Abstract
The bacterial diversity of a hot spring in Bakreshwar, India, was investigated by a culture-independent approach. 16S ribosomal DNA clones derived from the sediment samples were found to be associated with gamma-Proteobacteria, cyanobacteria, and green nonsulfur and low-GC gram-positive bacteria. The first of the above phylotypes cobranches with Shewanella, a well-known iron reducer. This phylogenetic correlation has been exploited to develop culture conditions for thermophilic iron-reducing microorganisms.
Microbial metal reduction has become a subject of intensive investigation as a result of its overwhelming environmental significance (4). This has led to the isolation and identification of a number of metal-reducing bacteria. Bacterial species identified as dissimilatory metal reducers include facultative anaerobes such as Shewanella oneidensis and Shewanella alga as well as strict anaerobes like Geobacter metallireducens and Desulfovibrio sp. (18). Shewanella, a gram-negative bacterium, can use a wide range of electron acceptors, including fumarate, trimethylamine N-oxide, dimethyl sulfoxide, nitrate, nitrite, thiosulfate, and sulfite, as well as insoluble acceptors, such as metal oxides or oxyhydroxides (14, 16). It is widely distributed in freshwater and marine environments (18, 19, 21, 23). A psychrophilic and moderately barophilic strain of Shewanella violacea has been discovered in deep-sea sediments (20). However, to date no thermophilic strain related to Shewanella has been reported.
A number of thermophiles and hyperthermophiles have been isolated from samples of hot sediments, mud, rocks, soils, and waters. Hot environments have been searched also for metal reducers. Certain hyperthermophiles such as Thermotoga maritima are known to grow as respiratory organisms when Fe(III) is provided as an electron acceptor (27). Furthermore, Pyrobaculum islandicum has been shown to reduce U(VI), Tc(VII), Cr(VI), Co(III), and Mn(IV) at 100°C (10).
Several hot springs in different regions of the Indian subcontinent have been known to geologists for many years (6, 9, 12). However, their microbial diversity has not been explored by molecular phylogenetic approaches. In the present study, we have applied the 16S rRNA methodology (2) to determine the bacterial community structure of Agnikunda, a hot spring in Bakreshwar, situated in the state of West Bengal in India. The investigation has revealed the presence, among a few other bacteria, of novel nonmarine, thermophilic relatives of Shewanella (formerly Alteromonas), a well-known iron reducer. Subsequently, iron-reducing enrichment culture to cultivate such organisms has been developed.
Hot spring sediment DNA.
Samples were collected from the hot spring Agnikunda at Bakreshwar, in the Birbhum district of West Bengal in India. The surface temperature of the sediment varied between 66 and 69°C. The pH of the water was measured as 9.1 to 9.3. The sediment contained 1.1 to 1.5% organic carbon and, interestingly, 280 to 422 μmol of reducible Fe(III) and 280 to 600 μmol of reduced iron per g of wet sediment. Initially, DNA extracted from the hot spring sediments by a direct lysis procedure (3, 29) could not be amplified in a PCR with Taq DNA polymerase, possibly due to the presence of high humic acid contamination in the samples (30). Successive washes with buffers differing in EDTA concentration prior to lysis (28) enabled the purification of DNA having A260/A280 between 1.67 and 1.79. Such DNA could be amplified successfully. The EDTA wash procedure, however, yielded only 1 to 3 μg of DNA from 4 g of wet sediment.
Analysis of 16S rRNA genes.
In order to analyze the bacterial diversity in Agnikunda sediments, 16S ribosomal DNA (rDNA) libraries were constructed. Fifty nanograms of the total community DNA was amplified with bacterium-specific forward primer 5′-AGA GTT TGA ACA TGG CTG-3′ (S-D-Bact-0027-a-S-18) and reverse primer 5′-CTA GCG ATT CCG ACT TCA-3′ (S-D-Bact-1327-a-A-18) (1). The numbers refer to the positions in the Escherichia coli 16S rRNA (5). Reaction mixtures were incubated in a thermal cycler (GeneAmp 2400 PCR system; PE Applied Biosystems, Norwalk, Conn.) for an initial denaturation at 94°C for 2 min followed by 40 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 2 min. Amplified DNAs were purified by the spin column method (Wizard PCR Prep DNA purification system; Promega Corp., Madison, Wis.). The purified DNAs were cloned directly by the TA cloning method (13) with a pGEM-T Easy Vector System II kit (Promega). Twenty-five positive clones were sequenced using either vector-specific primers or the same PCR primers. Homology search with the BLAST system showed that the hot spring sediment clones could be classified into four major phylotypes. Ten of the clones corresponded to the gamma subdivision of the Proteobacteria, eight were affiliated with cyanobacteria, three belonged to the green nonsulfur group, and four were low-GC gram-positive bacteria (Table 1). All the sequences were checked for the presence of chimeric sequences by using the CHECK_CHIMERA program (available at http://rdp.cme.msu.edu). No such chimeras were, however, detected.
TABLE 1.
Summary of the bacterial 16S rDNA clone sequences identified in Agnikunda (AKB) sediment
| Affiliation | 16S rDNA clone type (GenBank accession no.) | No. of clones | % of clones in the library | Closest relative according to BLAST search (accession no.) | % of identity with the closest relative |
|---|---|---|---|---|---|
| Cyanobacteria | AKB01 (AF350246) | 4 | 32 | Synechococcus elongates (D83715) | 94 |
| AKB02 (AF345900) | 1 | Synechococcus elongates (D83715) | 96 | ||
| AKB04 (AY028969) | 2 | Synechococcus elongates (D83715) | 93 | ||
| AKB14 (AF490373) | 1 | Synechococcus elongates (D83715) | 94 | ||
| Proteobacteria (gamma subdivision) | AKB03 (AY028968) | 3 | 40 | Shewanella alga BrY (X81621) | 88 |
| AKB06 (AF465626) | 1 | Shewanella alga (U91544) | 87 | ||
| AKB07 (AF465627) | 1 | Shewanella alga BrY (X81621) | 87 | ||
| AKB08 (AF465827) | 1 | Shewanella alga BrY (X81621) | 88 | ||
| AKB09 (AF465828) | 1 | Shewanella alga BrY (X81621) | 86 | ||
| AKB10 (AF490369) | 1 | Shewanella alga (U91545) | 86 | ||
| AKB11 (AF490370) | 1 | Aeromonas sp. (AF099027) | 91 | ||
| AKB13 (AF490372) | 1 | Shewanella alga (U91544) | 95 | ||
| Green nonsulfur bacteria (Thermus group) | AKB05 (AY028970) | 3 | 12 | Thermus thermophilus (AJ251940) | 88 |
| Low-GC gram-positive bacteria | AKB12 (AF490371) | 4 | 16 | Desulfotomaculum luciae (AF069293) | 93 |
Phylogenetic analysis.
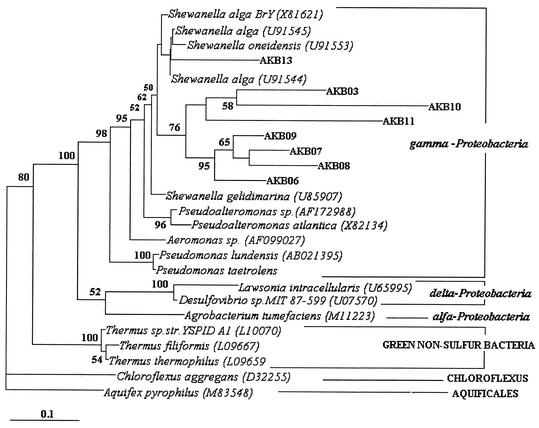
The gamma-proteobacterial sequences retrieved from Agnikunda sediment were aligned with sequences in the small-subunit rRNA database of the RDP server by using the Clustal W 1.6 program (26). Phylogenetic analysis was restricted to nucleotide positions that could be unambiguously aligned in all the sequences. A phylogenetic tree was constructed using a maximum likelihood algorithm (7) with 100-bootstrap resampling (8) (Fig. 1). Among the 10 proteobacterial clones there were eight unique sequences. Three of the 16S rRNA sequences are represented by AKB03. In the tree, one clone (AKB13) clusters within the Shewanella genus. This clone shows 95% nucleotide identity to S. alga (U91544) (Table 1). The result is consistent with the phylogenetic position of this sequence assigned by the RDP at http://rdp.cme.msu.edu/cgis/hierarchy_preview.cgi. The seven other clones (AKB03, AKB06, AKB07, AKB08, AKB09, AKB10, and AKB11) group separately and form a cobranch with species in the Shewanella cluster originating from a common node with a bootstrap value of 50 (Fig. 1). The sequence identity of these clones with different Shewanella species varied between 86 and 91% (Table 1). It may be noted here that the RDP has placed at least AKB11 in the same family as Shewanella, Alteromona-daceae (http://rdp.cme.msu.edu/cgis/hierarchy_preview.cgi).The results obtained with the maximum likelihood algorithm were also verified by using the neighbor joining DNA distance (22) and maximum parsimony (25) treeing methods (data not shown).
FIG. 1.
Phylogenetic tree deduced from the gamma-proteobacterial 16S rDNA of hot spring sediment clones by maximum likelihood algorithm. Reference sequences were chosen to represent the broadest diversity of Bacteria. Aquifex pyrophilus and Chloroflexus aggregans were used as outgroups for the analysis. Division- and subdivision-level groupings are either bracketed or marked with a horizontal line at the right of the figure. Branch points supported (bootstrap values) are indicated at the branch points of the nodes of the tree. A scale is shown below the tree to indicate the branch length.
Enrichment culture and Fe(III) reduction.
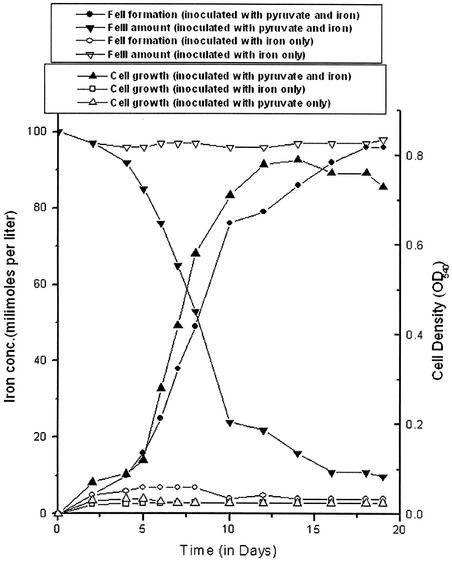
A number of Shewanella strains are known to use for anaerobic respiration a wide variety of electron acceptors including metal oxides and hydroxides (16, 17). In an attempt to cultivate the Shewanella-like iron-reducing microorganisms present in the hot spring sediments, enrichment medium containing (per liter of deionized water) 0.33 g of KH2PO4, 0.33 g of NaCl, 0.33 g of KCl, 0.6 g of NaH2PO4, and 2.5 g of Na2CO3 was prepared. Yeast extract (0.01%, wt/vol) was added as a vitamin supplement, and 10 mM pyruvate was added as an electron donor. The pH was adjusted to 7.0 at 25°C with 10% (wt/vol) NaOH. Amorphous Fe(III) oxyhydroxide, used as the electron acceptor at ca. 90 mmol of Fe(III) per liter, was synthesized by titrating a solution of FeCl3 with 10% (wt/vol) NaOH to pH 9.0 (24). Cultures were grown in 10 ml of medium in Hungate tubes under an atmosphere of CO2 (100%) (24) at 66°C. Such conditions favored growth as well as Fe(III) reduction with time (Fig. 2). The growth continued until the 14th day. There was a concomitant increase in the concentration of reduced iron in the medium. The maximum Fe(III) reduction was found to be 96% in 19-day-old cultures. No growth could be observed when either Fe(III) or pyruvate was omitted (Fig. 2) or pyruvate was replaced with citrate or acetate (data not shown).
FIG. 2.
Growth of Shewanella-like consortium at 66°C and Fe(III) reduction in a medium with pyruvate as the electron donor and poorly crystalline Fe(III) oxide as the electron acceptor. The medium also contained 0.01% yeast extract. The results are the means of triplicate cultures. OD540, optical density at 540 nm.
Twelve 16S rDNA clones were retrieved from the enrichment culture by using the same set of PCR primers as above (27F and 1327R). From these, six unique sequences were revealed. These sequences were aligned with those retrieved directly from the hot spring sediment. Two, four, two, and one enrichment clone completely matched with four proteobacterial clones, AKB03, AKB06, AKB07, and AKB13, respectively. In the cases of the other two sequences (representing three clones) there were some deviations which could be due to statistical error or bias caused by PCR. Our results thus validate the earlier suggestions made by Lovley and his coworkers (27) that isolation of as-yet-uncultured thermophiles or hyperthermophiles with medium containing Fe(III) as the electron acceptor could be a productive strategy for culturing these organisms. The relative abundance of gamma-Proteobacteria in both sediment and enrichment culture samples was determined by hybridization to a fluorescently labeled oligonucleotide probe specific for 23S rRNA of the subdivision. Approximately 30% of the microorganisms in the sediment were gamma-Proteobacteria, whereas in the enrichment culture the proportion was nearly 90% (data not shown).
Concluding remarks.
The discovery of Shewanella-like thermophilic bacteria is interesting from the standpoints of both the understanding of molecular genetics of metal reduction at higher temperatures and its biotechnological applications (11). Although a number of dissimilatory metal reducers, mesophilic and thermophilic, are already known, the molecular genetics of metal reduction by different Shewanella strains has been extensively studied. Cell fractionation studies of S. oneidensis MR-1 demonstrated the presence of ferric reductase activity in the outer membrane as well as the inner membrane of anaerobically grown cells (15). mtrB, a gene that encodes an outer membrane involved in Fe(III) and Mn(IV) reduction, has been isolated and sequenced (4). Myers and Myers (17) have further discovered in S. oneidensis MR-1 outer membrane cytochrome genes omcA and omcB mutations which decrease the cells' ability to reduce Mn(IV) and not Fe(III). Identification and a comparative analysis of similar genes in the thermophilic counterpart may provide a molecular insight into the mechanism of metal reduction at high temperatures. Work is in progress to isolate and characterize pure cultures of such microorganisms.
Acknowledgments
We express our deep gratitude to Binayak Dutta-Roy, who has been the main inspiration behind this work. We also thank Diti Chatterjee and Brajadulal Chattopadhyay of Jadavpur University and Ranjan Datta of the Central Forensic Science Laboratory for their technical help and cooperation.
This work was supported by a grant from the BRNS, Department of Atomic Energy, Government of India (sanction no. 97/37/31-BRNS/996), and a grant from the Department of Science and Technology, Government of India (SR/FIST/LS II-093/2000).
REFERENCES
- 1.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beliaev, A. S., and D. A. Saffarini. 1998. Shewanella putrefaciens mtrB encodes an outer membrane protein required for Fe(III) and Mn(IV) reduction. J. Bacteriol. 180:6292-6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosius, J., J. L. Palmer, J. P. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee, G. C., and S. K. Guha. 1968. The problems of origin of high temperature springs of India, p. 141-149. In Proceedings of the 23rd International Geological Congress, vol. 17.
- 7.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 9.Gupta, M. L., H. Narain, and V. K. Saxena. 1975. Geochemistry of thermal waters from various geothermal provinces of India, p. 47-58. In Proceedings of the Grenoble Symposium of the International Association of Hydrological Sciences. IAHS Press, Institute of Hydrology, Wallingford, Oxfordshire, United Kingdom.
- 10.Kashefi, K., and D. R. Lovley. 2000. Reduction of Fe(III), Mn(IV), and toxic metals at 100°C by Pyrobaculum islandicum. Appl. Environ. Microbiol. 66:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lovley, D. R., and J. D. Coates. 2000. Novel forms of anaerobic respiration of environmental relevance. Curr. Opin. Microbiol. 3:252-256. [DOI] [PubMed] [Google Scholar]
- 12.Majumdar, R. K., N. Majumdar, and A. L. Mukherjee. 2000. Geoelectric investigations in Bakreswar geothermal area, West Bengal, India. J. Appl. Geophys. 45:187-202. [Google Scholar]
- 13.Marchuk, D., M. Drumm, A. Saulino, and F. C. Collins. 1991. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 19:1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moser, D., and K. Nealson. 1996. Growth of the facultative anaerobe Shewanella putrefaciens by elemental sulfur reduction. Appl. Environ. Microbiol. 62:2100-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myers, C., and J. Myers. 1993. Ferric reductase is associated with the membranes of anaerobically grown Shewanella putrefaciens MR-1. FEMS Microbiol. Lett. 108:15-22. [Google Scholar]
- 16.Myers, C., and K. Nealson. 1988. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240:1319-1321. [DOI] [PubMed] [Google Scholar]
- 17.Myers, J. M., and C. R. Myers. 2001. Role of outer membrane cytochromes OmcA and OmcB of Shewanella putrefaciens MR-1 in the reduction of manganese dioxide. Appl. Environ. Microbiol. 67:260-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nealson, K. H., and D. Saffarini. 1994. Iron and manganese in anaerobic respiration: environmental significance, physiology, and regulation. Annu. Rev. Microbiol. 48:311-343. [DOI] [PubMed] [Google Scholar]
- 19.Nealson, K. H., C. R. Myers, and B. Wimpee. 1991. Isolation and identification of manganese reducing bacteria, and estimates of microbial manganese reducing potential in the Black Sea. Deep Sea Res. 38:S907-S920. [Google Scholar]
- 20.Nogi, Y., C. Kato, and K. Horikoshi. 1998. Taxonomic studies of deep-sea barophilic Shewanella strains and description of Shewanella violacea sp. nov. Arch. Microbiol. 170:331-338. [DOI] [PubMed] [Google Scholar]
- 21.Obuekwe, C., D. Westlake, J. Plambeck, and F. Cook. 1981. Corrosion of mild steel in cultures of ferric iron reducing bacteria isolated from crude oil. II. Mechanism of anodic depolarization. Corrosion (Houston) 37:632-637. [Google Scholar]
- 22.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 23.Semple, K., and D. Westlake. 1987. Characterization of iron-reducing Alteromonas putrefaciens strains from oil field fluids. Can. J. Microbiol. 33:366-371. [Google Scholar]
- 24.Slobodkin, A. I., T. P. Tourova, B. B. Kuznetsov, N. A. Kostrikina, N. A. Chernyh, and E. A. Bonch-Osmolovskaya. 1999. Thermoanaerobacter siderophilus sp. nov., a novel dissimilatory Fe(III)-reducing, anaerobic, thermophilic bacterium. Int. J. Syst. Bacteriol. 49:1471-1478. [DOI] [PubMed] [Google Scholar]
- 25.Swofford, D. L., G. J. Olsen, P. J. Waddell, and D. M. Hillis. 1996. Phylogenetic inference, p. 407-514. In D. M. Hillis, C. Moritz, and B. K. Mable (ed.), Molecular systematics. Sinauer Associates, Inc., Sunderland, Mass.
- 26.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vargas, M., K. Kashefi, E. L. Blunt-Harris, and D. R. Lovley. 1998. Microbiological evidence for Fe(III) reduction on early Earth. Nature 395:65-67. [DOI] [PubMed] [Google Scholar]
- 28.Watson, R. J., and B. Blackwell. 2000. Purification and characterization of a common soil component which inhibits the polymerase chain reaction. Can. J. Microbiol. 46:633-642. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto, H., A. Hiraishi, K. Kato, H. X. Chiura, Y. Maki, and A. Shimizu. 1998. Phylogenetic evidence for the existence of novel thermophilic bacteria in hot spring sulfur-turf microbial mats in Japan. Appl. Environ. Microbiol. 64:1680-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou, J., M. A. Bruns, and J. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]