Abstract
The detection and identification of pathogens from water samples remain challenging due to variations in recovery rates and the cost of procedures. Ultrafiltration offers the possibility to concentrate viral, bacterial, and protozoan organisms in a single process by using size-exclusion-based filtration. In this study, two hollow-fiber ultrafilters with 50,000-molecular-weight cutoffs were evaluated to concentrate microorganisms from 2- and 10-liter water samples. When known quantities (105 to 106 CFU/liter) of two species of enteric bacteria were introduced and concentrated from 2 liters of sterile water, the addition of 0.1% Tween 80 increased Escherichia coli strain K-12 recoveries from 70 to 84% and Salmonella enterica serovar Enteritidis recoveries from 36 to 72%. An E. coli antibiotic-resistant strain, XL1-Blue, was recovered at a level (87%) similar to that for strain K-12 (96%) from 10 liters of sterile water. When E. coli XL1-Blue was introduced into 10 liters of nonsterile Rio Grande water with higher turbidity levels (23 to 29 nephelometric turbidity units) at two inoculum levels (9 × 105 and 2.4 × 103 per liter), the recovery efficiencies were 89 and 92%, respectively. The simultaneous addition of E. coli XL1-Blue (9 × 105 CFU/liter), Cryptosporidium parvum oocysts (10 oocysts/liter), phage T1 (105 PFU/liter), and phage PP7 (105 PFU/liter) to 10 liters of Rio Grande surface water resulted in mean recoveries of 96, 54, 59, and 46%, respectively. Using a variety of surface waters from around the United States, we obtained recovery efficiencies for bacteria and viruses that were similar to those observed with the Rio Grande samples, but recovery of Cryptosporidium oocysts was decreased, averaging 32% (the site of collection of these samples had previously been identified as problematic for oocyst recovery). Results indicate that the use of ultrafiltration for simultaneous recovery of bacterial, viral, and protozoan pathogens from variable surface waters is ready for field deployment.
Waterborne outbreaks of enteric diseases are a major public health concern, yet monitoring and identifying the disease-causing pathogens from water samples remain difficult. One of the biggest problems is the lack of a consistent method to simultaneously concentrate multiple organisms from a single water sample. Another common difficulty is the broad variation in recoveries, especially from water samples with high turbidity levels (1, 15, 16, 23). Additionally, cost is an important factor in the detection, monitoring, and identification of pathogenic microorganisms because different methods of concentration are frequently used for viruses, protozoan parasites, and bacteria. Some of these methods use disposable filters which are expensive because they are designed for one-time use.
To preserve the public health, water treatment facilities must monitor the source and the finished water. In addition, in order to evaluate the risk of exposure to waterborne pathogens, monitoring the occurrence and distribution of enteric pathogens in water is considered indispensable. Large volumes of water (10 to 100 liters of raw water and up to 1,000 liters of finished water) should be tested to ensure adequate protection (10, 14).
Some approaches have been developed to concentrate multiple microorganisms, but there is variation in the rates of recovery of different types of pathogens (6, 7, 13). The properties of microbial particles, such as size, shape, composition of the outermost layer, and stability, have been shown to influence the concentration efficiency (6, 18, 21, 25). The recovery of organisms is also affected by water quality parameters such as turbidity, pH, and the levels of salts and organics (4, 5, 21, 26).
Ultrafiltration offers important advantages over other filtration systems by simultaneously concentrating parasites, viruses, and bacteria in the initial step. Ultrafiltration uses a size-exclusion-based mode of concentration, where molecules smaller than the pore size of the filter pass through the membrane and out of the system and larger particles are concentrated in the retentate. The cross-flow circulation pattern with recirculation of the retentate reduces fouling of the membrane and makes it possible to filter large volumes of turbid water while maintaining the organisms in suspension (6, 9, 19, 26).
The purpose of this study was to determine the feasibility of two reusable hollow-fiber filter models (surface areas, 0.017 and 0.2 m2) to efficiently concentrate bacteria from water. In addition, simultaneous recoveries of other organisms (Cryptosporidium parvum, T1 phage, and PP7 phage) were compared by using environmental samples. This approach allowed multiple organisms to be recovered and the recovery rates from water with different turbidities (0.3 to 29 nephelometric turbidity units [NTU]) to be reproducibly quantified.
MATERIALS AND METHODS
Water samples.
Environmental samples (2 to 14 liters) were collected from the following resources: Las Cruces tap water, well water (New Mexico State University Fisheries and Wildlife Lab), and the Rio Grande (Las Cruces, N.Mex.). Other surface water samples were collected from Lake Erie (Silver Creek, N.Y.) and from the following reservoirs: Hetch Hetchy (Moccasin, Calif.), Charleroi (Charleroi, Pa.), Nottingham (Cleveland, Ohio), and Cobb County (Marietta, Ga.). Water samples were kept at 4°C until they were used, at which time a 200-ml sample was analyzed to determine turbidity (APHA method 2130B [3a]).
Initial filter preparation and sanitization.
The ultrafiltration setup consisted of a filter, a tubing system, two reservoirs, and a pump that were connected as described previously (8).
Two polyacrylonitrile, 50,000-molecular-weight-cutoff, hollow-fiber ultrafilters (AHP-0013 Microza; Pall Corp., Glen Cove, N.Y.) with surface areas of 0.017 and 0.2 m2 were used for 2- and 10-liter volumes of water, respectively. The filter preparation and sterility controls were the same for both filters and are outlined in Fig. 1. The filter preparation consisted of sanitization and blocking. The membrane was sanitized by recirculating a 500-ml solution of 200 mg of sodium hypochlorite/liter for 30 min, after which a sample was taken and plated on nutrient agar plates (sterility control sample 1) (Fig. 1). Then, a 0.1% sodium thiosulfate solution was recirculated across the filter for 30 min. The filter was then blocked with 5% calf serum (500 ml) for 60 min, followed by a second block with 20 ml of the same agent. After overnight blocking with agitation at 4°C, a second sample was taken and plated (sterility control sample 2). A third blocking step was done for 60 min before filtration.
FIG. 1.
Procedure for the evaluation of the reusable hollow-fiber ultrafilter for the concentration of viral, bacterial, and protozoal pathogens from 10-liter water samples.
Filtration.
The small-scale ultrafiltration system (filter surface area, 0.017 m2) was evaluated by using 2 liters of sterile, deionized water and phosphate-buffered saline (PBS; pH 7) with and without 0.1% Tween 80 as the liquid medium. The 2-liter samples were processed at a transmembrane pressure of 80 kPa and were filtered as previously reported (9, 26). The 10-liter volumes were processed by using the larger, 0.2-m2 filter and filtered as outlined in Fig. 1. PBS, 0.1% Tween 80, and inocula were introduced and mixed manually. A screw-down pressure regulator was then partially closed to produce a permeate flow of ∼300 ml/min, while maintaining a cross-flow of ∼3,600 ml/min. The water sample was allowed to circulate through the filtration system in the cross-flow mode for 10 min (with the permeate port closed) to further mix the sample. An initial sample was taken to enumerate inocula (input, subsample I). Filtration was continued until ∼250 ml of sample remained in the retentate beaker, at which time the peristaltic pump was shut off and the entire retentate (subsample II) volume was collected. To remove additional microorganisms that may have adhered to the filter, glycine was added to the retentate to give a final concentration of 0.05 M. The retentate was circulated in the cross-flow mode for 30 min, and then a sample was taken for enumeration of bacteria or viruses (retentate-eluate, subsample III). In order to detect the low numbers of oocysts, the 250-ml retentate-eluate sample was centrifuged at 1,200 × g for 20 min at 4°C. The resulting pellet was resuspended in 10 ml of sterile water. A 10-ml sample was taken to confirm sterility in the permeate (subsample IV). Viruses were quantified from the supernatant (subsample V), and bacteria were quantified from the resuspended pellet (subsample VI).
Postfiltration filter sanitization.
After filtration, the filter was sanitized as described above. In addition, the concentration of free sodium hypochlorite was determined at the end of each sanitization by measuring the absorbance at 530 nm using N,N-diethyl-p-phenylenediamine (DR/2010 method ID #80; Hach, Loveland, Colo.). The filter module was flushed with sterile filtered water until the residual concentration of free sodium hypochlorite was below 0.02 mg/liter. A 500-ml solution of 10% acetic acid was recycled for 60 min, and then the solution was neutralized with 500 ml of 2× PBS in the cross-flow mode. A third sample was plated on nutrient agar (sterility control sample 3). When bacterial growth was observed, an additional cycle of sanitization was done. After sanitization of the filter, sterile 10% sodium dodecyl sulfate was added and the filter was stored at 4°C for the next experiment (Fig. 1).
Microorganisms.
Escherichia coli XL1-Blue (2) was transformed with a plasmid (pBSK; Stratagene, La Jolla, Calif.) harboring the gene for ampicillin resistance (2, 20), whereas E. coli K-12 and Salmonella enterica serovar Enteritidis were obtained from the New Mexico State University Biology Department Culture Collection. E. coli K-12 and Salmonella serovar Enteritidis were grown in nutrient broth for 16 h at 37°C on a shaker platform operating at 200 rpm. E. coli XL1-Blue was grown in nutrient broth with 50 μg of ampicillin per ml and 10 μg of tetracycline per ml for 16 h at 37°C. These bacteria were assayed for CFU on nutrient agar by drop and spread plate techniques using 0.03 and 0.10 ml, respectively, from the concentrated water sample (10 liters). No ampicillin- or tetracycline-resistant bacteria were recovered from 10 liters of native Rio Grande water.
C. parvum oocysts (human and mouse strain AZ-I) were purchased from Parasitology Research Laboratories, LLC (Neosho, Mo.). Oocysts were purified by density gradient centrifugation and resuspended in an antibiotic solution for overnight shipment to the laboratory. Prior to use, oocysts were enumerated via fluorescent-antibody assay as described by Kuhn and Oshima (8).
Two model viruses, bacteriophages TI and PP7, were used for this study. E. coli (ATCC 11303) was utilized as the host strain for the growth and assay of bacteriophage T1 (ATCC 11303-B1), and Pseudomonas aeruginosa (ATCC 15692) was used for the growth and assay of bacteriophage PP7 (ATCC 15692-B2). The viruses were enumerated by plaque assay (12).
Sample dilution and calculations.
The recovery efficiency of each organism was calculated by the following equation:
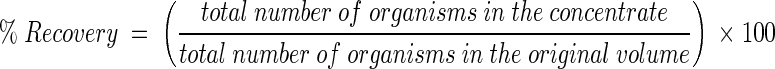 |
Organisms included viruses, bacteria, and protozoa. Viruses were assayed from the retentate-eluate (subsample III) and from the supernatant (subsample V). Bacteria were assayed from the retentate-eluate (subsample III) and from the centrifuged pellet (subsample VI). Protozoa were assayed from the centrifuged pellet (subsample VI).
RESULTS
Ultrafilter sterility.
Consistent with previous work (8, 9, 26), the washing procedures were effective in completely removing protozoa and viruses. When bacterial growth was observed, additional cycles of sanitization were done until the plate counts were zero.
Recovery of bacteria from 2- and 10-liter samples.
When E. coli strain K-12 and Salmonella serovar Enteritidis were introduced into 2 liters of PBS-buffered sterile water, the two bacteria were recovered at 70 and 36% of their respective input values (Table 1). When Tween 80 (0.1%) was added to the initial suspension, the average recovery of E. coli K-12 increased to 84% and recovery of serovar Enteritidis doubled to 72% (Table 1). Recovery rates of E. coli K-12 for the 10- and 2-liter samples were similar. There was similarly very little difference between recoveries of strain K-12 and the antibiotic-resistant strain XL1-Blue.
TABLE 1.
Recovery efficiencies of E. coli (2 strains) and S. enterica serovar Enteritidis from sterile water buffered with PBS (pH 7)
| Vol of water (liters) | E. coli strain | na | Input (cells/liter)
|
Mean % recovery (SD)
|
||
|---|---|---|---|---|---|---|
| E. coli | Serovar Enteritidis | E. coli | Serovar Enteritidis | |||
| 2b | K-12 | 3 | 5.0 × 105 | 1.0 × 106 | 70 (13.4) | 36 (1.0) |
| K-12 | 4 | 5.0 × 105 | 3.5 × 105 | 84 (20.8) | 72 (48.0) | |
| 10 | K-12 | 3 | 2.0 × 106 | NDc | 87 (2.3) | ND |
| XL1-B | 4 | 5.0 × 105 | ND | 80 (18.0) | ND | |
n, number of replicate experiments.
This experiment was the only one that did not include the addition of Tween 80 to the water.
ND, not determined.
Ninety-six percent of E. coli strain XL1-Blue was recovered from 10 liters of groundwater, and when similar quantities were introduced into the higher-turbidity Rio Grande surface waters, recoveries remained high at 89% (Table 2). Similar recoveries (92%) were also observed when input numbers of XL1-Blue were reduced to 2.4 × 103 liter−1. In the challenge experiments shown in Tables 1 and 2, bacterial recovery numbers were assayed using samples taken from the retentate-eluate (Fig. 1, filtration step III).
TABLE 2.
Recovery efficiencies of E. coli (XL1-Blue) from 10 liters of groundwater and Rio Grande surface water
| Water typea | Avg turbidity (NTU)b | E. coli input (cells/liter) | Mean % recovery (SD) |
|---|---|---|---|
| Groundwater | 0.3 | 2.0 × 106 | 96 (5.6) |
| Rio Grande water | 29.2 | 9.0 × 105 | 89 (6.7) |
| 22.8 | 2.4 × 103 | 92 (5.6) |
Each experiment was repeated four times.
Values are averages of four different water samples.
Recovery of multiple organisms from 10-liter Rio Grande samples.
Cochallenge experiments with model viruses (T1 phage and PP7 phage), bacteria (E. coli XL1-Blue), and protozoa (C. parvum) were carried out with 10 liters of Rio Grande surface waters. Recovery numbers for bacteria and viruses were assayed using samples taken from either the retentate-eluate filtration step (as shown in Tables 1 and 2) or from the additional centrifugation step. Because of the low input numbers of C. parvum, recovery numbers could be assayed only after centrifugation (Fig. 1, filtration step VI). From the retentate-eluate samples, E. coli recovery was 95% and T1 phage and PP7 phage recoveries were 73 and 62%, respectively (Table 3). The elution step decreased the variability and increased the recovery, especially of the viruses. The average viral recovery before elution was 42% (standard deviation [SD], 38), and after elution recovery was 68% (SD, 11). The centrifugation step did not affect E. coli recovery, while viral recovery decreased slightly. The recovery of C. parvum after centrifugation was consistently around 54% (Table 3).
TABLE 3.
Recovery efficiencies with ultrafiltration and centrifugation of E. coli (XL1-Blue), Cryptosporidium, T1 phage, and PP7 phage from 10 liters of Rio Grande surface watera
| Microorganism | Input (cells/liter) | Mean % recovery (SD)
|
|||
|---|---|---|---|---|---|
| Ultrafiltration
|
Centrifugation
|
||||
| Retentate | Retentate-eluate | Supernatant | Pellet | ||
| E. coli | 9 × 105 | 86 (12.0) | 95 (7.8) | 1 (0.5) | 96 (11.2) |
| Cryptosporidium | 1 × 101 | NDb | ND | ND | 54 (1.5) |
| T1 phage | 1 × 105 | 38 (22) | 73 (17) | 59 (22) | ND |
| PP7 phage | 1 × 105 | 45 (55) | 62 (5) | 46 (7) | ND |
Each experiment was repeated three times. Average pH and NTU of surface water samples were 7.2 and 22.8, respectively.
ND, not determined.
Recovery of multiple organisms from 10 liters of surface waters with histories of poor oocyst recovery.
Five samples from surface waters from around the United States, including Lake Erie, Hetch Hetchy, Charleroi, Nottingham, and Cobb County, were inoculated with similar levels of microbes as shown in Table 3. All recovery efficiencies were calculated from samples after centrifugation, with the viral numbers derived from the supernatant (Fig. 1, filtration step V) and the bacterial and protozoal numbers derived from the resuspended pellet (Fig. 1, filtration step VI). Bacterial recoveries from the five surface water samples were consistently high, with recovery rates ranging from 87 to 97% (Table 4). Viral recoveries for these samples were similar to recoveries from the Rio Grande samples, whereas protozoal recoveries from these samples were lower than those from the Rio Grande samples, particularly in two of the samples where only 19% of the introduced protozoa were recovered.
TABLE 4.
Recovery efficiencies of E. coli (XL1-Blue), Cryptosporidium, T1 phage, and PP7 phage from 10 liters of surface waters from different locations around the United Statesa
| Surface water source | Turbidity (NTU) | % Recoveryb
|
|||
|---|---|---|---|---|---|
| E. coli | Crypto- sporidium | T1 phage | PP7 phage | ||
| Lake Erie | 56.2 | 87 | 45 | 61 | 55 |
| Hetch Hetchy | 1.4 | 97 | 19 | 60 | 61 |
| Charleroi | 10.4 | 89 | 38 | 74 | 65 |
| Nottingham | 4.4 | 94 | 19 | 68 | 71 |
| Cobb County | 9.4 | 91 | 37 | 31 | 62 |
| Mean | 91.6 (4.0) | 31.6 (11.9) | 58.8 (16.5) | 62.8 (5.8) | |
Water samples were inoculated with levels of microbes similar to those shown in Table 3.
SD are given in parentheses.
DISCUSSION
In previous studies, the ultrafiltration process to recover and detect viruses (11, 26) and protozoa (8, 9) from environmental surface waters has been optimized. In this study, we took a similar approach in first optimizing for the ultrafiltration recovery of representative enteric bacteria and then optimizing the procedure to simultaneously concentrate all three groups of pathogens.
The addition of Tween 80 to the initial suspension resulted in an increase in the recovery of bacteria, principally serovar Enteritidis. Previous work has shown that Tween 80 stabilizes virus and improves elution (9, 11), and the nonionic detergent probably prevented the adhesion of cells to the ultrafilter membrane surface in our study. However, though serovar Enteritidis recoveries doubled in the presence of Tween 80, recoveries remained highly variable, especially in contrast to the consistent recoveries of the two E. coli strains used.
There was little difference between E. coli strain K-12 recovery rates from 2 liters (84%) or 10 liters (87%) of deionized water. In order to track the recovery of the E. coli introduced into Rio Grande samples, which we have shown to harbor significant levels of coliforms (G. B. Smith, unpublished data), we used an antibiotic-resistant strain of E. coli and found recovery of this strain to be very similar to that of the K-12 strain. Interestingly, in comparisons of recoveries from groundwater (turbidity, 0.3 NTU) and Rio Grande (turbidity, 29.2 NTU) samples, water turbidity had little or no effect on the recoveries of strain XL1-Blue. In a final optimization test for bacterial recoveries, it was found that lowering the input numbers of E. coli by two orders of magnitude had no effect on the recovery percentage (92%) from the Rio Grande samples. Similarly, in a previous study, different concentrations of protozoa in water samples did not influence recovery efficiencies of Cryptosporidium (9).
When we introduced representative viruses, bacteria, and protozoa simultaneously into water with higher turbidity, specifically, the Rio Grande surface water samples, bacterial recoveries remained greater than 90%, while the recoveries of viruses (46 to 59%) and C. parvum (54%) were consistent with recoveries observed previously (8, 9, 26). Other studies have examined the use of ultrafiltration to concentrate a single type of microorganism from water (7, 8, 19). However, there are few studies that describe the successful recovery of multiple organisms from a single water sample. For example, Juliano and Sobsey (6) used raw water and a 10-liter disposable hollow-fiber ultrafilter and reported recovery efficiencies of 34% for viruses, 27% for E. coli, and 64% for C. parvum. The recoveries reported here are similar to their results for C. parvum but higher for E. coli and viruses. In addition, constant recoveries were observed from different environmental water samples. In contrast to previous work (6, 19), the present procedure took only 45 min to filter multiple target pathogens from higher-turbidity water.
Water samples from widely different geographical areas were tested to determine whether differences in water quality may affect recovery efficiency. Some of these sites were selected because low recovery efficiencies for C. parvum have been reported (3). Compared to C. parvum recoveries from the Rio Grande samples, which ranged from 50 to 55%, the recoveries from the other surface waters with a history of poor oocyst recovery ranged from 19.0 to 44.5%. Interestingly, the worst oocyst recoveries (19%) were from the two water sources having the lowest turbidities, and oocyst recoveries from both of these samples were also poor in other studies (3, 8, 9). In contrast to the variable recoveries of C. parvum, recoveries of bacteria (92%) and viruses (59 and 63%) from these water sources remained high and consistent with recoveries from the Rio Grande samples.
The disinfection of the membrane before and after each filtration is particularly important for bacteria because of their potential for rapid reproduction under diverse environmental conditions. The disinfection procedure outlined here allows for multiple reuses of the ultrafilter; we have commonly reused one filter more than 40 times. Though the sanitation and disinfection process is time-consuming, sampling on a daily basis is feasible when multiple filters are maintained.
The procedure we have outlined here, based on previously published ultrafiltration procedures for concentrating viruses (26) and protozoa (8), has demonstrated the feasibility of simultaneously recovering viral, bacterial, and protozoan pathogens and therefore represents an important contribution to rapid, consistent detection procedures currently needed to protect water supplies.
Acknowledgments
This work was supported by Southwest Consortium for Environmental Research and Policy (SCERP) and Paso Del Norte Health Foundation.
REFERENCES
- 1.Bukhari, Z., R. M. McCuin, C. R. Fricker, and J. C. Clancy. 1998. Immunomagnetic separation of Cryptosporidium parvum from source water samples of various turbidities. Appl. Environ. Microbiol. 64:4495-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 5:376-378. [Google Scholar]
- 3.Connel, K., J. Scheller, K. Miller, and C. C. Rodgers. 2000. Performance of methods 1622/23 in the ICR supplemental surveys. In Proceedings of the Water Quality Technology Conference 2000. American Water Works Association, Denver, Colo. [CD-ROM.]
- 3a.Eaton, A. D., L. S. Clesceri, and A. E. Greenberg (ed.). 1995. Standard methods for the examination of water and wastewater. American Public Health Association, Washington, D.C.
- 4.Gerba, C. P., S. R. Farrah, S. M. Goyal, C. Wallis, and J. L. Melnik. 1978. Concentration of enteroviruses from large volumes of tap water, treated sewage, and seawater. Appl. Environ. Microbiol. 35:540-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guttman-Bass, N., and J. Catalano-Sherman. 1985. Effects of humic materials on virus recovery from water. Appl. Environ. Microbiol. 49:1260-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juliano, J., and M. D. Sobsey. 1998. Simultaneous concentration of Cryptosporidium, bacteria, and viruses from water by hollow-fiber ultrafiltration. In Proceedings of the Water Quality Technology Conference 1998. America Water Works Association, Denver, Colo. [CD-ROM.]
- 7.Kfir, R., C. Hilner, M. Du Preez, and B. Bateman. 1995. Studies evaluating the applicability of utilizing the same concentration techniques for the detection of protozoan parasites and viruses in water. Water Sci. Technol. 31:417-423. [Google Scholar]
- 8.Kuhn, R. C., and K. H. Oshima. 2001. Evaluation and optimization of a reusable hollow fiber ultrafilter as a first step in concentrating Cryptosporidium parvum oocysts from water. Water Res. 35:2779-2783. [DOI] [PubMed] [Google Scholar]
- 9.Kuhn, R. C., and K. H. Oshima. 2002. Hollow-fiber ultrafiltration of Cryptosporidium parvum oocysts from a wide variety of 10-L surface water samples. Can. J. Microbiol. 48:542-549. [DOI] [PubMed]
- 10.Lopez-Pila, J. M., H. Dizer, and D. Wolfgang. 1996. Wastewater treatment and elimination of pathogens: new prospects for an old problem. Microbiologia 12:525-536. [PubMed] [Google Scholar]
- 11.Olszewisky, J., L. Winona, and K. H. Oshima. 2001. Hollow-fiber ultrafiltration to concentrate viruses from environmental waters. In Proceedings of the Water Quality Technology Conference 2001. American Water Works Association, Denver, Colo. [CD-ROM.]
- 12.Oshima, K. H., T. T. Evans-Strickfaden, A. K. Highsmith, and E. W. Ades. 1995. The removal of phages T1 and PP7 and poliovirus from fluids with hollow-fiber ultrafilters with molecular weight cutoffs of 50,000, 13,000 and 6,000. Can. J. Microbiol. 41:316-322. [DOI] [PubMed] [Google Scholar]
- 13.Payment, P., A. Berube, D. Perreault, R. Armon, and M. Trudel. 1989. Concentration of Giardia lamblia cysts, Legionella pneumophila, Clostridium perfringens, human enteric viruses, and coliphages from large volumes of drinking water, using a single filtration. Can. J. Microbiol. 35:932-935. [DOI] [PubMed] [Google Scholar]
- 14.Rochelle, P. A., R. De Leon, A. Johnson, M. H. Stewart, and R. L. Wolfe. 1999. Evaluation of immunomagnetic separation for recovery of infectious Cryptosporidium parvum oocysts from environmental samples. Appl. Environ. Microbiol. 65:841-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sartory, D. P., A. C. Parton, J. Roberts, and K. Bergmann. 1998. Recovery of Cryptosporidium oocysts from small and large volume water samples using a compressed foam filter system. Lett. Appl. Microbiol. 27:318-322. [DOI] [PubMed] [Google Scholar]
- 16.Sefcova, H. 1998. Hygiene aspects of drinking water ultrafiltration. Cent. Eur. J. Public Health 6:314-316. [PubMed] [Google Scholar]
- 17.Shields, P. A., T. F. Ling, V. Tjatha, D. O. Shah, and S. R. Farrah. 1986. Comparison of positively charged membrane filters and their use in concentrating bacteriophage in water. Water Res. 20:145-151. [Google Scholar]
- 18.Simmons, O., J. J. Juliano, C. D. Heaney, D. S. Francy, F. W. Schaefer III, and M. D. Sobsey. 1999. Comparison of filtration methods for primary recovery of Cryptosporidium parvum from water. In Proceedings of the International Symposium on Waterborne Pathogens. American Water Works Association, Denver, Colo. [CD-ROM.]
- 19.Simmons, O. D., III, M. D. Sobsey, C. D. Heaney, F. W. Schaefer III, and D. S. Francy. 2001. Concentration and detection of Cryptosporidium oocysts in surface water samples by method 1622 using ultrafiltration and capsule filtration. Appl. Environ. Microbiol. 67:1123-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, G. B., and J. M. Tiedje. 1992. Isolation and characterization of a nitrite reductase gene and its use as a probe for denitrifying bacteria. Appl. Environ. Microbiol. 58:376-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobsey, M. D. 1976. Methods for detection of enteric viruses in water and wastewater, p. 89-127. In G. Berg, H. L. Bodily, E. H. Lennette, J. L. Melnick, and T. G. Metcalf (ed.), Viruses in water. American Health Association, Washington, D.C.
- 22.Swabby-Cahill, K. D., G. W. Clark, and A. R. Cahill. 1998. Adherence of Giardia cysts and Cryptosporidium oocysts to plastic and glass. In Proceedings of the Water Quality Technology Conference 1998. American Water Works Association, Denver, Colo. [CD-ROM.]
- 23.Tutunjian, R. 1983. Ultrafiltration process in biotechnology. Ann. N. Y. Acad. Sci. 413:238-253. [DOI] [PubMed] [Google Scholar]
- 24.U.S. Environmental Protection Agency. 1999. Method 1622: Cryptosporidium in water by filtration/IMS/FA. EPA 821-R-99-01. Office of Water, U.S. Environmental Protection Agency, Washington, D.C.
- 25.Venczel, L. V., M. Arrowood, M. Hurd, and M. D. Sobsey. 1997. Inactivation of Cryptosporidium parvum oocysts and Clostridium perfringens spores by a mixed-oxidant disinfectant and by free chlorine. Appl. Environ. Microbiol. 63:1598-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winona, L. J., A. W. Ommani, J. Olszewsky, J. B. Nuzzo, and K. H. Oshima. 2001. Efficient and predictable recovery of viruses from water by small scale ultrafiltration systems. Can. J. Microbiol. 47:1033-1041. [PubMed] [Google Scholar]



