Abstract
Norwalk virus and other human caliciviruses (noroviruses) are major agents of gastroenteritis, and water is a major route of their transmission. In an effort to control Norwalk virus in drinking water, Norwalk virus reduction by bench-scale ozone disinfection was determined using quantitative reverse transcription (RT)-PCR for virus assays. Two other enteric viruses, poliovirus 1 and coliphage MS2, were included for comparison, and their reductions were assayed by infectivity assays as well as by RT-PCR. Virus reductions by ozone were determined using a dose of 0.37 mg of ozone/liter at pH 7 and 5°C for up to 5 min. Based on two RT-PCR assays, the reductions of Norwalk virus were >3 log10 within a contact time of 10 s, and these were similar to the reductions of the other two viruses determined by the same assay methods. Also, the virus reductions detected by RT-PCR assays were similar to those detected by infectivity assays, indicating that the RT-PCR assay is a reliable surrogate assay for both culturable and nonculturable viruses disinfected with ozone. Overall, the results of this study indicate that Norwalk virus as well as other enteric viruses can be reduced rapidly and extensively by ozone disinfection and that RT-PCR is a useful surrogate assay for both culturable and nonculturable viruses disinfected with ozone.
Norwalk virus (NV) and other human caliciviruses (noroviruses) are major agents of epidemic gastroenteritis, and water is an important route of their transmission (5). These viruses are also suspected to be important agents of endemic gastroenteritis caused by fecally contaminated drinking water (8). Furthermore, a previous study suggested that NV is very resistant to free-chlorine disinfection (6) because a virus suspension in water was still infectious after administration of a dose of 3.75 mg of free chlorine/liter and a contact time of 30 min. Poliovirus and rotaviruses were completely inactivated at the same disinfection condition. Considering the apparent high infectivity (low infectious dose) and widespread occurrence in the population, the resistance of NV to chlorination could pose a high risk to the public who are served by conventionally treated, chlorinated drinking water.
Because NV cannot be grown or assayed for infectivity in any known laboratory host (5), reverse transcription (RT)-PCR is the only sensitive and specific assay system currently available. However, it was not certain whether the RT-PCR assay could accurately predict the loss of virus infectivity by disinfection. Therefore, two other enteric viruses, poliovirus 1 (PV1) and coliphage MS2, were included in this study to allow comparison of RT-PCR assay data with infectivity assay data for these viruses and thereby determine whether the assays provide equivalent information. Also, these two viruses have been widely used as indicator viruses for disinfection efficiency (13). A previous study reported that RT-PCR, especially RT-PCR for small (<300 nucleotides) targets, may not reliably quantify virus infectivity because the RNA of inactivated viruses may be amplified (14). To increase the agreement between virus reduction data obtained with infectivity assays and that obtained with RT-PCR assays, new primer sets that can amplify larger regions of viral RNA genomes were developed and used in this study. This approach was used to increase the size of the genomic target sites examined for ozone-induced damage to genomic RNA that would prevent successful amplification.
Ozone has been used for decades as a primary disinfectant for drinking water in many European countries and received more interest in other countries after the discovery of potentially harmful by-products of chlorine disinfection. Ozone is a very strong oxidant and an effective disinfectant against most waterborne pathogens (13). In fact, most enteric viruses studied, such as rotaviruses, parvoviruses, and hepatitis A virus, are substantially inactivated by ozone (with CT99 [concentration × exposure time to achieve 99% inactivation of a microorganism] values being much less than 1 mg/liter · min). Despite its growing use and proven effectiveness against other waterborne pathogens, ozone has not been studied for its ability to disinfect NV or other human caliciviruses. Therefore, the objectives of this study were to determine the NV reduction caused by bench-scale (semibatch) ozone disinfection in buffered, oxidant demand-free water by using quantitative RT-PCR for virus assays and to compare this reduction with those of two other important public health-related viruses previously studied for their inactivation by ozone.
MATERIALS AND METHODS
Viruses and preparation.
The prototype NV strain 8FIIa was obtained in stool samples taken from infected human volunteers and stored at −80°C. The samples were then suspended in phosphate-buffered saline (10 to 20% [wt/vol]) and extracted by homogenization in an equal volume of chloroform. The supernatant was recovered by low-speed (5,000 × g) centrifugation for 20 min at 4°C. PV1 strain LSc was propagated in Buffalo green monkey kidney (BGMK) cells. Infected-cell lysates were frozen and thawed three times and extracted by homogenization in an equal volume of chloroform. The supernatant was recovered by low-speed (5,000 × g) centrifugation for 20 min at 4°C. F-specific coliphage MS2 was grown in Escherichia coli C3000 by the double-agar-layer plaque technique (12). The top agar layer having confluent lysis of the host cells was harvested by scraping into a small amount of phosphate-buffered saline, and the virus was extracted with an equal volume of chloroform. The supernatant was recovered by low-speed (4,000 × g) centrifugation for 30 min at 4°C. These virus extracts were further purified by centrifugal ultrafiltration (Centricon 100) and dispersed by serial filtration through 0.2- and 0.08-μm-pore-size polycarbonate filters pretreated with 0.1% Tween 80 solution in water.
Glassware and reagents.
Glassware for experiments was prepared by soaking in an ozone solution (>1 mg/liter) for 1 h, thorough rinsing with ozone demand-free (ODF) water, and baking in an oven for 2 h at 200°C. All solutions and buffers were made with ODF water that was generated by ozonating and purging Dracor reagent grade water as described in Standard Methods for the Examination of Water and Wastewater, 18th ed. (1).
Ozone generation and delivery and measurement of residual ozone.
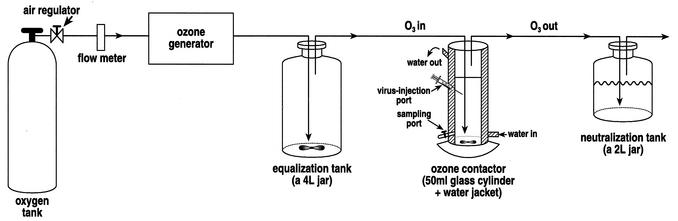
Ozone was generated by passing dry oxygen gas through a corona discharge type ozone generator (Sander Ozonizer model 200), and the generated ozone was directed successively through an equilibrium tank, the ozone contactor, and an ozone destruction system (Fig. 1). The flow rate was ∼50 ml/min, and the ozone dosage was adjusted by a valve on the generator. Residual ozone in water was determined by the Indigo method as described in Standard Methods (1).
FIG. 1.
Ozonation apparatus. Ozone was generated by passing dry O2 gas through a corona discharge type ozone generator (Sander Ozonizer model 200), and the generated ozone was directed successively through an equilibrium tank, the ozone contactor, and an ozone destruction system. The flow rate was ∼50 ml/min, and the ozone dosage was adjusted by a valve on the generator.
Experimental protocol.
Ozonation of test viruses in water was done in a specially constructed reactor consisting of a water-jacketed, borosilicate glass graduated cylinder fitted with a syringe port for injection of virus suspension and with gravity feed sampling ports. The reactor contents of 0.01 M ODF phosphate buffer (pH 7; 30 to 35 ml) were magnetically stirred and maintained at 5°C by circulating chilled water through the reactor water jacket. Ozone was supplied continuously to the reactor through a fritted glass bubble diffuser tube to achieve the target concentration in the contactor. The concentration of ozone inside the reactor was controlled by the gas flow and the voltage of the generator. The time needed to reach a steady state of ozone concentration in a batch of test water in the reactor varied from experiment to experiment but was usually between 20 to 30 min. The virus disinfection experiments were done after water conditioning with ozone for at least 60 min. After a steady-state ozone concentration was reached, and with ozone continuing to be delivered to the test water in the reactor, a virus suspension (3.3 to 3.7 ml) was injected into the reactor through the syringe port at zero time. Prior to real disinfection experiments, NaCl tracer tests were done to estimate the time needed to achieve complete mixing of injected materials inside the reactor. These experiments showed that it took ∼10 s to reach complete mixing of injected NaCl solution, and therefore the first sampling time for injected viruses was set at 10 s. Samples of 0.6 ml were withdrawn from the reactor via the gravity outlet into vials, which contained a volume of 0.1% sterile sodium thiosulfate or ODF phosphate buffer equal to the collected sample volume. These samples were collected at various times following virus injection to determine remaining virus (10, 30, 60, and 600 s) or ozone concentrations (20, 90, 180, and 330 s) as a function of contact time. Viruses were assayed by both cell culture assays and two RT-PCR assays, short target (ST) and long target (LT). Ozone concentration was determined by the Indigo method as described in Standard Methods (1).
PCR primers.
The oligonucleotide primers used in this study for ST RT-PCR for NV, PV1, and MS2 have been previously described (2, 10, 11). The oligonucleotide primers for LT RT-PCR for the viruses were developed for this study. The highly conserved RNA polymerase region of the NV was used as the target for the synthesis of 1,024 bp of NV cDNA (5′ primer, 5′-CCG GAG TAT ATG AGC CAG CAT; 3′ primer, 5′-CCT CAC TTG TAT TGG TCC TCC TTC TGT T-3′). Also, the highly conserved RNA polymerase region of PV1 was used as the target for the synthesis of an 866-bp PV1 cDNA (5′ primer, 5′-CGA TCC CAG GCT TAA GAC AGA CTT TGA G-3′; 3′ primer, 5′-GGT AGG AAG CAA TTA CAT CAT CAC CAT AGG C-3′). The highly structured replicase region of coliphage MS2 was used as the target for the synthesis of a 494-bp MS2 cDNA (5′ primer, 5′-ATG AGG ATT ACC CAT GTC GAA G-3′; 3′ primer, 5′-TCC CTA CAA CGA GCC TAA ATT C-3′).
Infectivity assays.
PV1 was assayed by the plaque technique on confluent layers of BGMK cell cultures grown in 60-mm-diameter petri dishes, and coliphage MS2 was assayed by the double-agar-layer plaque technique on host E. coli C3000 as previously described (12).
RT-PCR assays. (i) ST RT-PCR.
Viruses were assayed by RT-PCR using a Perkin-Elmer RNA core kit as previously described (10). RT was performed at 42°C for 60 min by using Moloney murine leukemia virus reverse transcriptase with specific antisense primers for NV and PV1 or random hexamers for MS2. PCR was performed by using Taq polymerase with additional specific sense primers for NV and PV1 and both antisense and sense primers for MS2. A total of 40 cycles of PCR was carried out using the following program: denaturation for 1.5 min at 95°C, annealing for 1.5 min at 55°C, and extension for 1.5 min at 72°C. A 15-μl portion of RT-PCR product was analyzed by gel electrophoresis on 2% agarose, the electrophoresed gel was stained with ethidium bromide, and resolved PCR products were visualized by UV light using a transilluminator.
(ii) LT RT-PCR.
Viruses were assayed by RT-PCR using a Titan One Tube RT-PCR kit (Boehringer Mannheim Biochemicals) according to the manufacturer's instructions with some minor modifications. RT was performed at 50 to 55°C for 30 min by using avian myeloblastosis virus reverse transcriptase with specific antisense primers for NV, PV1, and MS2. PCR was performed by using a Taq-Pwo polymerase mixture with additional specific sense primers for each virus. A total of 35 cycles of PCR was carried out using the following program: first round (10 cycles), denaturation for 30 s at 94°C, annealing for 30 s at 55 to 60°C depending on the primers used, and extension for 1 min at 68°C; second round (25 cycles), denaturation for 30 s at 94°C, annealing for 30 s at 55 to 60°C depending on the primers used, extension for 1 min at 68°C with an additional 5 s for the second cycle, and a final elongation step at 68°C for 7 min. A 15-μl portion of RT-PCR product was analyzed by gel electrophoresis on 1.3% agarose, the electrophoresed gel was stained with ethidium bromide, and resolved PCR products were visualized by UV light using a transilluminator.
Data presentation.
Virus disinfection data, expressed as PFU per milliliter for PV1 and MS2 or PCR units per milliliter for NV as well as PV1 and MS2, are average values from either duplicate cultures or RT-PCR assays. For each experiment, the virus concentrations of the virus control samples at zero time were computed and expressed as N0, the initial virus concentration. For each test sample, the average concentration of each virus was computed. The proportion of initial viruses remaining at each test time (t) was computed for each virus by dividing the virus concentration at each test time (Nt) by the initial virus concentration (N0). These values were then log10 transformed [log10 (N/N0)], and the values from duplicate experiments were averaged. These mean data for log10 (Nt/N0) were then paired with the data for sampling time (t) and plotted.
RESULTS
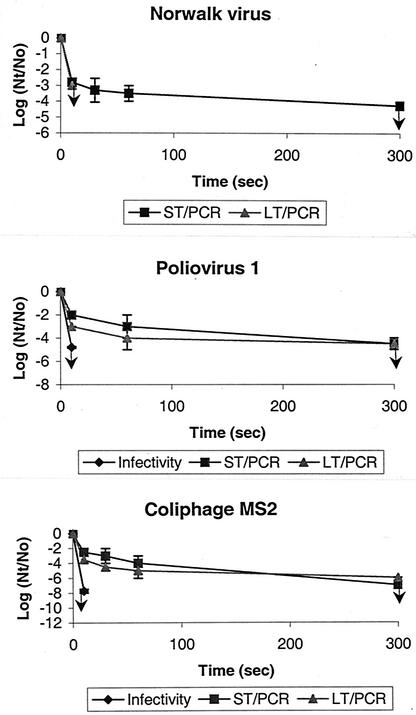
Figure 2 shows the reduction kinetics of the three viruses resulting from a 0.37-mg/liter dose of ozone in buffered ODF water at pH 7 and 5°C. Based on RT-PCR, viruses were reduced very rapidly during early contact times (2 to 4 log10 reductions by 10 s), but virus reduction was relatively slow during later contact times (0 to 1 log10 additional reductions by 60 s). Viruses were still reduced at a relatively low rate during later contact times (1 to 4 log10 additional reductions by 300 s). These disinfection kinetics are consistent with changes in the residual ozone concentrations in the (semibatch) disinfection system, which declined upon virus addition and then increased again as the ozone demand of the added virus suspensions was satisfied (Table 1, footnote a).
FIG. 2.
Virus inactivation by ozone (initial dose, 0.37 mg/liter; pH 7; 5°C). Arrows indicate the detection limits.
TABLE 1.
Reductions of NV, PV 1, and coliphage MS2 by ozone in buffered demand-free watera
| Virus | Virus reduction (log10 value)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Infectivity assay
|
ST PCR
|
LT PCR
|
||||||
| Expt 1 | Expt 2 | Mean | Expt 1 | Expt 2 | Mean | Expt 1 | Expt 2 | |
| NV | NAb | NA | NA | >4 | >4.5 | >3 | >3 | |
| PV 1 | >4.7 | >4.9 | 5 | 4 | 4.5 | >5 | 4 | |
| MS2 | >7.5 | >8.0 | >7 | >6 | >7 | >6 | ||
Ozone dose, 0.37 mg/liter; pH 7; 5°C; 5-min contact time. The residual ozone concentrations (mg/liter) at each sampling time were as follows: experiment 1, 0.38 (0 s), 0.14 (20 s), 0.03 (90 s), and 0.28 (330 s); experiment 2, 0.36 (0 s), 0.06 (20 s), 0.03 (90 s), and 0.16 (330 s).
NA, not applicable.
Table 1 shows the overall virus reductions by ozone at a dose of 0.37 mg/liter at pH 7 and 5°C after 5 min. Reductions of all viruses reached detection limits by both the infectivity and the two RT-PCR assays, except the ST PCR for PV1, within a contact time of 5 min. The lower detection limits of log10 virus reductions based on ST and LT PCR assays approached those of infectivity assays for PV1 and MS2. Although the rate and extent of infectivity reduction were greater than the rate and extent of reduction of viral genomic RNA as detected by the two RT-PCR assays, virus reductions based on LT PCR assays were generally 1 log10 higher than those based on ST PCR assays.
DISCUSSION
NV appears to be at least as sensitive to a 0.37-mg/liter dose of ozone as the other viruses tested, PV1 and male-specific RNA coliphage MS2. The reduction of NV detected by LT PCR was >3 log10 after a contact time of 10 s, which is at least similar to that of PV1 (3 log10) and MS2 (3.5 log10). Also, the reduction of NV detected by ST PCR was ∼3 log10 after a contact time of 10 s, which is greater than that of PV1 (2 log10) and MS2 (2.5 log10).
The reductions of PV1 and MS2 based on both ST and LT PCR assays corresponded relatively well with those based on infectivity assays in the initial phase of inactivation, where first-order reduction kinetics can be followed. This may be because the mechanism of virus inactivation is the oxidative degradation that occurs rapidly under these ozonation conditions and also perhaps because viral nucleic acid is a key and perhaps primary target of ozone. Kim et al. (7) observed that RNA of bacteriophage f2 is released from phage particles after the phage capsid is degraded into protein subunits by ozonation. They suggested that ozone alters the protein capsid first to liberate RNA and that the naked RNA may be secondarily inactivated by ozone. They also reported this phenomenon to be concentration dependent, with degradation of virus capsids occurring faster at higher dosages of ozone. However, Roy et al. (9) reported that the rate-determining step in inactivation of poliovirus was a mass transfer diffusional process and suggested that the primary mode of inactivation by ozone appears to be nucleic acid damage instead of viral capsid damage. Therefore, it is possible that the agreement between loss of infectivity and loss of RT-PCR titers in this study resulted because an important and perhaps primary target of ozone is the viral nucleic acid. In any event, the results of this study suggest that RT-PCR is a useful infectivity surrogate assay for nonculturable viruses in ozone disinfection studies, especially at higher doses of ozone, as was the case in this study.
Finally, the results of this study indicate that NV contamination of drinking water can be controlled by adequate ozone disinfection practice. The reduction of NV by a 0.37-mg/liter dose of ozone was at least 3 log10 by 10 s based on both ST and LT PCR and >4 log10 in 5 min based on ST PCR. Comparing the reductions of viruses by ozone in these semibatch experiments with reported virus reductions by ozone in previous simple batch experiments is difficult because the ozone exposure and reaction conditions are different. However, NV appears to be no more resistant to ozone than are other human enteric viruses such as rotavirus (15) and hepatitis A virus (4). (The reductions of PV1 and MS2 detected by infectivity assay were more than 5 and 7 log10, respectively, with the same dosage of ozone in 10 s.) Therefore, the treatment requirement for virus removal from drinking water of 4 log10 specified in the Surface Water Treatment Rule (3) appears to be readily achievable by adequate ozonation in the case of NV contamination as well as contamination by other enteric viruses such as enteroviruses, hepatitis A virus, and rotaviruses.
Acknowledgments
This research was funded by the National Water Research Institute (HRA 699-512-92 and WQI 699-527-95).
REFERENCES
- 1.American Public Health Association. 1992. Standard methods for the examination of water and wastewater, 18th ed. American Public Health Association, Washington, D.C.
- 2.De Leon, R., S. M. Matsui, R. S. Baric, J. E. Herrmann, N. R. Blackow, H. B. Greenberg, and M. D. Sobsey. 1992. Detection of Norwalk virus in stool specimens by reverse transcriptase-polymerase chain reaction and nonradioactive oligoprobes. J. Clin. Microbiol. 30:3151-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Federal Register. 1989. Drinking water; national primary drinking water regulations; filtration, disinfection; turbidity, Giardia lamblia, viruses, Legionella, and heterotrophic bacteria. 54:27486-27526.
- 4.Hall, R. M., and M. D. Sobsey. 1993. Inactivation of hepatitis A virus and MS2 by ozone and ozone-hydrogen peroxide in buffered water. Wat. Sci. Technol. 27:371-378. [Google Scholar]
- 5.Kapikian, A. Z., M. K. Estes, and R. M. Chanock. 1996. Norwalk group of viruses. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, New York, N.Y.
- 6.Keswick, B. H., T. K. Satterwhite, P. C. Johnson, H. L. Dupont, S. L. Secor, J. A. Bitsula, G. W. Gary, and J. C. Hoff. 1985. Inactivation of Norwalk virus in drinking water by chlorine. Appl. Environ. Microbiol. 50:261-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim, C. K., D. M. Gentile, and O. J. Sproul. 1980. Mechanism of ozone inactivation of bacteriophage f2. Appl. Environ. Microbiol. 39:210-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Payment, P., L. Richardson, J. Siemiatycki, R. Dewar, M. Edwardes, and E. Franco. 1991. A randomized trial to evaluate the risk of gastrointestinal disease due to consumption of drinking water meeting current microbiological standards. Am. J. Public Health 81:703-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy, D., P. K. Y. Wong, R. S. Engelbrecht, and E. S. K. Chian. 1981. Mechanism for enteroviral inactivation by ozone. Appl. Environ. Microbiol. 41:718-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwab, K. J., R. De Leon, and M. D. Sobsey. 1995. Concentration and purification of beef extract mock eluates from water samples for the detection of enteroviruses, hepatitis A virus, and Norwalk virus by reverse transcription-PCR. Appl. Environ. Microbiol. 61:531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin, G., and M. D. Sobsey. 1998. Reduction of Norwalk virus, poliovirus 1, and coliphage MS2 by monochloramine disinfection of water. Wat. Sci. Technol. 38:151-154. [Google Scholar]
- 12.Sobsey, M. D., T. Fuji, and P. A. Shields. 1988. Inactivation of hepatitis A virus and model viruses in water by free chlorine and monochloramine. Wat. Sci. Technol. 20:385-391. [Google Scholar]
- 13.Sobsey, M. D. 1989. Inactivation of health-related microorganisms in water by disinfection processes. Wat. Sci. Technol. 21:179-195. [Google Scholar]
- 14.Sobsey, M. D., D. A. Battigelli, G. Shin, and S. Newland. 1998. RT-PCR amplification detects inactivated viruses in water and wastewater. Wat. Sci. Technol. 38:91-94. [Google Scholar]
- 15.Vaughn, J. M., Y.-S. Chen, K. Lindberg, and D. Morales. 1987. Inactivation of human and simian rotaviruses by ozone. Appl. Environ. Microbiol. 53:2218-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]