial isolates from the fermentor, revealing a similarity of 99.8% between the two isolates. Comparative 16S rDNA sequence analysis showed that the closest related sequence in the data bank is from Paenibacillus sp. strain SAFN-007 (99.3% similarity with F2001-L and 99.4% with F2001-O) from a study on the microbial diversity of spacecraft assembly facilities. The next closely related Paenibacillus spp., P. agarexedens, P. kobensis, and P. curdlanolyticus (Fig. 1), had 16S rDNA similarities of 93.2, 93.5, and 93.3%, respectively, to isolate F2001-L and 93.4, 93.6, and 93.2%, respectively, to isolate F2001-O.
FIG. 1.
Phylogenetic relationship of isolates F2001-L and F2001-O. The 16S rDNA-based tree is based on the results of a maximum likelihood analysis. Only sequences which share common residues in at least 50% of all available sequences from gram-positive organisms with low GC content were included for the calculation of this tree. The tree topology was evaluated and corrected according to the results of distance and maximum parsimony analyses. The bar indicates 10% estimated base changes.
Design and evaluation of probe specific for bacteria isolated from L. bicolor S238N culture produced in a fermentor.
A specific 16S rRNA-targeted oligonucleotide probe for FISH was designed on the basis of a consensus of the 16S rDNA sequences of the two bacterial strains isolated from the L. bicolor S238N fermentor culture, F2001-L and F2001-O. Probe design and specificity analysis were performed with an ARB sequence database containing a total of about 16,000 complete and partial 16S rRNA sequences, and the ARB tools Probe Design and Probe Match. Positions comprising ambiguities between the target isolates were avoided. The Probe Design tool suggested 16S rRNA positions 216 to 233 as a diagnostic region for a probe (alignments presented in Table 1); the probe corresponding to this position was named BIF216. It perfectly matched F2001-L and F2001-O but also Paenibacillus sp. strain SAFN-007, which was included in the database later.
TABLE 1.
Sequence alignment of 16S rRNAs in the vicinity of the oligonucleotide target region (boxed), performed with the ARB softwarea
| Strain | Sequence name | Sequence |
|---|---|---|
| Probe BIF216 | 3′-ACAATGAGCCCCTACCCG-5′ | |
| Target region 16S | 5′-UGUUACUCGGGGAUGGGC-3′ | |
| Paenibacillus sp. strain F2001-L | -==================- | |
| Paenibacillus sp. strain F2001-O | -==================- | |
| Paenibacillus sp. strain SAFN-007 | AY167820 | -==================- |
| Paenibacillus sp. strain EJP73 | AJ302333 | -===C===u==========- |
| Paenibacillus sp. | PnbSpe16 | -===C===u==========- |
| Paenibacillus glucanolyticus | PnbGluca | -===C===u==========- |
| Paenibacillus amylolyticus | PnbAmylo | -===C===u==========- |
| Paenibacillus pabuli | PnbPabul | -===C===u==========- |
| Paenibacillus sp. strain SAFN-022b | AY167825 | -===C===u==========- |
| Paenibacillus borealis | PSP011322 | -==NC===u==========- |
| Paenibacillus lautus | D78472 | -===C===u========N=- |
| Bacillus longisporus | BacLongi | -===C===u==========- |
| Bacillus lautus 4 | BacLautu | -===C===u==========- |
| Thermoactinomyces peptonophilus | ThcPepto | -==C====u=N========- |
| Bacteroides sp. | BctSpeci | -g=============A===- |
The mismatches with the target region (upper row) are indicated by capital (strong mismatches) and lowercase (weak mismatches) letters. The bacteria used for the evaluation of probe BIF216 specificity are in boldface.
The optimal hybridization conditions of probe BIF216 were determined by whole-cell hybridization on pure cultures of F2001-L. In comparison, EJP73 served as a nontarget organism, because its 16S rRNA sequence presents the two above-mentioned mismatches; the stringency necessary to discriminate between the target organism and EJP73 is the same for all organisms presenting these two mismatches (26). A very strong probe-conferred fluorescence was observed for F2001-L at between 10% and 50% formamide, whereas the signal intensity with EJP73 stayed in the background at these stringencies. As a consequence, the BIF216 probe was used at a stringency of 35% formamide. Five other nontarget bacterial strains chosen according to their ecological relation to fungi and having more than two mismatches with the probe were not detected by BIF216 (data not shown).
Detection and identification of bacteria associated with mycelium of L. bicolor S238N in collection subcultures.
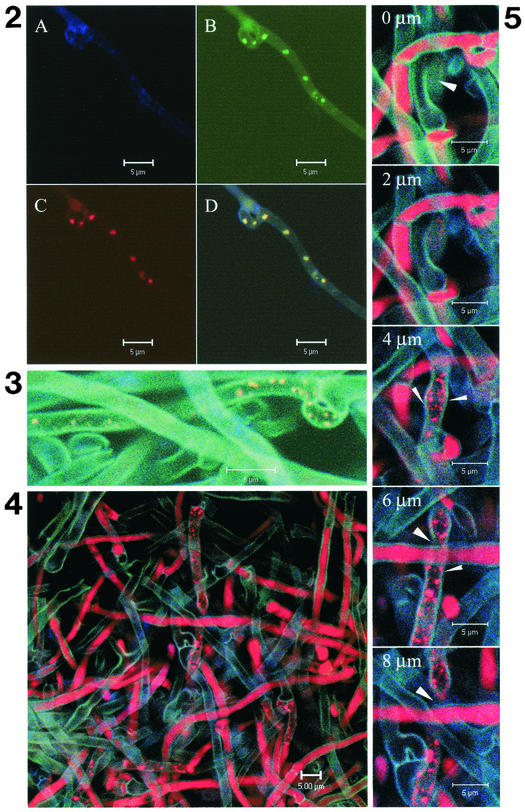
The presence of bacterial rRNA in the mycelium of L. bicolor S238N was first investigated with the probe specific for eubacteria, EUB338mix. Small (≤0.5 μm) spherical bacteria were detected in five collection subcultures of L. bicolor S238N aged 1 to 6 months, grown on solid and liquid P5 and P25 modified media. Some bacteria were found in the terminal cell of some hyphae (data not shown) and in several clamp connections (Fig. 2, Fig. 3, and Fig. 4). Generally speaking, the bacteria were variable in number, up to 50 per fungal cell, and occurred in only a few fungal cells. In sample P5c7 especially, very elongated cells containing many bacteria were observed repeatedly (Fig. 4).
FIG. 2.
FISH of fungal culture P5c7 with group-specific 16S rRNA oligonucleotide probes: (A) no probe, (B) EUB338mix-FITC, specific for the domain bacteria, (C) LGC354mix-Cy3, specific for Firmicutes. Round bacteria measuring less than 0.5 μm wide were detected with the probes EUB338mix-FITC and LGC354mix-Cy3, indicating that the bacteria belong to the Firmicutes. The superimposition of the images (D) shows an exact colocalization of the signals obtained. No autofluorescence of the bacteria was observed without probe (A).
FIG. 3.
FISH of fungal culture P5c7 without probe in Cy5, with the species-specific oligonucleotide probe BIF216-Cy3 (probe specific for the Paenibacillus isolates from the fermentor), and the Firmicutes-specific probe LGC354mix-FITC. The superimposition of the images results in a yellow color for the bacteria, indicating colabeling by LGC354mix-FITC (green) and BIF216-Cy3 (red).
FIG. 4.
FISH of fungal culture P5c7 without probe in Cy5, with nonEUB338-FITC, antisense probe, and with the Firmicutes-specific probe LGC354mix-FITC. After superimposing the images, the bacteria fluoresce red, which corresponds to hybridization of the LGC354mix-Cy3 probe, whereas they would have fluoresced yellow (green plus red) in the case of nonspecific hybridization with nonEUB338-FITC (green).
Among the group-specific probes tested (ALF968, BET 42a, GAM42a, CF319, HGC69a, and LGC354mix), similar structures were observed with the LGC354mix probe only, which is specific for Firmicutes, in colocalization with EUB338mix (Fig. 2). The bacteria detected with LGC354mix also hybridized with BIF216, which is specific for Paenibacillus spp. (Firmicutes) isolates from the L. bicolor S238N culture in a fermentor (Fig. 3). In contrast, no bacterial signal was detected with probe nonEUB338; in Fig. 4, the spherical bacteria were labeled with the LGC354mix-Cy3 probe alone. Optical sectioning through the hyphae clearly showed that the bacterial cells were intracellular (Fig. 5).
FIG. 5.
Selection of a gallery of CLSM pictures corresponding to FISH of fungal culture P5c7. Optical sections were realized every micrometer (one picture presented every 2 μm) in a sample hybridized with nonEUB338-FITC (control for the specificity of the hybridization) and LGC354mix-Cy3 (Firmicutes). Bacteria are expected to appear as red dots. The fungal cell wall fluoresces turquoise nonspecifically, delimiting the fungal cell compartment. Thick arrowheads indicate plain fungal cell wall when the optical section is above or beneath the hypha, and thin arrowheads indicate fungal cell wall flanking the fungal cytoplasm when the optical section occurs inside the hypha. Bacteria appear inside the fungal cell concomitantly with their appearance in the cytoplasm (sections 2 to 8 μm), between the two turquoise lines corresponding to the fungal cell wall.
DISCUSSION
Fluorescence in situ hybridization combined with CLSM allowed the detection of rare, small intracellular spherical objects (≤0.5 μm) in several collection subcultures of L. bicolor S238N. These objects hybridized exclusively with three probes, EUB338mix, LGC354mix, and BIF216, specific for eubacteria, low GC-content gram-positive bacteria, and for Paenibacillus isolates from the L. bicolor S238N culture produced in a fermentor, respectively. The specificity of the hybridizations performed here is attested to by the following observations: the coherent labeling of the bacterial structures obtained in the different colocalization experiments with these probes, the nonhybridization of probes specific for other phyla, and the nonhybridization of an antisense probe. This demonstrates the bacterial nature of the intrafungal structures. Moreover, with FISH, only bacteria containing enough ribosomes produce detectable fluorescence; therefore, dead bacteria, lacking rRNA, and starved or dormant bacteria, usually with very low rRNA levels, cannot be detected (3, 11). In agreement with this correlation between rRNA content and cellular activity, the probe-conferred fluorescence levels of the intracellular bacteria detected in this study were high enough to indicate physiologically active bacteria. In contrast, this study provides no information concerning the viability of the colonized fungal cells; this will have to be assessed in order to confirm that the intracellular bacteria are endosymbionts, not saprobes.
We demonstrated that these intrafungal bacteria are paenibacilli and that they are at the least very closely related to the L. bicolor S238N fermentor culture isolates. Indeed, BIF216, the 16S rRNA-directed probe designed for these bacterial isolates, also hybridized on the intracellular bacteria of the L. bicolor S238N collection subcultures. Interestingly, according to the 16S rDNA sequence similarities of these Paenibacillus isolates, together with Paenibacillus sp. strain SAFN-007, a strain isolated from spacecraft assemblies, they may form a new species.
With all the results taken together, the question which arises is whether the intrafungal paenibacilli and the paenibacilli isolated from the L. bicolor S238N fermentor culture are very closely related bacteria or a single bacterial strain. The use of the 16S rRNA-directed probe BIF216 was not sufficient to conclude this; only observation of exocytosis of the bacteria would do so. For the pure fungal cultures, this has not been witnessed yet; so far, all the bacteria that we observed were inside intact fungal cells, as confirmed by optical sectioning by CLSM, and no bacteria were found in the buffer in which the mycelium was suspended. In contrast, concerning the fermentor culture, the fragmenting of the fungus before inoculation could have allowed some bacteria to escape and to develop extracellularly. Thus, the Paenibacillus isolates F2001-L and F2001-O that we purified from this fermentor culture could have originated from the intracellular compartment of the fungus. However, this hypothesis needs confirmation.
In this study, a prominent trait of the intrafungal paenibacilli was their sporadic occurrence. Indeed, only some subcultures of L. bicolor S238N examined here presented intracellular bacteria, and the majority of the hyphae in the samples observed were free of bacteria. This questions the age of this fungal-bacterial association. The fact that intracellular bacteria were detected in fungal samples subcultured from our 4°C L. bicolor S238N collection, which has been maintained axenically since isolation, suggests that this fungal-bacterial association is at least 25 years old. The presence of intracellular bacteria now has to be investigated in other axenic strains of L. bicolor S238 from our collection, all related to S238N (13), in order to determine the generality and the stability of this association. For this purpose, environmental samples of L. bicolor also need to be examined. Indeed, paenibacilli are rather common soil (4), rhizosphere (36), and mycosphere inhabitants (37), and some of them are mycorrhizal helper bacteria (29). Therefore, a possible origin for the intracellular bacteria of L. bicolor S238N would be the mycosphere of this fungus. Interestingly, other Paenibacillus species (P. validus and P. burgondia) were also found associated with pure cultures of the endomycorrhizal fungus Glomus spp., to which they are beneficial (10, 15), but they are clearly different phylogenetically from the L. bicolor S238N fermentor isolates F2001-L and F2001-O.
In conclusion, this work has demonstrated the sporadic presence of cryptic intracellular bacteria in pure subcultures of an ectomycorrhizal fungus, L. bicolor S238N. These bacteria were identified as Paenibacillus spp. by FISH analysis with a specific 16S rRNA-directed oligonucleotide probe and were clearly localized inside the hyphae by CLSM. Further experiments will have to investigate the stability and the generality of this fungus-bacterium association. In any case, the discovery of intracellular bacteria inside a pure fungal culture which was conserved axenically for 25 years is important. Moreover, this fungal strain, which is being commercially used for controlled mycorrhization, the role of these intracellular bacteria, and their possible impact on the ectomycorrhizal symbiosis have to be studied thoroughly.
Acknowledgments
We acknowledge F. Martin (Institut National de la Recherche Agronomique, Nancy, France) for critical review of the manuscript, S. Kulakauskas and J. Tremblay (Institut National de la Recherche Agronomique, Jouy-en-Josas, France) for technical advice, and P. Hutzler (GSF, Neuherberg, Germany) for helpful advice on CLSM. We are also grateful to G. Bending (Horticulture International, Wellesbourne, United Kingdom) for providing the bacterial strain EJP73 and to P. Bertaux (IUFM, Lorraine, France) for proofreading the manuscript.
The collaboration between the French and German laboratories was supported by Cost 830, which provided short-term mission funding to J. Bertaux.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., L. Krumboltz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Axelrood, P. E., M. L. Chow, C. S. Arnold, K. Lu, J. M. McDermott, and J. Davies. 2002. Cultivation-dependant characterization of bacterial diversity from British Columbia forest soils subjected to disturbance. Can. J. Microbiol. 48:643-654. [DOI] [PubMed] [Google Scholar]
- 5.Barbieri, E., L. Potenza, I. Rossi, D. Sisti, G. Giomaro, S. Rossetti, C. Beinfohr, and V. Stocchi. 2000. Phylogenetic characterisation and in situ detection of a Cytophaga-Flexibacter-Bacteroides phylogroup bacterium in Tuber borchii Vittad. ectomycorrhizal mycelium. Appl. Environ. Microbiol. 66:5035-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbieri, E., J. Bertaux, P. Frey-Klett, J. Garbaye, A. Hartmann, M. Schmid, and V. Stocchi. 2002. New evidence for in situ detection of a Cytophaga-Flexibacter-Bacteroides bacterium associated to the ectomycorrhizal Tuber borchii Vittad. mycelium. Sixth European Conference on Fungal Genetics, Pisa, Italy.
- 7.Baumann, P., L. Baumann, C.-Y. Lai, D. Rouhbakhsh, N. A. Moran, and M. A. Clark. 1995. Genetics, physiology, and evolutionnary relationships of the genus Buchnera: intracellular symbiont of aphids. Annu. Rev. Microbiol. 62:55-94. [DOI] [PubMed] [Google Scholar]
- 8.Bianciotto, V., C. Bandi, D. Minerdi, M. Sironi, H. V. Tichy, and P. Bonfante. 1996. An obligately endosymbiotic mycorrhizal fungus itself harbors obligately intracellular bacteria. Appl. Environ. Microbiol. 62:3005-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianciotto, V., E. Lumini, L. Lanfranco, D. Minerdi, P. Bonfante, and S. Perotto. 2000. Detection and identification of bacterial endosymbionts in arbuscular mycorrhizal fungi belonging to the family Gigasporaceae. Appl. Environ. Microbiol. 66:4503-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Budi, S. W., D. van Tuinen, G. Martinotti, and S. Gianinazzi. 1999. Isolation from the Sorghum bicolor mycorrhizosphere of a bacterium compatible with arbuscular mycorrhiza development and antagonistic towards soil-borne fungal pathogens. Appl. Environ. Microbiol. 65:5148-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen, H., M. Hansen, and J. Sørensen. 1999. Counting and size classification of active soil bacteria by fluorescence in situ hybridization with an rRNA oligonucleotide probe. Appl. Environ. Microbiol. 65:1753-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daims, H., A. Brühl, R. Amann, K.-H. Schleifer, and M. Wagner. 1999. Probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:438-448. [DOI] [PubMed] [Google Scholar]
- 13.Di Battista, C., M.-A. Selosse, D. Bouchard, E. Stenström, and F. Le Tacon. 1996. Variations in symbiotic efficiency, phenotypic characters and ploidy level among different isolates of the ectomycorrhizal basidiomycete Laccaria bicolor strain S238. Mycol. Res. 100:1315-1324. [Google Scholar]
- 14.Distel, D. L., E. F. DeLong, and J. B. Waterbury. 1991. Phylogenetic characterization and in situ localization of the bacterial symbiont of shipworms (Teredinidae: Bivalvia) by with 16S rRNA sequence analysis and oligodeoxynucleotide probe hybridization. Appl. Environ. Microbiol. 57:2376-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hildebrandt, U., K. Janetta, and H. Bothe. 2002. Towards growth of arbuscular mycorrhizal fungi independently of a host plant. Appl. Environ. Microbiol. 68:1919-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kempf, V. A., K. Trebesius, and I. B. Autenrieth. 2000. Fluorescent in situ hybridization allows rapid identification of microorganisms in blood cultures. J. Clin. Microbiol. 38: 830-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 133. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., London, England.
- 18.Lathe, R. 1985. Synthetic oligonucleotide probes deduced from amino acid sequence data. Theoretical and practical considerations. J. Mol. Biol. 183:1-12. [DOI] [PubMed] [Google Scholar]
- 19.Le Tacon, F., D. Mousain, J. Garbaye, D. Bouchard, J.-L. Churin, C. Argillier, J.-M. Amirault, and B. Généré. 1997. Mycorhizes, pépinières et plantations forestières en France. Rev. For. Fr. No. Spéc. 1997:131-154.10319519 [Google Scholar]
- 20.MacDonald, R. M., and M. R. Chandler. 1981. Bacterium-like organelles in the vesicular-arbuscular mycorrhizal fungus Glomus caledonius. New Phytol. 89:241-246. [Google Scholar]
- 21.MacDonald, R. M., M. R. Chandler, and B. Mosse. 1981. The occurrence of bacterium-like organelles in vesicular-arbuscular mycorrhizal fungi. New Phytol. 90:659-663. [Google Scholar]
- 22.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 23.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K.-H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 24.Meier, H., R. Amann, W. Ludwig, and K.-H. Schleifer. 1999. Specific oligonucleotide probes for in situ detection of a major group of gram-positive bacteria with low DNA G+C content. Syst. Appl. Microbiol. 22:186-196. [DOI] [PubMed] [Google Scholar]
- 25.Moran, N., and J. Wernegreen. 2000. Lifestyle evolution in symbiotic bacteria: insights from genomics. Trends Ecol. Evol. 15:321-326. [DOI] [PubMed] [Google Scholar]
- 26.Neef, A., A. Zaglauer, H. Meier, R. Amann, H. Lemmer, and K. H. Schleifer. 1996. Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl. Environ. Microbiol. 62:4329-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neef, A. 1997. Anwendung der in situ Einzelzell-Identifizierung von Bakterien zur Populationsanalyse in komplexen mikrobiellen Biozönosen. Ph.D. thesis. Technische Universität, Munich, Germany.
- 28.Pachlewski, R., and J. Pachlewska. 1974. Studies on symbiotic properties of mycorrhizal fungi of pine (Pinus sylvestris) with the aid of the method of mycorrhizal synthesis in pure culture on agar. Forest Research Institute, Warsaw, Poland.
- 29.Poole, E. J., G. D. Bending, J. M. Whipps, and D. J. Read. 2001. Bacteria associated with Pinus sylvestris-Lactarius rufus ectomycorrhizas and their effects on mycorrhiza formation in vitro. New Phytol. 151:743-751. [DOI] [PubMed] [Google Scholar]
- 30.Rai, A. N., E. Söderback, and B. Bergman. 2000. Cyanobacterium-plant symbioses. New Phytol. 147:449-481. [DOI] [PubMed] [Google Scholar]
- 31.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K.-H. Schleifer. 1994. In situ probing of Gram-positive bacteria with high DNA G+C content with 23S rRNA-targeted oligonucleotides. Microbiology 140:2849-2858. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Scannerini, S., and P. Bonfante-Fasolo. 1991. Bacteria and bacteria-like objects in endomycorrhizal fungi (Glomaceae), p. 273-287. In L. Margulis and R. Fester (ed.), Symbiosis as a source of evolutionary innovation. MIT Press, Cambridge, Mass.
- 34.Schüßler, A., and M. Kluge. 2000. Geosiphon pyriforme, an endocytosymbiosis between fungus and cyanobacteria, and its meaning as a model system for AM research, p. 151-161. In B. Hock (ed.), The mycota IX. Springer Verlag, Berlin, Germany.
- 35.Schüβler, A., D. Schwarzott, and C. Walker. 2001. A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol. Res. 105:1413-1421. [Google Scholar]
- 36.Seldin, L., A. S. Rosado, D. W. Da Cruz, A. Nobrega, J. D. Van Elsas, and E. Paiva. 1998. Comparison of Paenibacillus azotofixans strains isolated from rhizoplane, rhizosphere, and non-root-associated soil from maize planted in two different Brazilian soils. Appl. Environ. Microbiol. 64:3860-3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timonen, S., K. S. Jørgensen, K. Haahtela, and R. Sen. 1998. Bacterial community structure at defined locations of Pinus sylvestris-Suillus bovinus and Pinus sylvestris-Paxillus involutus mycorrhizospheres in dry pine forest humus and nursery peat. Can. J. Microbiol. 44:499-513. [Google Scholar]
- 38.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]