Abstract
The crtYB locus was used as an integrative platform for the construction of specific carotenoid biosynthetic mutants in the astaxanthin-producing yeast Xanthophyllomyces dendrorhous. The crtYB gene of X. dendrorhous, encoding a chimeric carotenoid biosynthetic enzyme, could be inactivated by both single and double crossover events, resulting in non-carotenoid-producing transformants. In addition, the crtYB gene, linked to either its homologous or a glyceraldehyde-3-phosphate dehydrogenase promoter, was overexpressed in the wild type and a β-carotene-accumulating mutant of X. dendrorhous. In several transformants containing multiple copies of the crtYB gene, the total carotenoid content was higher than in the control strain. This increase was mainly due to an increase of the β-carotene and echinone content, whereas the total content of astaxanthin was unaffected or even lower. Overexpression of the phytoene synthase-encoding gene (crtI) had a large impact on the ratio between mono- and bicyclic carotenoids. Furthermore, we showed that in metabolic engineered X. dendrorhous strains, the competition between the enzymes phytoene desaturase and lycopene cyclase for lycopene governs the metabolic flux either via β-carotene to astaxanthin or via 3,4-didehydrolycopene to 3-hydroxy-3′-4′-didehydro-β-ψ-caroten-4-one (HDCO). The monocylic carotenoid torulene and HDCO, normally produced as minority carotenoids, were the main carotenoids produced in these strains.
During the last decades fast progress has been made within the field of molecular biology of carotenoid biosynthesis in bacteria, fungi, and plants (reviewed in references 6 and 23). Although more than 600 different carotenoids have been identified in nature, only a few are used industrially. The acyclic carotenoid lycopene, the bicyclic carotenoid β-carotene, and the oxygenated bicyclic carotenoids (xanthophylls) canthaxanthin and astaxanthin are used as food colorants, animal feed additives, and in pharmaceuticals and cosmetics (15). The potential commercial interest for the production of carotenoids and the cloning of genes encoding biosynthetic enzymes has led to all kinds of examples of metabolic pathway engineering. These examples include the overexpression of a gene encoding a rate-limiting enzyme (14, 17), the expression of carotenogenic genes in noncarotenogenic heterologous hosts (12, 18, 20, 32), the increase of the carbon flux into the carotenoid biosynthetic pathway (1, 12, 17, 32), and the combination of genes and modification of catalytic activities in order to improve and/or modify carotenoid biosynthetic pathways (18, 24-26, 32).
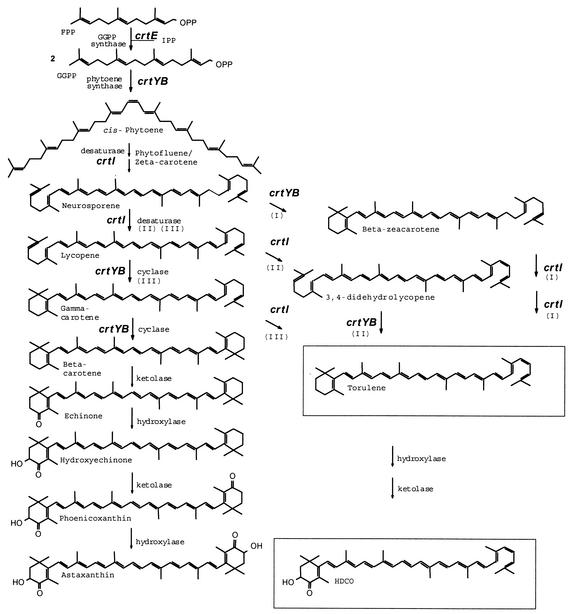
So far, the green microalga Haematococcus pluvialis and the heterobasidiomycetous yeast Xanthophyllomyces dendrorhous, the perfect state of Phaffia rhodozyma, are the only microbial systems with commercial potentials for the production of astaxanthin. This oxygenated carotenoid is used as a feed additive in aquaculture to obtain the desired degree of pigmentation of flesh from salmon and trout. Furthermore, when astaxanthin was applied as a nutraceutical, several positive actions on degenerative diseases have been reported (8, 19, 33). The pathway for astaxanthin biosynthesis, as proposed by Andrewes and coworkers (5) is shown in Fig. 1. Several genes involved in the astaxanthin biosynthetic pathway of X. dendrorhous have been cloned and characterized recently (28-31; T. Hoshino, K. Ojima, and Y. Setoguchi, September 2000, Astaxanthin synthetase, European patent application EP 1 035 206 A1; J. C. Verdoes, J. Wery, and A. J. J. van Ooyen, July 1997, Improved methods for transforming Phaffia strains, transformed Phaffia strains so obtained, and recombinant DNA in said methods, International patent application WO 97/23633) and a transformation system has been developed (35, 36).
FIG. 1.
The astaxanthin biosynthetic pathway in X. dendrorhous proposed by Andrewes et al. (5). The main carotenoids found after the introduction of additional gene copies of the phytoene desaturase-encoding gene (crtI) in the X. dendrorhous strains CBS 6938 and PR-1-104 (12) are boxed (this study). Roman numbers (I, II, and III) indicate three potential routes for the formation of torulene from neurosporene.
In this paper, we describe the engineering of the astaxanthin biosynthetic pathway of X. dendrorhous by two different approaches. By specific gene inactivation, the accumulation of intermediates is demonstrated. Furthermore, overexpression of carotenogenic genes led to altered carotenoid production levels and carotenoid compositions.
MATERIALS AND METHODS
Molecular techniques and gene cloning.
Standard methods were used, unless otherwise indicated, according to Sambrook et al. (22). DNA was treated with restriction enzymes and other nucleic acid-modifying enzymes according to the specifications of the manufacturers. Plasmid DNA from Escherichia coli was isolated by using Qiagen columns (Westburg BV, Leusden, The Netherlands). DNA fragments were purified by using a QIAEX II gel extraction kit. Determination of nucleotide sequences was performed with a Taq DYE primer cycle sequencing kit (Applied Biosystems, Nieuwerkerk aan de IJssel, The Netherlands). The digoxigenin (DIG) nonradioactive labeling and detection kit from Roche Diagnostics (Mannheim, Germany) was used in Southern blot analysis. DNA probes were DIG labeled with the DIG PCR labeling kit (Roche Diagnostics). Chromosomal DNA was isolated from sodium dodecyl sulfate-lysed protoplasts of X. dendrorhous as described previously (34).
PCR conditions and primers.
The PCRs were carried out in an automated thermal cycler (Perkin-Elmer Nederland, Nieuwerkerk aan de IJssel, The Netherlands) by using SUPER Taq (HT Biotechnology Ltd., Cambridge, England) under the conditions recommended by the supplier. The standard PCR cycle profile was 5 min at 94°C; 25 to 30 cycles consisting of 1 min at 94°C, 2 min at 50°C, and 2 to 3 min at 72°C; and a final step consisting of 10 min at 72°C. In the recombinant PCR, 0.1 μg of each fragment was used and the total number cycles in the second PCR was reduced to 20. The following primers were used: 5′ Pgpd, 5′-dCCCGGATCCGCGGCCGCGAATTCCTGGTGGGTGCATGTATGTA-3′; 3′ crtYB-Pgpd, 5′-dTATGCGAGAGCCGTCATGATGGTAAGAGTGTTAG; 5′ Pgpd-crtYB, 5′-dTCTTACCATCATGACGGCTCTCGCATATTACC-3′; 3′ crtYB, 5′-dGAGTCCCATGGTGTGGTTGC-3′; 5′ crtYB-Nter, 5′-dCGCAATGACGGCTCTCGC; 3′ crtYB-Cter, 5′-dTTACTGCCCTTCCCATCCG; 5′ PcrtI, 5′-dCGCGGATCCACTGACGTGCCTCTGCGG-3′; 3′ PcrtI, 5′-dGTTCTTTCCCCATCGAGTATAG-3′; 5′ crtI, 5′-dACTCTTACCATCATGGGAAAAGAACAAGATCAGG-3′; 3′ crtI, 5′-dCGCGGATCCGAAGGCGGTCCATAACAGTCATG-3′. In these primer sequences, the coding regions are indicated by bold letters and the start codons are underlined. Restriction sites, introduced to facilitate subcloning, are double underlined.
Plasmids, strains, and cultivation conditions.
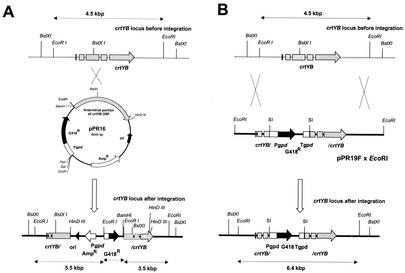
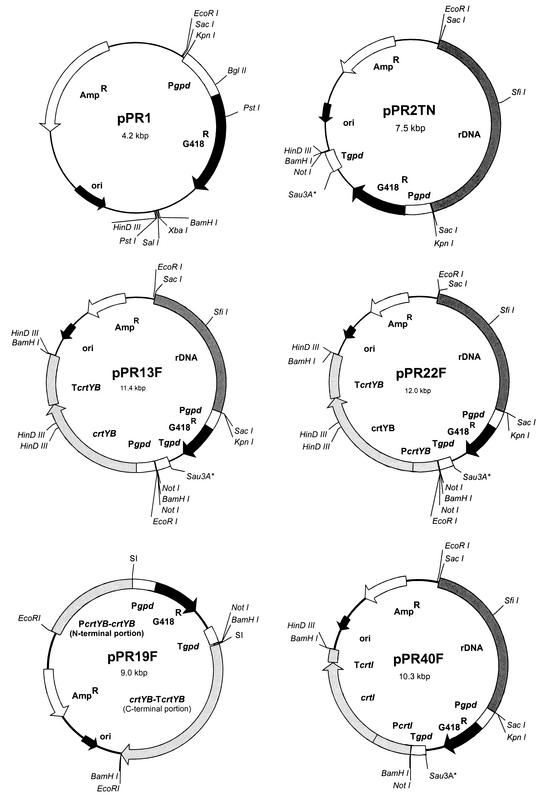
All plasmids and strains used in this study are summarized in Table 1. The E. coli strain XL1-Blue MRF′ was used in all cloning experiments, and E. coli strain DM1 was used for plasmid propagation of transformation vectors of X. dendrorhous. To construct pPR16 (Fig. 2), the 1.9-kb BamHI-HindIII fragment derived from pPRcrtYB (31), containing the N-terminal portion of the crtYB open reading frame, was cloned in the corresponding sites of pPR1 (35) (Fig. 3). To construct pPR19F (forward) (Fig. 3) and pPR19R (reverse), a 1.8-kb EcoRI fragment containing the G418 resistance expression cassette (Pgpd G418r Tgpd) was inserted in the BstXI site of pPR10F (31). Prior to this insertion, EcoRI and BstXI fragments were blunted with the Klenow fragment of E. coli DNA polymerase I and bacteriophage T4 DNA polymerase, respectively. By subcloning, two BamHI sites were added to the genomic 4.5-kb EcoRI DNA fragment, containing the crtYB gene with flanking regions. Then this BamHI fragment was cloned into vector pPR2TN, yielding pPR22F or pPR22R, depending on the orientation of the inserted fragment (Fig. 3). To express the crtYB gene under the control of the promoter region of the glyceraldehyde-3-phosphate dehydrogenase-encoding gene (gpd), a recombinant PCR strategy was used. The promoter region of gpd was amplified as a fragment of approximately 425 bp by using the primers 5′ Pgpd and 3′ crtYB-Pgpd and with chromosomal DNA as the template. In addition, the 5′ end of the crtYB gene was synthesized by using the primers 5′ Pgpd-crtYB and 3′ crtYB. Both fragments were purified, and the small overlap between the fragments was used to link Pgpd to the 5′ end of crtYB in a second PCR with the primers 5′ Pgpd and 3′ crtYB. The expected fragment of 0.55 kb was purified from the PCR mixture and restricted with BamHI and NcoI. The BamHI-NcoI fragment in pPR10F, encoding PcrtYB-5′ crtYB, was replaced by the BamHI-NcoI fragment synthesized by PCR, yielding pPR11. The HindIII-BstXI fragment, containing Pgpd-5′ crtYB, and the BstXI-BamHI fragment, containing 3′ crtYB-TcrtYB, were isolated from pPR11 and pPR10R, respectively, and cloned in the BamHI and HindIII sites of pMTL22P. From this plasmid, designated pPR12, the expression cassette (Pgpd crtYB TcrtYB) could be released as a 3.9-kb BamHI fragment. This fragment was cloned in the BamHI site of pPR2TN, yielding pPR13F and pPR13R, depending on the orientation of the inserted fragment (Fig. 3). The crtI cDNA fragment, encoding phytoene desaturase, was fused to the promoter region of the crtI gene (PcrtI) to achieve crtI overexpression. The fusion product of approximately 2.8 kb (PcrtI crtI TcrtI) was isolated from the PCR mixture, digested with BamHI, and cloned in the BamHI site of pPR2TN. Depending on the orientation of the expression cassettes, the plasmids were named pPR40F and pPR40R (Fig. 3). Linear plasmid DNA molecules, which had to be introduced into X. dendrorhous, were purified from the restriction mixture by phenol extraction and concentrated by an ethanol precipitation, and the DNA pellet was dissolved in ultrapure H2O.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Sources or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| XL1-Blue-MRF′ | Δ(mcrA) 183Δ(mcrCB hsdSMR mrr) 173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac[F′ proAB laqqZΔM15 Tn10] | Gibco BRL |
| DM1 | F−dam−13::Tn9 (Cmr) dcm− mcrB HsdR− M−gal1 gal2 Ara− Lac− Thr− Leu− Tonr Tsxrsup0 | Gibco BRL |
| X. dendrorhous (P. rhodozyma) | ||
| CBS 6938 | Wild-type strain (DNA donor strain) | |
| PR-1-104 | β-Carotene-accumulating mutant of CBS 6938 | 13 |
| Plasmids | ||
| pUC19 | General cloning vector for E. coli (Ap) | 22 |
| pMTL22P | General cloning vector for E. coli (Ap) | 31 |
| pPRGDH6 | 5.5-kb EcoRI fragment carrying the gpd gene of X. dendrorhous in pUC19 (Ap) | 28 |
| pPR1 | General transformation vector for X. dendrorhous | 35 |
| pPR2TN | Derivative of pPR1 with terminator sequences of the gpd gene of X. dendrorhous and additional unique cloning sites (Ap, G418) | 31 |
| pPRcrtYB | cDNA encoding the phytoene synthase-lycopene cyclase of X. dendrorhous in pBluescript (Ap) | 31 |
| pPR10F/R | 4.5-kb EcoRI fragment carrying the genomic copy of the crtYB gene in pUC19 (Ap) | 31 |
| pPR11 | BamHI-NcoI PCR fragment containing Pgpd-crtYB cloned in the 6.3-kb BamHI-NcoI site of pPR10F (Ap) | This study |
| pPR12 | Expression cassette (Pgpd crtYB TcrtYB) as BamHI fragment in pMTL22P (Ap, G418) | This study |
| pPR13F/R | BamHI fragment (Pgpd crtYB TcrtYB) cloned in corresponding site of pPR2TN (Ap, G418) | This study |
| pPR16 | BamHI-HindIII (crtYBΔ) fragment cloned in corresponding sites of pPR1 (G418) | This study |
| pPR19F/R | Insertion of selection cassette (Pgpd G418 Tgpd) on blunt ended (Klenow) EcoRI fragment in the T4 DNA polymerase-treated BstXI site of pPR10F (crtYB::Pgpd G418 Tgpd) (Ap, G418) | This study |
| pPR21 | crtYB gene of X. dendrorhous as a 4.5-kb BamHI fragment in pMTL22P (Ap) | 31 |
| pPR22F/R | 4.5-kb BamHI fragment from pPR21 in BamHI site of pPR2TN (Ap, G418) | 31 |
| pPRpd2 | cDNA encoding phytoene desaturase of X. dendrorhous in pBluescript (Ap) | 30 |
| pPRpdcos1 | Phytoene desaturase-positive cosmid clone from a genomic library of X. dendrorhous (Ap) | 30 |
| pPR40F/R | 2.8-kb BamHI PCR fragment containing the expression cassette for phytoene desaturase (Pcrti crtl Tcrtl) cloned in BamHI site of pPR2TN (Ap, G418) | This study |
The selective antibiotics are indicated in parentheses.
FIG. 2.
Schematic representation of the specific crtYB gene inactivation approach by single (A) and double (B) crossover. Prior to the introduction in X. dendrorhous by electrotransformation, the plasmids pPR16 and pPR19F were linearized with the endonucleases BstXI and EcoRI, respectively. SI, site of insertion (the insert was a blunted EcoRI fragment in a blunted BstXI site) (Table 1).
FIG. 3.
Graphical presentation of the transformation vectors (pPR1 and pPR2TN) and carotenogenic expression vectors (pPR13F, pPR22F, pPR19F, and pPR40F). Depending on the orientation of the carotenogenic expression cassette, the vectors were denominated F (forward) or R (reverse) when the carotenogenic gene was transcribed in the same or opposite direction, respectively, as the G418 marker gene. The Sau3A* (not unique) this site was created by the ligation of BamHI and BglII sites. SI, site of insertion (the insert was a blunted EcoRI fragment in a blunted BstXI site; details are given in Table 1).
E. coli was cultivated in Luria-Bertani medium at 37°C. When appropriate, ampicillin was added to a final concentration of 50 μg/ml. Strains of X. dendrorhous were cultivated in YM broth (3.0 g of yeast extract, 3.0 g of malt extract, and 10.0 g of dextrose per liter; Difco). Normally, one colony taken from a fresh agar plate was used to inoculate 30 ml of medium in a 250-ml Erlenmeyer shake flask. Flasks were incubated for 96 h in a cooled rotary shaker (New Brunswick Scientific, Nijmegen, The Netherlands) with a rotation speed of 250 rpm at 21°C. To select for G418 resistance, 40 μg of Geneticin (G-418 sulfate, Invitrogen Life Technology, The Netherlands)/ml was added to the medium.
Carotenoid extraction and analysis.
An optimized protocol, based on the dimethyl sulfoxide method of Sedmak et al. (27), was used to isolate the carotenoids from X. dendrorhous cells. After cultivation, cells were collected by centrifugation and washed twice with demineralized water and the cell pellet was freeze-dried. Freeze-dried cell material (10 mg) was extracted with dimethyl sulfoxide (3 ml) for 15 min at 60°C. After centrifugation, the total carotenoid content can be determined from the supernatant by recording its absorbance at 470 or 450 nm. The total extract was transferred into a separator funnel, 3 ml of diethyl ether was added, and the mixture was kept on ice for 1 to 3 min before 0.5 ml of water was added. The lower phase was removed, 5 ml of acetone and then 5 ml of 10% (vol/vol) ether-petrol were added. Finally, 8 to 10 ml of water was added to obtain phase separation. The upper phase was collected, washed with water (10 ml), dried in a stream of N2, and resuspended in acetone. High-performance liquid chromatography (HPLC) separation was on a 25-cm by 3-μm Nucleosil C18 column (Macherey-Nagel, Düren, Germany) with acetonitrile-methanol-water (50:48:2, by volume). Spectra were recorded online with a photodiode array detector 440 (Kontron, Straubenhard, Germany). Carotenoid identification was carried out with authentic standards. Different keto-hydroxy-β-carotene derivatives were obtained by combinatorial biosynthesis in E. coli as previously described (7).
RESULTS
Specific inactivation of the crtYB gene.
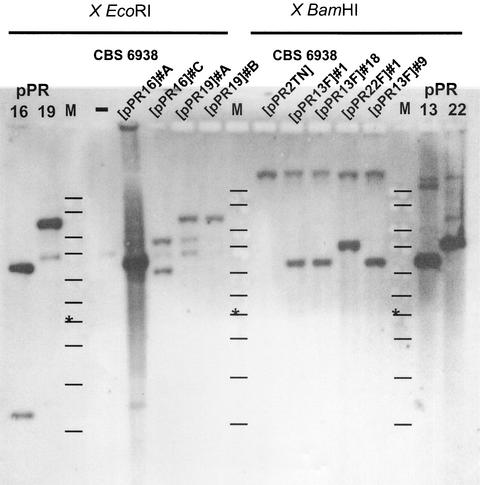
We wanted to establish a specific gene inactivation approach, which is applicable to the construction of strains which are able to accumulate specific carotenoid intermediates of the astaxanthin biosynthetic pathway. For this study, the crtYB gene, encoding a bifunctional enzyme involved in the condensation of two geranylgeranyl diphosphate molecules in phytoene and the cyclization of lycopene into β-carotene, was used (31). Previously, an integrative DNA transformation system was developed to introduce multiple copies of a transformation vector, which is selected for by resistance to G418, into the ribosomal DNA of X. dendrorhous (35, 36). To inactivate the endogenous crtYB gene of X. dendrorhous by homologous recombination, two types of knockout vectors were designed (Fig. 2). Vector pPR16 was constructed to inactivate the crtYB gene by a single crossover event within the N-terminal portion of this gene. Vector pPR19F was used to disrupt the crtYB gene by a double crossover at the crtYB locus. After transformation and selection for G418 resistance, both plasmids yielded three white-colored colonies and several colonies with a color phenotype identical to the recipient strain. Southern blot analysis was used to show that, in these latter transformants, the plasmids had integrated in the genome but outside the crtYB locus (results not shown). Two white-colored colonies of each transformation experiment were subjected to Southern blot analysis to demonstrate site-specific integration in the endogenous crtYB locus (Fig. 4). The transformants CBS 6938(pPR16) C, CBS 6938(pPR19) A, and CBS 6938(pPR19) B showed the expected hybridization pattern. The intensity of the hybridization signal suggested that, in CBS 6938(pPR16) A, multiple DNA fragments were integrated at the crtYB locus. As expected, no accumulation of carotenoids was observed when carotenoid extracts from the transformants were analyzed by HPLC. Additionally, 11 red-colored colonies were found among pPR16 transformants. Analysis of the carotenoid composition of some of these colonies indicated the accumulation of low amounts of astaxanthin. Furthermore, the plasmid integrated at the crtYB locus, as was found by Southern blot analysis. Apparently, the integration event had resulted in a truncated crtYB gene copy (Fig. 2) which retained some of its phytoene synthase and lycopene cyclase activity. With E. coli, it was found that the enzymatic activities of such a truncated enzyme are reduced to, respectively, 30 and 15% (31).
FIG. 4.
Autoradiogram of a Southern blot of chromosomal DNA isolated from several transformants of X. dendrorhous strain CBS 6938. Plasmids and chromosomal DNA were digested with the endonucleases indicated at the top of the panels. The blot was hybridized with a DIG-labeled cDNA probe encoding CrtYB. A DNA ladder containing fragments of 10, 8, 6, 5, 4, 3.5, 3 (marked with an asterisk), 2.5, 2, and 1 kb was used as a marker (lanes M). The expected hybridization patterns for transformants obtained after the integration of pPR16 or pPR19F at the crtYB locus are depicted in Fig. 2.
Overexpression of the crtYB gene in X. dendrorhous CBS 6938.
To study the effect on astaxanthin biosynthesis and carotenoid composition, the phytoene synthase-lycopene cyclase-encoding gene (crtYB) was overexpressed in X. dendrorhous wild-type strain CBS 6938. The regulation of carotenoid biosynthesis in X. dendrorhous is largely unknown. Some preliminary data (16) suggest that the pathway is regulated by feedback inhibition of the end product astaxanthin. The promoter region of crtYB (PcrtYB) might contain binding sites of regulatory proteins, interaction with which can regulate the transcription of crtYB. Therefore, the crtYB gene was also linked to the promoter region of the glyceraldehyde-3-phosphate dehydrogenase-encoding gene (Pgpd) of X. dendrorhous. The plasmids pPR2TN (control), pPR13F, and pPR22F (Fig. 3) were linearized in their ribosomal DNAs with SfiI and were introduced in X. dendrorhous CBS 6938. The color of a colony of X. dendrorhous CBS 6938 or CBS 6938(pPR2TN) is pink. The color of all transformants of X. dendrorhous CBS 6938 carrying integrated copies of pPR13F or pPR22F, hereafter referred to as CBS 6938(pPR13F) and CBS 6938(pPR22F), was orange. No visual differences in color were observed when the different CBS 6938(pPR13F) and CBS 6938(pPR22F) colonies were compared. This indicates that the carotenoid composition in these strains has been changed due to the introduction of additional copies of the crtYB gene. One transformant of CBS 6938(pPR2TN) and several transformants of CBS 6938(pPR13F) and CBS 6938(pPR22F) were randomly selected. Southern blot analysis demonstrated the integration of multiple copies of the transformation vector in these transformants (Fig. 4). Initially, the spectra of the carotenoid extracts were recorded from 250 to 600 nm. In CBS 6938 and CBS 6938(pPR2TN), the absorbance peak was at 471 nm, whereas the absorbance peak for CBS 6938(pPR13F) and CBS 6938(pPR22F) shifted to 465 to 467 and 468 nm, respectively. The carotenoid composition was further analyzed by HPLC. In both types of transformants, an increase in the total amount of carotenoids was found (Table 2). This increase is mainly caused by higher amounts of both β-carotene and echinone. In transformant CBS 6938(pPR13F) no. 18, an almost twofold increase in the total amount of carotenoids and a slight increase in the specific astaxanthin production were observed. However, in all transformants analyzed, the relative amount of astaxanthin was reduced compared to that of the control.
TABLE 2.
Carotenoid composition of X. dendrorhous strains CBS 6938 and PR-1-104 overexpressing the phytoene synthase-lycopene cyclase-encoding gene by using homologous (pPR22) and heterogeneous (pPR13) expression signalsa
| Carotenoid | Specific amt ± SD (μg/g [dry weight]) (% relative distribution) of carotenoid in strainb:
|
||||
|---|---|---|---|---|---|
| CBS 6938(pPR2TN) no. 1 | CBS 6938(pPR22F) no. 1 | CBS 6938(pPR13F) no. 1 | CBS 6938(pPR13F) no. 9 | CBS 6938(pPR13F) no. 18 | |
| Astaxanthin | 84 ± 11 (33) | 78 ± 22 (24) | 48 ± 4 (16) | 70 ± 13 (26) | 95 ± 29 (21) |
| Phoenicoxanthin | 50 ± 7 (19) | 43 ± 30 (15) | 43 ± 1 (14) | 43 ± 11 (16) | 59 ± 21 (13) |
| HO-echinone | 20 ± 2 (8) | —c (0) | 4 ± 5 (1) | — (0) | — (0) |
| Echinone | 31 ± 3 (12) | 88 ± 7 (33) | 114 ± 4 (38) | 78 ± 24 (28) | 181 ± 41 (39) |
| β-Carotene | 28 ± 7 (11) | 40 ± 3 (15) | 69 ± 9 (23) | 50 ± 3 (18) | 113 ± 31 (24) |
| HDCO | 39 ± 2 (15) | 23 ± 1 (9) | 18 ± 2 (6) | 31 ± 11 (11) | 17 ± 2 (4) |
| Torulene | 5 ± 3 (5) | — (0) | 7 ± 10 (2) | — (0) | — (0) |
| Total | 256 ± 12 (100) | 271 ± 42 (106) | 302 ± 13 (118) | 273 ± 13 (107) | 465 ± 24 (182) |
In PR-1-104 and its derivatives, only β-carotene is produced. Total amounts of β-carotene were as follows (results are in micrograms per gram [dry weight], with % relative distribution in parentheses): PR-1-104(pPR2TN) no. 1, 308±28 (100); PR-1-104(pPR22F) no. 1, 407±8 (132); PR-1-104(pPR22F) no. 2, 418±3 (136); PR-1-104(pPR13F) no. 1, 412±26 (134); PR-1-104(pPR13F) no. 2, 372±11 (121).
All strains were cultivated in duplicate, and the indicated numbers are the averages of two cultures ± standard deviations. Relative amounts are indicated in parentheses (the value for the strain with the empty cloning vector [pPR2TN] is set at 100).
—, not detected.
Overexpression of crtYB expression cassettes in PR-1-104.
The plasmids pPR2TN, pPR13F, and pPR22F were also introduced in a carotenoid biosynthetic mutant of X. dendrorhous CBS 6938. This strain, PR-1-104, does not produce xanthophylls but accumulates β-carotene (13). After introduction of the plasmids in strain PR-1-104, several G418-resistant colonies were isolated. The color phenotype of all pPR13F- and pPR22F-derived colonies was different from that of the host strain and PR-1-104(pPR2TN) transformants, indicating that the carotenoid composition had changed. All PR-1-104(pPR13F) and PR-1-104(pPR22F) transformants displayed a bright yellow color, and therefore, two colonies of each were randomly selected for further analysis. In all PR-1-104 transformants, only the accumulation of one carotenoid, β-carotene, was observed (Table 2). No significant differences in production levels were determined between the two expression signals.
Overexpression of the phytoene desaturase-encoding gene (crtI) in X. dendrorhous strains.
The data presented in Table 2 suggest that an increase in the conversion of lycopene, due to the overexpression of the crtYB gene encoding lycopene cyclase, results in a decrease in the formation of monocyclic carotenoids, e.g., torulene and 3-hydroxy-4-keto-3′,4′-didehydro-β-carotene (HDCO). The crtI gene product of X. dendrorhous is a dehydrogenase that introduces four additional double bonds into phytoene, yielding lycopene (30). To study the effect of higher lycopene levels, the crtI gene of X. dendrorhous, encoding phytoene desaturase, was overexpressed. To achieve this, the plasmids pPR40F and pPR40R (Fig. 3) were introduced in X. dendrorhous CBS 6938. The color phenotype of the transformants, transformed with either pPR40F or pPR40R, varied from pink to dark red. The transformants CBS 6938(pPR40F) no. 3 and CBS 6938(pPR40F) no. 4 were selected for further analysis based on the color intensity of the colonies on YM broth agar plates and on the spectrophotometric analysis of carotenoid extracts. The results of the HPLC analysis of the different carotenoid extracts are summarized in Table 3. The total carotenoid production of the crtI-overexpressing strains is lower than that of the control strain. In the control strain, 85% of the carotenoids are bicyclic carotenoids with astaxanthin as the major component. Under the applied cultivation conditions, an accumulation of the intermediates echinone and β-carotene was observed in CBS 6938(pPR2TN). In transformants CBS 6938(pPR40F) no. 3 and CBS 6938(pPR40F) no. 4, a fourfold increase in the sum of all monocyclic carotenoids and a 50% reduction of the astaxanthin content was observed (Table 3). In these transformants, the major component was HDCO, a monocyclic carotenoid that is normally detected as a minor compound.
TABLE 3.
Carotenoid composition of X. dendrorhous strains CBS 6938 and PR-1-104 overexpressing the phytoene desaturase-encoding gene (crtI)a of X. dendrorhous
| Carotenoid | Specific amt ± SD (μg/g [dry weight]) (% distribution) of carotenoid in strainb:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| CBS 6938(pPR2TN) no. 1 | CBS 6938(pPR40F) no. 3 | CBS 6938(pPR40F) no. 4 | PR-1-104(pPR2TN) no. 1 | PR-1-104(pPR40F) no. 4 | PR-1-104(pPR40F) no. 11 | PR-1-104(pPR40R) no. 3 | PR-1-104(pPR40R) no. 14 | |
| Astaxanthin | 132 ± 17 (50) | 48 ± 14 (25) | 44 ± 7 (24) | |||||
| Phoenicoxanthin | ||||||||
| HO-echinone | 18 ± 9 (7) | 15 ± 12 (8) | 16 ± 5 (9) | |||||
| Echinone | 42 ± 9 (16) | 11 ± 7 (6) | 12 ± 5 (6) | |||||
| β-Carotene | 30 ± 7 (11) | 7 ± 5 (3) | 6 ± 4 (3) | 371 ± 2 (90) | 344 ± 15 (84) | 136 ± 17 (65) | 88 ± 11 (53) | 164 ± 2 (70) |
| HDCO | 42 ± 8 (16) | 100 ± 22 (54) | 101 ± 17 (55) | |||||
| HO-ketotorulene | ||||||||
| Torulene | 2 ± 2 (2) | 7 ± 2 (4) | 9 ± 7 (5) | 40 ± 1 (10) | 63 ± 4 (16) | 71 ± 1 (35) | 76 ± 3 (47) | 68 ± 6 (30) |
| Total | 265 ± 22 (100) | 187 ± 18 (71) | 185 ± 24 (70) | 410 ± 1 (100) | 406 ± 18 (99) | 207 ± 18 (50) | 164 ± 8 (40) | 233 ± 4 (57) |
The expression of the introduced crtI gene is regulated by the promoter and terminator regions of the crtI gene.
The reported data represent the mean values ± standard deviations of at least two cultures. The determinations were performed in duplicate for each culture. The relative production levels compared to that of the control strain CBS 6938(pPR2TN) (100%) are indicated in parentheses. For the PR-1-104 strains, the relative production levels are compared to those of the control strain PR-1-104(pPR2TN) and are indicated in parentheses.
The vectors pPR40F and pPR40R were also introduced in PR-1-104, a β-carotene-accumulating X. dendrorhous strain. Transformants of PR-1-104 displayed a color phenotype ranging from yellow to red, including orange. Out of 28 transformants of PR-1-104, four strains, hereafter referred to as PR-1-104(pPR40F) no. 4 and no. 11 and PR-1-104(pPR40R) no. 3 and no. 14, were selected. The results of the analysis of carotenoid extracts are shown in Table 3. In most of the transformants, a decrease in the total carotenoid content and the specific β-carotene content was observed. The relative torulene content increased from 10 to 47%. There seemed to be a negative correlation between the total amount of the monocyclic carotenoid torulene and the total carotenoid content.
DISCUSSION
The potential of X. dendrorhous as a microbiological source of natural astaxanthin was recognized soon after the isolation of this yeast by Hermann Phaff and coworkers (21). So far, strategies to improve astaxanthin production in X. dendrorhous were based on classical mutagenesis and selection (2, 9, 13) and/or the improvement of fermentation conditions (4, 11). Here, we describe the use of recombinant DNA technology for metabolic engineering of the astaxanthin biosynthetic pathway in X. dendrorhous. The crtYB gene of X. dendrorhous encoding the chimeric phytoene synthase-lycopene cyclase was used to construct specific carotenoid biosynthetic mutants. The combination of a white phenotype and the resistance towards G418 and the results of a Southern blot analysis (Fig. 4) indicated a successful inactivation of the endogenous crtYB gene, by either a single or a double crossover event. From the difference in transformation efficiency between the vectors pPR16, pPR19F, and pPR2TN, it can be concluded that integration at the crtYB gene locus in X. dendrorhous, and most probably in all other single-gene loci, is 100-fold less efficient than integration at the loci of the ribosomal DNA (pPR2TN). The main disadvantage of mutagenesis, with UV or chemical compounds, for the isolation of biosynthetic mutants is the chance of introducing multiple mutations. Although this method has been used successfully (2, 13), the specific gene inactivation approach presented in this paper is much more defined and has an additional advantage. To our knowledge, no lycopene-accumulating strain of X. dendrorhous has been isolated by classical mutagenesis yet (13). An explanation for this phenomenon is the crtYB gene product, as it possesses both phytoene synthase and lycopene cyclase activity (31). Two white strains, PR-1-120 and PR-1-139, were classified as phytoene synthase-negative mutants (13). A heterologous phytoene desaturase-encoding gene or a mutated crtYB gene copy of X. dendrorhous that lacks lycopene cyclase activity can be introduced in order to construct a lycopene-accumulating X. dendrorhous strain. However, we have found that these mutants have retained some of the lycopene cyclase activity (J. C. Verdoes, unpublished data). Engineered strains such as CBS 6938(pPR16) or CBS 6938(pPR19F), with inactivated crtYB genes and displaying no lycopene cyclase activity, are much more defined and are therefore better as starting material for such an approach.
One possibility for the improvement of the metabolic productivity of an organism is genetic modification. This strategy can be successful when an increase of the flux through a pathway is achieved by, e.g., the overproduction of the rate-limiting enzyme, an increase of precursors, or the modification of the regulatory properties of enzymes. The isolation of several carotenogenic genes of X. dendrorhous enabled us to study the effect of their overexpression on carotenoid biosynthesis. Overexpression of the chimeric crtYB gene either under control of the gpd or crtYB promoter leads to a different carotenoid composition in both the wild type and a β-carotene-accumulating X. dendrorhous strain (Table 2). In transformant CBS 6938(pPR13F) no. 18, the total number of carotenoids increased by 82%. Although the absolute astaxanthin content was higher than in the wild-type strain, the relative amount decreased. This is the result of a 270% increase in the amount of β-carotene and echinone. A similar increase was observed in all transformants of CBS 6938 carrying additional crtYB gene copies. Furthermore, in the transformants of CBS 6938, the relative amount of monocyclic carotenoids, e.g., torulene and HDCO, is reduced by at least 50%. No accumulation of carotenoids other than β-carotene was observed when the crtYB gene was overexpressed in a β-carotene-accumulating X. dendrorhous strain. Compared to the control strain, a small but significant increase in the total amount of β-carotene was observed in all transformants.
Overexpression of the phytoene desaturase-encoding gene (crtI) of X. dendrorhous affected the ratio between bicyclic and monocyclic carotenoids in both CBS 6938 and PR-1-104 (Table 3). In the control strain, more than 84% consisted of bicyclic carotenoids. However, this number is reduced to less than 50% in the transformants. In transformants of CBS 6938, the main carotenoid is HDCO (Fig. 1). Furthermore, the relative astaxanthin content is decreased twofold and that of β-carotene and echinone is decreased by a factor 3. Introduction of crtI gene copies in the β-carotene-accumulating strain PR-1-104 has a negative effect on the β-carotene production and the total carotenoid production. In PR-1-104(pPR40R) no. 3, there was an increase in the specific and relative amounts of the monocyclic carotenoid torulene by factors of 2 and 5, respectively (Table 3).
Recently, An et al. (3) proposed the presence of a monocyclic carotenoid pathway in X. dendrorhous in addition to the dicyclic one proposed by Andrewes et al. (5). The fact that torulene is the end product in a β-carotene-accumulating strain carrying multiple copies of the phytoene desaturase-encoding gene (crtI) suggests that the enzymes that convert β-carotene into astaxanthin are the same ones that convert torulene into HDCO (Fig. 1). Apparently, these enzymes have a broad substrate range and can accept both monocyclic and bicyclic carotenoids. In some of the carotenoid-hyperproducing mutants studied by An et al. (3), one or more mutations affecting phytoene synthase activity and/or crtI gene expression may explain the observed increased levels of monocyclic carotenoids.
The carotenoid biosynthetic enzymes of X. dendrorhous are specific only to certain regions of the substrate molecule. It was shown previously that neurosporene is also a substrate for the cyclase moiety of crtYB in a heterologous genetic background (31). The data presented in this paper showed that a single desaturase is responsible for the introduction of up to five double bonds into phytoene.
Overexpression of the crtYB gene, encoding the bifunctional carotenogenic enzyme, in CBS 6938 resulted in the accumulation of the intermediates β-carotene and echinone (Table 2). When the flux towards β-carotene was reduced by the introduction of additional copies of the phytoene desaturase-encoding gene, a decrease in the amounts of these two compounds was observed (Table 3). These results indicate that, under overexpression of the crtYB gene, the oxygenation reactions (e.g., of β-carotene and echinone) are limiting in the pathway to astaxanthin. The increase of lycopene cyclase activity also resulted in a decrease of the carotenoids derived from 3,4-didehydrolycopene like torulene and HDCO.
A decisive reaction for the formation of monocyclic or bicyclic products is the desaturation sequence to lycopene and further on to 3,4-didehydrolycopene. In the nontransformed strain, cyclization of lycopene, which directs the metabolic flux towards astaxanthin, is the dominating reaction. However, when the gene encoding phytoene desaturase is overexpressed, the five-step desaturation to 3,4-didehydrolycopene is intensified, resulting in an accumulation of torulene and HDCO as subsequent products (Table 3). Apparently, the strength of crtI expression, i.e., the amounts and activities of phytoene desaturase present, determine the number of double bonds to be introduced by the desaturase. It can be concluded from the results of the crtI and crtYB transformants that, in X. dendrorhous, the competition between desaturase and cyclase for lycopene governs the metabolic flux either via β-carotene to astaxanthin or via 3,4-didehydrolycopene to HDCO. This indicates that a change in the ratios of carotenogenic enzymes in X. dendrorhous by either induced mutations or metabolic engineering may affect the amounts and composition of carotenoids. We propose that, like in Phycomyces blakesleeanus (10), the carotenogenic enzymes of X. dendrorhous are present in a complex (Fig. 5). Increased levels of the phytoene desaturase might alter the sequence of reactions and therefore the end products that are formed. From this viewpoint, it might be important, in order to optimize astaxanthin production, to overexpress multiple carotenogenic genes in such a way, e.g., by coregulated expression, that the ratios are not affected.
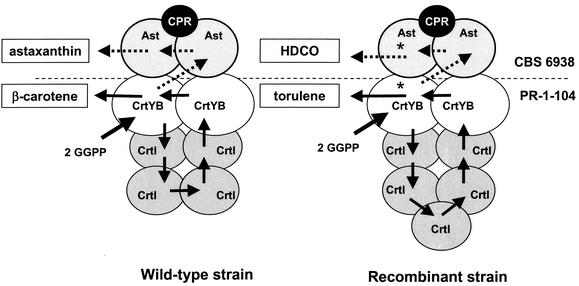
FIG. 5.
Hypothetical representation of the carotenogenic complex in X. dendrorhous (left) and a recombinant strain overexpressing the phytoene desaturase (CrtI)-encoding gene (right). Increased levels of a carotenogenic enzyme might alter the sequence of reactions. In the β-carotene-accumulating strain PR-1-104, the astaxanthin synthetase (Ast) enzyme is inactive or absent (above dotted line). The main carotenoids under each specific condition are indicated in boxes. *, the 3,4-didehydro ends of torulene and HDCO are not substrates for lycopene cyclase (CrtYB) and astaxanthin synthetase (Ast), respectively. The number and stoichiometry of enzymes are speculative and based on the number of different enzymatic steps. CPR, cytochrome P450 reductase.
It is anticipated that ultimately, by using the methods presented in this study and by a combination of overexpression and deletion of specific carotenoid biosynthetic genes, the carotenoid content in X. dendrorhous can be altered significantly and can be directed to produce a specific carotenoid in higher amounts.
Acknowledgments
We thank Stephan Hesse for contributions to the construction of the different expression vectors. Martin de Wit is kindly acknowledged for assistance with HPLC analysis.
Part of this work was supported by a grant from the European Commission (Fifth Framework).
REFERENCES
- 1.Albrecht, M., N. Misawa, and G. Sandmann. 1999. Metabolic engineering of the terpenoid biosynthetic pathway of Escherichia coli for the production of the carotenoids β-carotene and zeaxanthin. Biotechnol. Lett. 21:791-795. [Google Scholar]
- 2.An, G.-H., J. Bielich, R. Auerbach, and E. A. Johnson. 1991. Isolation and characterization of carotenoid hyperproducing mutants of yeast by flow cytometry and cell sorting. Bio/Technology 9:70-73. [DOI] [PubMed] [Google Scholar]
- 3.An, G.-H., M.-H. Cho, and E. A. Johnson. 1999. Monocyclic carotenoid biosynthetic pathway in the yeast Phaffia rhodozyma (Xanthophyllomyces dendrorhous). J. Biosci. Bioeng. 88:189-193. [DOI] [PubMed] [Google Scholar]
- 4.An, G.-H., C.-H. Kim, E.-S. Choi, and S.-K. Rhee. 1996. Medium optimization for cultivation of carotenoid hyperproducing Phaffia rhodozyma mutant HT-5FO1C. J. Ferment. Bioeng. 82:515-518. [Google Scholar]
- 5.Andrewes, A. G., H. J. Phaff, and M. P. Starr. 1976. Carotenoids of Phaffia rhodozyma, a red-pigmented fermenting yeast. Phytochemistry 15:1003-1007. [Google Scholar]
- 6.Armstrong, G. A. 1997. Genetics of eubacterial carotenoid biosynthesis: a colorful tale. Annu. Rev. Microbiol. 51:629-659. [DOI] [PubMed] [Google Scholar]
- 7.Breitenbach, J., N. Misawa, S. Kajiwara, and G. Sandmann. 1996. Expression in Escherichia coli and properties of the carotene ketolase from Haematococcus pluvialis. FEMS Microbiol. Lett. 140:241-246. [DOI] [PubMed] [Google Scholar]
- 8.Chew, B. P., J. S. Park, M. W. Wong, and T. S. Wong. 1999. A comparison of the anticancer activities of dietary beta-carotene, canthaxanthin and astaxanthin in mice in vivo. Anticancer Res. 19:1849-1853. [PubMed] [Google Scholar]
- 9.Chumpolkulwong, N., T. Kakazono, S. Nagai, and N. Nishio. 1997. Increased astaxanthin production by Phaffia rhodozyma mutants isolated as resistant to diphenylamine. J. Ferment. Bioeng. 83:429-434. [Google Scholar]
- 10.De La Guardia, M. D., C. M. G. Aragón, F. J. Murillo, and E. Cerdá-Olmedo. 1971. A carotenogenic enzyme aggregate in Phycomyces: evidence from quantitative complementation. Proc. Natl. Acad. Sci. USA 68:2012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang, T. J., and Y.-S. Cheng. 1993. Improvement of astaxanthin production by Phaffia rhodozyma through mutation and optimization of culture conditions. J. Ferment. Bioeng. 75:466-469. [Google Scholar]
- 12.Farmer, W. R., and J. C. Liao. 2000. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat. Biotechnol. 18:533-537. [DOI] [PubMed] [Google Scholar]
- 13.Girard, P., B. Falconnier, J. Bricout, and B. Vladescu. 1994. β-Carotene producing mutants of Phaffia rhodozyma. Appl. Microbiol. Biotechnol. 41:183-191. [Google Scholar]
- 14.Hoshino, T., R. Fujii, and T. Nakahara. 1994. Overproduction of carotenoids in Thermus thermophilus. J. Ferment. Bioeng. 77:423-424. [Google Scholar]
- 15.Johnson, E. A., and W. A. Schroeder. 1995. Microbial carotenoids, p. 119-178. In A. Fiechter (ed.), Advances in biochemical engineering biotechnology, vol. 53. Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 16.Johnson, E. A., and W. A. Schroeder. 1995. Astaxanthin from the yeast Phaffia rhodozyma. Stud. Mycol. 38:81-89. [Google Scholar]
- 17.Kajiwara, S., P. D. Fraser, K. Kondo, and N. Misawa. 1997. Expression of an exogenous isopentenyl diphosphate isomerase gene enhances isoprenoid biosynthesis in Escherichia coli. Biochem. J. 324:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mann, V., M. Harker, I. Pecker, and J. Hirschberg. 2000. Metabolic engineering of astaxanthin production in tobacco flowers. Nat. Biotechnol. 18:888-892. [DOI] [PubMed] [Google Scholar]
- 19.Mayne, S. T. 1996. Beta-carotene, carotenoids and disease prevention in humans. FASEB J. 10:690-701. [PubMed] [Google Scholar]
- 20.Misawa, N., and H. Shimada. 1998. Metabolic engineering for the production of carotenoids in non-carotenogenic bacteria and yeasts. J. Biotechnol. 59:169-181. [DOI] [PubMed] [Google Scholar]
- 21.Phaff, H. J., M. W. Miller, M. Yoneyama, and M. Soneda. 1972. A comparative study of the yeast florae associated with trees on the Japanese islands and on the west coast of North America, p. 759-774. In G. Terui (ed.), Proceedings of the 4th IFS: fermentation technology today. Society of Fermentation Technology, Kyoto, Japan.
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Sandmann, G. 1994. Carotenoid biosynthesis in microorganisms and plants. Eur. J. Biochem. 223:7-24. [DOI] [PubMed] [Google Scholar]
- 24.Sandmann, G., M. Albrecht, G. Schnurr, O. Knörzer, and P. Böger. 1999. The biotechnological potential and design of novel carotenoids by gene combination in Escherichia coli. TIBTECH 17:233-237. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt-Dannert, C., D. Umeno, and F. H. Arnold. 2000. Molecular breeding of carotenoid biosynthetic pathways. Nat. Biotechnol. 18:750-753. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt-Dannert, C. 2000. Engineering novel carotenoid in microorganisms. Curr. Opin. Biotechnol. 11:255-261. [DOI] [PubMed] [Google Scholar]
- 27.Sedmak, J. J., D. K. Weerasinghe, and S. O. Jolly. 1990. Extraction and quantification of astaxanthin from Phaffia rhodozyma. Biotechnol. Technol. 4:107-112. [Google Scholar]
- 28.Verdoes, J. C., J. Wery, T. Boekhout, and A. J. J. van Ooyen. 1997. Molecular characterization of the glyceraldehyde-3-phosphate dehydrogenase gene of Phaffia rhodozyma. Yeast 13:1231-1242. [DOI] [PubMed] [Google Scholar]
- 29.Verdoes, J. C., and A. J. J. van Ooyen. 1999. Isolation of isopentenyl diphosphate isomerase encoding gene of Phaffia rhodozyma; improved carotenoid production in Escherichia coli. J. Bot. Gall. 146:43-53. [Google Scholar]
- 30.Verdoes, J. C., N. Misawa, and A. J. J. van Ooyen. 1999. Cloning and characterization of the astaxanthin biosynthetic gene encoding phytoene desaturase of Xanthophyllomyces dendrorhous. Biotechnol. Bioeng. 63:750-755. [DOI] [PubMed] [Google Scholar]
- 31.Verdoes, J. C., P. Krubasik, G. Sandmann, and A. J. J. van Ooyen. 1999. Isolation and functional characterisation of a novel type of carotenoid biosynthetic gene from Xanthophyllomyces dendrorhous. Mol. Gen. Genet. 262:453-461. [DOI] [PubMed] [Google Scholar]
- 32.Wang, C.-W., M.-K. Oh, and J. C. Liao. 1999. Engineered isoprenoid pathway enhances astaxanthin production in Escherichia coli. Biotechnol. Bioeng. 62:235-241. [DOI] [PubMed] [Google Scholar]
- 33.Wang, X., R. Willén, and T. Wadström. 2000. Astaxanthin-rich alga meal and vitamin C inhibit Helicobacter pylori infection in BALB/cA mice. Antimicrob. Agents Chemother. 44:2452-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wery, J., M. J. M. Dalderup, J. ter Linde, and A. J. J. van Ooyen. 1996. Structural and phylogenetic analysis of the actin gene from the yeast Phaffia rhodozyma. Yeast 12:641-651. [DOI] [PubMed] [Google Scholar]
- 35.Wery, J., D. Gutker, A. C. H. M. Renniers, J. C. Verdoes, and A. J. J. van Ooyen. 1997. High-copy-number integration into the ribosomal DNA of the yeast Phaffia rhodozyma. Gene 184:89-97. [DOI] [PubMed] [Google Scholar]
- 36.Wery, J., J. C. Verdoes, and A. J. J. van Ooyen. 1998. Efficient transformation of the astaxanthin-producing yeast Phaffia rhodozyma. Biotechnol. Technol. 12:339-405. [Google Scholar]