Abstract
Nuclear receptors are transcription factors that require multiple protein–protein interactions to regulate target gene expression. We have cloned a 27-kDa protein, termed NIX1 (neuronal interacting factor X 1), that directly binds nuclear receptors in vitro and in vivo. Protein–protein interaction between NIX1 and ligand-activated or constitutive active nuclear receptors, including retinoid-related orphan receptor β (RORβ) (NR1F2), strictly depends on the conserved receptor C-terminal activation function 2 (AF2-D). NIX1 selectively binds retinoic acid receptor (RAR) (NR1A) and thyroid hormone receptor (TR) (NR1B) in a ligand-dependent manner, but does not interact with retinoid X receptor (RXR) (NR2B) or steroid hormone receptors. Interestingly, NIX1 down-regulates transcriptional activation by binding to ligand-bound nuclear receptors. A 39-aa domain within NIX1 was found to be necessary and sufficient for protein–protein interactions with nuclear receptors. Northern blot analysis demonstrates low-abundance RNA messages only in brain and neuronal cells. In situ hybridization and immunohistochemistry revealed that NIX1 expression is restricted to the central nervous system and could be confined to neurons in the dentate gyrus of the hippocampus, the amygdala, thalamic, and hypothalamic regions. In summary, protein–protein interactions between the neuronal protein NIX1 and ligand-activated nuclear receptors are both specific and selective. By suppressing receptor-mediated transcription, NIX1 implements coregulation of nuclear receptor functions in brain.
Keywords: ligand-binding domain, activation function, ligand-dependent interacting protein, brain-specific expression
Nuclear receptors and steroid hormone receptors form a superfamily of sequence-specific transcription factors that regulate diverse biological processes, including cell growth and differentiation, development, homeostasis, and various organ functions in the adult, by stimulating or repressing target gene expression (for review see ref. 1). They all share a common modular structure composed of several domains. Receptor dimerization, ligand binding, repression in the absence of ligand, and ligand-dependent transactivation are mediated by the C-terminal region of nuclear receptors termed the ligand-binding domain (LBD). Crystal structures of unliganded (apo-) and liganded (holo-) LBDs reveal that ligand binding induces several conformational changes within the LBD structure, including the positional reorientation of the C-terminal helix 12 (2–5). Helix 12 contains the core motif of the activation function 2 (AF2-D) and is indispensable for transcriptional activation by nuclear receptor LBDs (6).
Intense study has focused on the molecular mechanisms of transcriptional regulation by nuclear receptors, especially on ligand-dependent protein–protein interactions occurring between the receptors and additional factors, designated corepressors and coactivators (for review see ref. 7).
Transcriptional repression mediated by unliganded nuclear receptors has been ascribed to the binding of corepressors N-CoR or SMRT (8, 9). Both N-CoR and SMRT recruit mSin3a, c-Ski, and histone deacetylases, thus forming multisubunit repressor complexes (10, 11).
Transcriptional activation by liganded nuclear receptors requires a functional AF2-D. Several related proteins of 160 kDa named SRC1 (12, 13), TIF2/GRIP1 (14, 15), AIB1/ACTR/p/CIP (16–18), and the bridging protein factor CBP/p300 (12, 19) have been shown to associate with nuclear receptors in an AF2-D-dependent fashion and augment the receptor's transcriptional activity. Recent findings support the notion that histone acetylation increases transcriptional activity (for reviews see refs. 20 and 21). SRC1, ACTR/p/CIP, and p300/CBP have been demonstrated to recruit and/or harbor intrinsic histone acetyltransferase activity (for review see ref. 7). Initial evidence for the physiological role of nuclear receptor cofactors has been provided by several studies: amplification and overexpression of AIB1 in many breast cancers suggest a role for AIB1 in tumorigenesis (16); a cold-inducible coactivator for PPARγ and thyroid hormone receptorβ (TRβ), named PGC1, has been shown to activate mitochondrial gene expression and thus could contribute to adaptive thermogenesis (22); targeted gene disruption of the SRC1 gene has been demonstrated to result in partial hormone resistance and compensatory expression of TIF2 mRNA (23). However, most studies on nuclear receptors and their cofactors reported to date indicate that their protein–protein interactions are promiscuous rather than selective. In addition, multiple cofactors are present in each cell, and their expression is neither tissue-specific nor cell type-restricted.
In this manuscript, we report the cloning of a 27-kDa neuronal-specific protein termed NIX1 (neuronal interacting factor X 1) that selectively down-regulates transcriptional activation by nuclear receptors. NIX1 directly binds to the holo-LBDs of distinct members of the NR1 and NR4 nuclear receptor subfamilies (24), and mutations within NIX1 define the platform for the protein–protein interaction with the holo-LBDs of nuclear receptors. In summary, selectivity and specificity of the protein–protein interaction combined with the highly restricted expression pattern imply that NIX1 regulates nuclear receptor functions in brain.
Materials and Methods
Yeast Two-Hybrid Screen and Isolation of NIX1 cDNA Clones.
A cDNA library derived from mouse brain poly(A)+ RNA in pGAD10 (CLONTECH) was screened for retinoid-related orphan receptor β (RORβ)-interacting proteins according to the manufacturer's instruction. Saccharomyces cerevisiae HF7c strain [MATa, ura3-52, his3-200, lys2-801, ade2-101, trp1-901, leu2-3,112, gal4-542, gal80-538, LYS2∷GAL1-HIS3, URA3∷(GAL4 17-mers)3-CYC1-lacZ] expressing Gal4 DNA-binding domain (DBD) fusion with RORβ76–459 was used for yeast two-hybrid screening. Library plasmids encoding positive clones that express both HIS3 and lacZ reporters were rescued and retransformed into SFY526 [MATa, ura3-52, his3-200, lys2-801, ade2-101, trp1-901, leu2-3,112, canr , gal4-542, gal80-538, URA3∷GAL1-lacZ], together with the original bait or other constructs, for testing the specificity of protein–protein interaction. Among five different cDNAs, a single specific clone contained a Gal4 activation domain fusion with the sequence coding for NIX1 amino acids 61–229. Full-length NIX1 cDNA was cloned by 5′-rapid amplification of cDNA ends (RACE)-PCR from mouse brain poly(A)+ RNA. Detailed information on gene-specific primer selection, cDNA modification, and cycling conditions are available on request. The putative 5′ end of NIX1 cDNA was confirmed by screening a brain cDNA library derived from male BALB/c mice (λZAPII; Stratagene).
Construction of Recombinant Vectors.
Detailed description on the cloning of all of the eukaryotic and prokaryotic expression vectors for RORβ and NIX1 described in the manuscript are available on request. All generated plasmids were verified by sequencing. The CMX expression plasmids encoding the human estradiol receptor (hERα), human retinoic acid receptor α (hRARα), human retinoid X receptor α (hRXRα), and human TRβ were described previously (25). pRSETB was purchased from Invitrogen.
Expression and Purification of Recombinant Proteins.
Glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli BL21lys according to ref. 25. After bacterial lysis, GST fusion proteins were bound to glutathione-Sepharose beads (Amersham Pharmacia) and either used directly for GST pulldown experiments or purified. His-tagged proteins were expressed from pRSETB expression plasmids using identical procedures, and purified from E. coli on Talon metal affinity resin according to the manufacturer's protocol (CLONTECH).
In Vitro Protein–Protein Interaction Assay.
GST pulldown assays.
Recombinant cDNAs encoding RORβ, hRARα, hRXRα, hTRβ, and hERα were transcribed and translated in rabbit TNT-coupled reticulocyte lysate (Promega) with [35S]methionine following the manufacturer's instructions and incubated either with specific ligands [all-trans-retinoic acid (RA), 1 μM; 4-[(E)-2-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl]benzoic acid (TTNPB), 1 μM; LG153, 0.1 μM; 3,5,3′-triiodothyronine (T3), 1 μM; estradiol, 0.1 μM] or vehicle for 30 min at 4°C. GST pulldown assays were performed according to ref. 25.
Electrophoretic mobility-shift assays (EMSAs).
Detailed experimental procedures for gel retardation assays were described by Greiner et al. (6) and Pfitzner et al. (25). The following oligonucleotides and their complements were used: P0, 5′-TAAGTAGGTCATGACCTACTTA-3′; DR5, 5′-AGCTTCAGGTCACCAGGAGGTCA-GAG-3′; GALp, 5′-GATCTGCCGGAGGACTGTCCTCCGAGG-3′. The protein-DNA complex was separated on 5% or 8% nondenaturing polyacrylamide gels at 4°C in 0.25 × TBE (RORβ), or 0.5× TBE (RAR/RXR).
Northern Blotting and in Situ Hybridization.
Total RNA was prepared from cultured cells or several mouse tissues by guanidine isothiocyanate extraction. mRNA was isolated by separating polyadenylated RNA from total RNA using streptavidin-immobilized biotin-labeled oligo(dT)20 probe (Roche Molecular Biochemicals). The poly(A+) mRNA was subjected to Northern blot analysis according to standard procedures. Hybridization was performed by using 32P-labeled NIX1 cDNA and ExpressHyb solution from CLONTECH following the manufacturer's instructions. Digoxigenin-labeled sense and antisense ribonucleotide probes were generated from pBSSK NIX1 by T3 and T7 polymerase using digoxigenin-11-UTP (Roche Molecular Biochemicals). Preparation of tissue sections and in situ hybridization were performed according to ref. 26.
Cell Culture and Transient Transfection Experiments.
HT22 and NIH 3T3 cells were cultured in DMEM. Neuro2A cells were cultured in Earl's modified Eagle's medium. Both media were supplemented with 10% FCS, penicillin, streptomycin, and glutamine. Transient transfection assays were carried out in 12-well plates (4 × 104 cells per well) using the standard calcium phosphate coprecipitation technique (25) or N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate (DOTAP) lipofection (Roche Molecular Biochemicals) according to the manufacturer's protocol. Luciferase activity was assayed as recommended by the manufacturer (Promega) in a Luminometer ML 3000 (Dynatech). Relative light units were normalized according to ref. 25. All experiments were repeated at least five times.
Immunofluorescence Studies.
NIH 3T3 cells (5 × 104) were seeded on glass coverslips in 12-well plates and transfected the following day by lipofection using DOTAP mixed with 1 μg of DNA per well. Twenty-eight hours after transfection, cells were washed with PBS, fixed with 5% paraformaldehyde, and permeabilized in 0.2% Triton X-100. After blocking with 0.2% porcine gelatin, mouse anti-NIX1 IgG (1:120) was applied for 1 h. FITC-conjugated goat anti-mouse immunoglobulins (Dako) were used (1:600) as a secondary antibody. Nuclear stainings were performed by using propidium iodide/RNase before mounting the specimen for immunofluorescence microscopy.
Results
Identification and cDNA Cloning of NIX1.
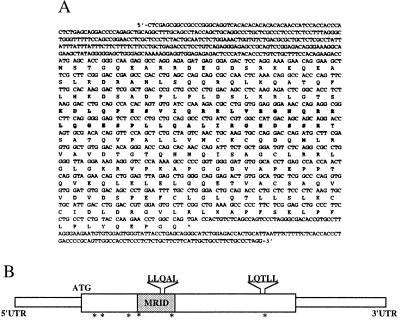
We used the yeast two-hybrid system to identify proteins interacting with the brain-specific nuclear orphan receptor RORβ. Five distinct cDNA clones with ORFs including a partial cDNA of 800 bp were identified. A full-length cDNA of the latter obtained by RACE-PCR and hybridization screening of a mouse brain λZAPII cDNA library was termed NIX1 cDNA. A 687-bp ORF without a consensus Kozak sequence surrounding the initiating AUG codon but with several in-frame stop codons within the 5′ untranslated region could be identified (Fig. 1).
Figure 1.
Analysis of NIX1 sequence. (A) The amino acid sequence of NIX1 has been submitted to the EMBL database. The minimal receptor interacting domain (amino acids 61–99) of NIX1 is highlighted (bold type). Note two nuclear receptor LXXLL interaction motifs (amino acids 87–91 and 192–196) and six consensus sites for phosphorylation by protein kinase C (amino acids 14–16, 21–23, 53–55, 59–61, 97–99, and 197–199). (B) Schematic representation of NIX1. Indicated are the minimal receptor interacting domain (gray box), the LXXLL motifs, and putative protein kinase C phosphorylation sites (asterisks).
The NIX1 cDNA encodes a protein of 229 amino acids with a predicted molecular mass of 27 kDa (Fig. 1). Sequence similarity searches reveal that NIX1 represents a protein with no homologs in any database except for a 429-bp clone reported in the expressed sequence tags (EST) database. However, several peptide motifs were recognized by performing prosite pattern searches including consensus sites for phosphorylation by protein kinase C.
NIX1 shares no similarity with protein domains of other nuclear receptor cofactors except for two nuclear receptor binding LXXLL motifs (L denotes leucine, X denotes any amino acid). Recent findings demonstrated LXXLL motifs to be critical for receptor–cofactor interactions (18, 27, 28). One LXXLL motif is found within the C terminus of NIX1 (amino acids 192–196), whereas the other is located in an opposite orientation within the central part of the protein (amino acids 87–91).
NIX1 Specifically Interacts with RORβ and Ligand-Activated Nuclear Receptors of the RAR and TR Subfamily in Vivo and in Vitro.
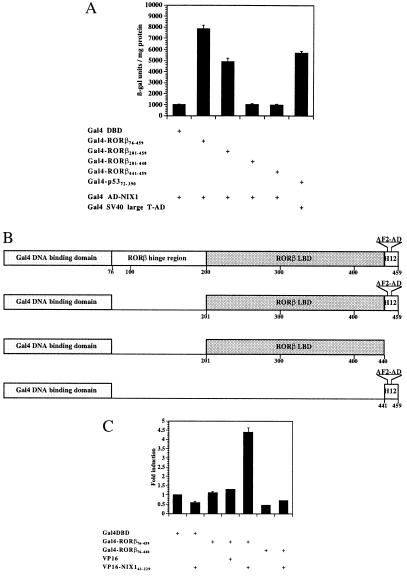
To quantify the in vivo interaction between NIX1 and RORβ observed in yeast, liquid β-galactosidase assays were performed. Various RORβ deletion mutants fused to the Gal4 DBD (GAL) were used as bait proteins, and full-length NIX1 linked to the Gal4 activation domain (NIX1-AD) was used as a prey (Fig. 2 A and B). According to the previously described transcriptional activation properties of RORβ (6), the AF2-D core motif of RORβ is required for the interaction with NIX1 in vivo but does not suffice. NIX1-AD associates with Gal4-RORβ201–459, thereby increasing β-galactosidase reporter gene activity 8-fold. Both Gal4-RORβ201–440 and Gal4-RORβ441–459 fail to interact with NIX1, which is reflected in basal β-galactosidase levels.
Figure 2.
Functional interaction between NIX1 and RORβ in vivo. (A) NIX1 interacts with RORβ in an AF2-D-dependent fashion. Plasmids expressing different deletion mutants of RORβ fused to the Gal4 DBD were introduced into the yeast reporter strain SFY526 together with NIX1 fused to the Gal4 activation domain. The transformants were grown in liquid dropout medium lacking tryptophan and leucine. Extracts were prepared and assayed for β-galactosidase activity, which is expressed in units per mg of protein. The stimulation in β-galactosidase activity resulting from the in vivo interaction between the Gal4-RORβ constructs and the NIX1-AD is indicated. Interaction between Gal4-p5373–708 and simian virus 40 large-T-AD served as an internal positive control. (B) Schematic representation of different RORβ deletion mutants fused to the Gal4 DBD domain. The AF2-D core (AF2-AD) motif is located at amino acid residues 445–451 within helix 12 (H12). (C) RORβ requires the AF2-D core for the interaction with NIX1 in mammalian cells. (GALp)3-TK LUC (1.25 μg) was cotransfected into CV1 cells with the indicated expression plasmids (200 ng each) coding for Gal4 DBD, Gal4 DBD fused to various RORβ deletion mutants, VP16 activation domain, and VP16-NIX1 amino acids 61–229. Fold induction averaged from seven independent experiments indicates the relative luciferase activity of the Gal4 DBD fusion proteins alone or in combination with the VP16 constructs over the Gal4 DBD vector.
In vivo interaction between NIX1 and various nuclear receptors, including RORβ, was further confirmed by a second independent method, the mammalian two-hybrid assay. Cotransfection of a VP16-NIX1 expression plasmid into CV1 cells increases the transcriptional activity of RORβ201–459 5-fold as compared with the control (Fig. 2C).
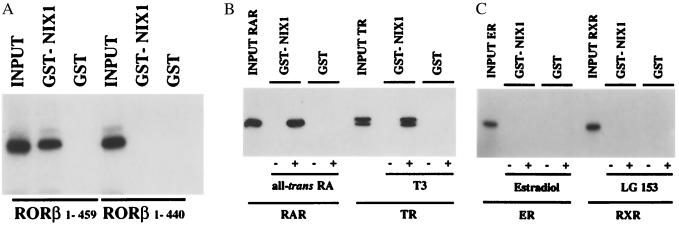
To investigate the interaction between NIX1 and nuclear receptors in vitro, GST pulldown experiments were performed with recombinant GST-NIX1 fusion proteins and various in vitro-translated [35S]methionine-labeled nuclear receptors. Fig. 3A demonstrates that full-length RORβ associates with GST-NIX1 in an AF2-D-dependent fashion, but fails to interact with the control GST protein. RORβ1–440 lacking the AF2-D core binds neither NIX1 nor the control GST protein, thus confirming that the specific interaction with NIX1 requires the AF2-D of RORβ in vitro. Additional GST pulldown experiments were addressed to test whether other nuclear receptors interact with NIX1 in a ligand- and AF2-D-dependent manner. Both RAR and TR only interact with NIX1 in the presence of their cognate ligands (Fig. 3B). Specific ligand-dependent interaction with NIX1 could also be detected for the vitamin D receptor and distinct nuclear receptors of the NR1 subfamily (data not shown). However, GST pulldown experiments show that NIX1 does not bind RXR either in the absence or in the presence of ligand (Fig. 3C). Steroid hormone receptors also fail to associate with GST-NIX1, as shown for the estrogen receptor in the presence and in the absence of estradiol (Fig. 3C). These results indicate that the ligand-dependent interaction with NIX1 is restricted to distinct nuclear receptors of the RAR and TR subfamily.
Figure 3.
Selective interaction between NIX1 and nuclear receptors in vitro. GST protein alone or NIX1 fused to GST was immobilized on glutathione-Sepharose beads, and GST-pulldown experiments were performed with in vitro translated [35S]methionine-labeled nuclear receptors and appropriate ligands or vehicles. Autoradiographs show NIX1-interacting nuclear receptors in comparison to 10% of the corresponding input. (A) Full-length RORβ in contrast to the AF2-D deletion mutant RORβ1–440 interacts with NIX1 in vitro. (B) NIX1 associates with RARα and TRβ in a ligand-dependent fashion. For RARα and TRβ, the ligands all-trans-retinoic acid (1 μM) and T3 (0.1 μM), respectively, were used. (C) ERα and RXRα fail to interact with NIX1 in the absence and in the presence of their ligands. For ERα, estradiol was used at 0.1 μM, for RXRα, LG153 was used at 1 μM.
NIX1 Interacts with DNA-Bound Nuclear Receptors.
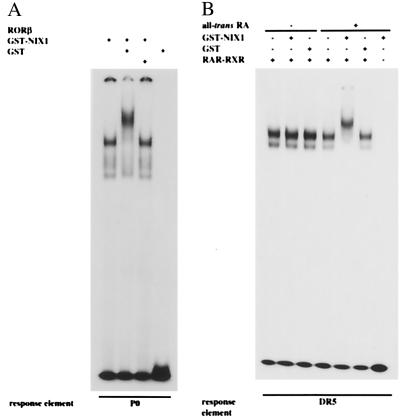
To establish ternary complex formation of DNA-bound nuclear receptors and NIX1, EMSAs were performed. DNA-protein complexes generated by incubating recombinant nuclear receptors with their radioactively labeled cognate response elements were challenged with recombinant NIX1. Fig. 4A illustrates that NIX1 specifically interacts with RORβ bound to a P0 response element, thus resulting in a clear supershift (Fig. 4A). To investigate the interaction with nuclear receptor heterodimers on DNA, NIX1 was incubated with RAR-RXR heterodimers bound to a DR5 retinoic acid response element. NIX1 specifically interacts with DNA-bound RAR-RXR heterodimers in a ligand-dependent fashion. Both the sequence of the response elements and the resulting heterodimeric organization are not decisive for the interaction with the cofactor (data not shown). Interestingly, NIX1 exclusively binds the DNA-protein complex in the presence of all-trans-retinoic acid (Fig. 4B) and other RAR agonists, but not in the presence of RAR antagonists or selective RXR agonists (data not shown). These data demonstrate that, on ligand-induced conformational changes, DNA-bound nuclear receptors associate with NIX1.
Figure 4.
NIX1 associates with DNA-bound nuclear receptors. EMSAs were performed by incubating purified recombinant nuclear receptor proteins with their 32P-labeled cognate response elements. DNA-protein complexes were subsequently challenged with recombinant GST alone or GST-NIX1 (see Materials and Methods for details). (A) His-tagged RORβ was incubated with the 32P-labeled palindromic P0 response element. Addition of recombinant GST-NIX1 (2 μg) in contrast to GST alone results in a clear supershift indicating ternary complex formation of DNA-bound RORβ and NIX1. GST-NIX1 by itself does not bind to the response element P0. (B) Purified recombinant GST-RARα and GST-RXRα were bound to the 32P-labeled cognate DR5 response element. Resulting heterodimeric receptor-DNA complexes are specifically retarded by GST-NIX1 only in the presence of all-trans-retinoic acid (1 μM), but not in the absence of ligand. GST-NIX1 does not bind the response element.
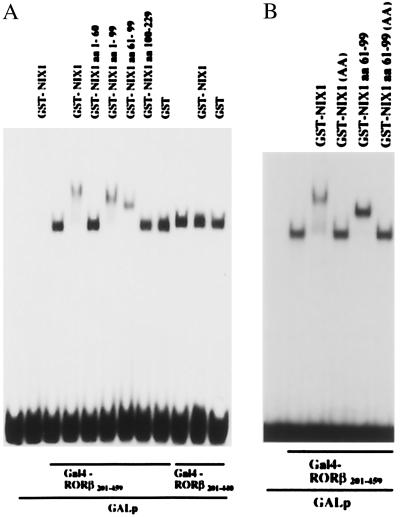
Identification of the Domain That Mediates the NIX1-RORβ Interaction.
To define the domains of NIX1 that mediate the interaction with nuclear receptors, different C- and N-terminal deletion mutants of NIX1 were generated and expressed as GST fusion proteins in E. coli. The EMSAs shown in Fig. 5A illustrate that deletion of the C-terminal amino acids 100–229 of NIX1 does not influence ternary complex formation. Conversely, GST-NIX1 amino acids 100–229 cannot bind the nuclear receptor although containing an LXXLL motif (amino acids 192–196). Further deletions identified a minimal protein fragment spanning the amino acids 61–99, that is both necessary and sufficient for the interaction between NIX1 and nuclear receptors such as RORβ (Fig. 5A) and RAR (data not shown). Interestingly, NIX1 amino acids 61–99 contain a nuclear receptor binding motif in an inverted orientation (LLQAL, amino acids 87–91) that might be critical for the receptor interaction. Replacement of both leucine 87 and leucine 88 with alanine in full-length NIX1 (NIX1 AA) or in NIX1 amino acids 61–99 produced proteins that fail to bind RORβ (Fig. 5B). These data indicate that the binding of nuclear receptors by NIX1 is mediated by the LLQAL motif within the minimal interaction domain spanning the amino acids 61–99.
Figure 5.
NIX1 contains a minimal receptor interacting domain spanning 39 amino acids. EMSAs were performed by incubating a 32P-labeled GALp response element with Gal4-RORβ201–459 or Gal4-RORβ201–440. Receptor-DNA complexes were challenged with purified GST, GST-NIX1, or various GST-NIX1 deletion mutants. (A) GST-NIX1 specifically interacts with DNA-bound Gal4-RORβ201–459, but does not bind to DNA alone or a DNA/Gal4-RORβ201–440 complex. Both GST-NIX1 amino acids 1–99 and GST-NIX1 amino acids 61–99 bind the receptor-DNA complex and mediate ternary complex formation comparable to GST-NIX1. GST alone, GST-NIX1 amino acids 1–60, or GST-NIX1 amino acids 100–229 do not associate with DNA-bound RORβ. (B) Gal4-RORβ201–459 bound to a 32P-labeled GALp oligonucleotide, was incubated with equal amounts of purified recombinant GST-NIX1 fusion proteins. Resulting protein-DNA complexes were separated on nondenaturing 6% polyacrylamide gels. GST-NIX1 and GST-NIX1 amino acids 61–99 specifically interact with DNA-bound RORβ. Replacement of both L87 and L88 with A in either full-length NIX1 or in NIX1 amino acids 61–99 resulted in proteins GST-NIX1 (AA) or GST-NIX1 amino acids 61–99 (AA), which do not associate with receptor-DNA complexes.
Neuronal-Specific Expression Pattern of NIX1.
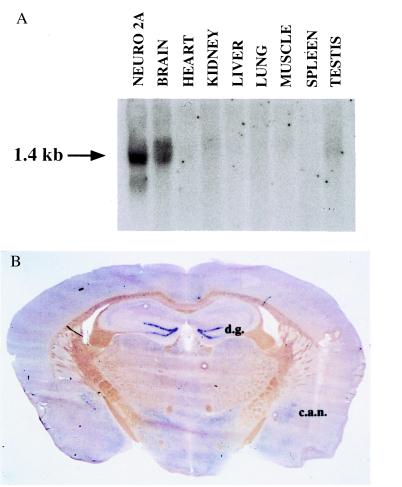
To investigate the expression pattern of NIX1, mRNA from distinct mouse tissues was isolated and subjected to Northern blot analysis. The Northern blot analysis in Fig. 6A identifies a 1.4-kb mRNA in brain and Neuro2A cells. NIX1 mRNA is not detected in heart, kidney, liver, lung, muscle, spleen, or testis, but is exclusively expressed in brain.
Figure 6.
Neuronal-specific expression of NIX1 mRNA. (A) Northern blot analysis of poly(A)+ RNA isolated from the indicated mouse tissues and Neuro2A cells. A total of 15.0 μg of poly(A)+ RNA was loaded in each lane. Hybridization using a 32P-labeled NIX1 cDNA probe identified a mRNA of approximately 1.4 kb. (B) In situ hybridization of NIX1 in mouse brain. NIX1 mRNA was exclusively found in neuronal cells. Digoxigenin-labeled anti-sense RNA was used to localize NIX1 mRNA in a coronal section to the dentate gyrus of the hippocampus (d.g.) and to the central nucleus of the amygdala (c.a.n.). NIX1 mRNA is also found in thalamic and hypothalamic regions, and in several brainstem nuclei.
To analyze the brain-specific expression of NIX1 mRNA in more detail, in situ hybridization studies were performed in mouse brain sections (Fig. 6B). In situ hybridization in a coronal section localizes significant expression of NIX1 mRNA to the dentate gyrus of the hippocampus (d.g.) and to the central nucleus of the amygdala (c.a.n.). NIX1 mRNA is also found in thalamic and hypothalamic regions and in several brainstem nuclei. NIX1 mRNA was exclusively found in neurons (data not shown). The data from the in situ hybridization experiments indicate that NIX1 mRNA is specifically expressed in distinct brain areas and can even be confined to subsets of neurons within these areas. Comprehensive studies on the localization of NIX1 expression will be described elsewhere.
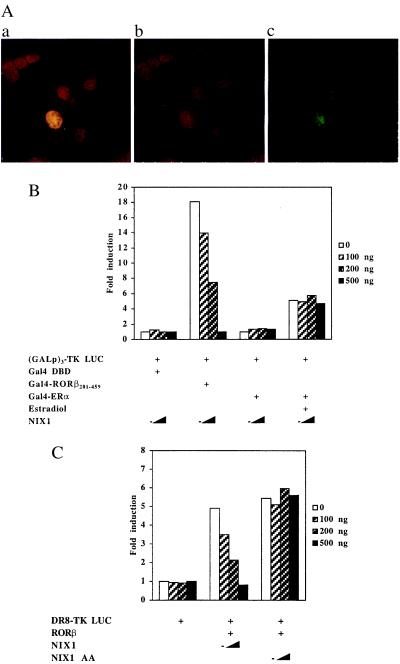
To visualize the localization of NIX1 protein within single cells, specific antibodies were raised and used in immunocytochemistry. Confocal laser microscopy images recorded in Fig. 7A reveal that NIX1 resides in the nucleus of transfected NIH 3T3 cells. NIX1 localization is indicated by the green fluorescence, whereas the cell nuclei are stained in red. Superimposing the images results in a yellow signal, indicating that NIX1 is located in the nucleus of the transfected fibroblasts.
Figure 7.
(A) NIX1 is located in the nucleus. NIH 3T3 fibroblasts and HT22 hippocampal cells were transfected with pCMX-NIX1. NIX1 immunodetection with affinity-purified anti-NIX1 antibody and FITC-labeled secondary antibody is shown in green. DNA staining by using propidium iodide and RNase is shown in red. The superimposed fluorescence microscopy images of both results in a yellow, nuclear stain. (B and C) NIX1 suppresses transactivation of distinct agonist-bound nuclear receptors. (B) (GALp)3-TK LUC was transfected into NIH 3T3 cells together with increasing amounts of pCMX-NIX1 (100, 200, and 500 ng) and expression plasmids (100 ng each) encoding for Gal4 DBD alone or the indicated Gal4 DBD-receptor fusions. For Gal4-ERα, transfected cells were treated with 0.1 μM estradiol or vehicle. (C) HT22 cells were transfected with expression plasmids (100 ng) for RORβ and increasing amounts pCMX-NIX1 or pCMX-NIX1 AA (100, 200, 500 ng). DR8-TK LUC (500 ng) was used as reporter plasmids.
NIX1 Inhibits Transcriptional Activation.
To investigate the transcriptional properties of NIX1, expression plasmids coding for NIX1 were tested in transient transfection experiments. Fig. 7 B and C illustrates that NIX1 specifically suppresses the transcriptional activation of RORβ in a dose-dependent manner. Cotransfection of increasing amounts of NIX1 expression plasmid specifically blocks transactivation by either Gal4-RORβ201–459 or wild-type RORβ when tested with their cognate luciferase reporter genes (6). However, neither the transcriptional activation by Gal4-ERα (Fig. 7B) nor by ERα (data not shown) is inhibited by NIX1. In addition, the interaction-deficient mutant NIX1 AA, which also resides in the nucleus (data not shown), fails to down-regulate the transactivation by nuclear receptors (Fig. 7C). These data indicate that NIX1 specifically interferes with the transcriptional activation of distinct nuclear receptors. Interestingly, no intrinsic transcriptional function is associated with NIX1 when fused to the Gal4 DBD (data not shown). These observations suggest that NIX1 blocks transcriptional activity of ligand-bound nuclear receptors, rather than containing its own transcriptional repression domain.
Discussion
Nuclear receptors control specific and selective target gene expression by serving as platforms for the assembly of multisubunit protein complexes. Unliganded nuclear receptors associate with corepressors N-CoR or SMRT (8, 9) which mediate gene repression by recruiting multisubunit repressor complexes. On ligand binding, nuclear receptors interact with various coactivators which provide and/or recruit histone acetyltransferase activity to stimulate receptor-dependent gene transcription (for review see ref. 7). Therefore, cofactors are ideal candidates to integrate signals from various pathways and regulate specific gene expression. However, most cofactors described to date interact with numerous receptors rather than selecting distinct partners. Except from the recently described coactivators, AIB1 (16), FHL2 (29), and PGC1 (22), they do not display restricted expression patterns and can be found in various tissues. Thus, additional cell type-specific cofactors may exist that select particular nuclear receptors and mediate specific functions in distinct tissues.
The data in this report describe the cloning and initial characterization of a neuronal-specific protein termed NIX1, which specifically interacts with holo-LBDs of certain nuclear receptors. NIX1 differs from previously reported nuclear receptor-associated proteins in several respects: it displays a neuronal specific expression pattern; it selectively interacts with distinct nuclear receptors of the RAR and TR subfamily, but does not bind steroid hormone receptors; it suppresses transcriptional activation by docking to activated nuclear receptors in an AF2-D-dependent fashion.
NIX1 displays a highly restricted expression pattern that is regulated spatially and temporally. mRNA encoding NIX1 cannot be detected in the newborn mouse, but is induced at around P7 and persists into adulthood (data not shown). In situ hybridization experiments and immunohistochemistry (data not shown) reveal that NIX1 expression can be found in neurons of the dentate gyrus, of the hippocampus, the amygdala, thalamic, hypothalamic regions, and several brainstem nuclei. Recently cloned nuclear receptor cofactors named AIB1, FHL2, and PGC1 have been reported to exhibit restricted expression patterns that are regulated in a cell type-specific fashion. NIX1 exclusively expressed in neurons represents the first neuronal-specific protein that selectively coregulates nuclear receptors in an AF2-D and ligand-dependent fashion.
NIX1 also differs from previously reported nuclear receptor interacting proteins in that it exclusively binds a distinct subset of activated nuclear receptors. With the exception of FHL2 (29), most coactivators described to date interact with various nuclear receptors, including steroid hormone receptors. In contrast, NIX1 exclusively binds distinct activated nuclear receptors of the RAR and TR subfamily, but clearly fails to associate with any steroid receptor, either in the presence or in the absence of their cognate ligands. Both corepressors, N-CoR and SMRT, have been reported to interact with unliganded nuclear receptors, including RAR, TR, and RXR (8, 9). In contrast, NIX1 specifically interacts with RAR, TR, and VDR in the presence of their ligand, but does not bind RXR. Thus, NIX1 displays a high degree of selectivity with respect to its nuclear receptor partner proteins.
Recent findings indicate that cofactors contain LXXLL nuclear receptor-binding motifs that are both necessary and sufficient for the interaction with the nuclear receptors (27, 28). NIX1 holds two copies of the LXXLL motif. For binding nuclear receptors, the LXXLL sequence located within the C terminus of NIX1 (amino acids 192–196) is dispensable. Positioned in an opposite orientation (amino acids 87–91), the second LXXLL motif is required for the highly selective interaction between NIX1 and nuclear receptors of the RAR and TR subfamily.
By definition, bona fide nuclear receptor coactivators/corepressors must harbor autonomous transcriptional activation/repression functions. NIX1 exerts neither function when fused to a Gal4 DBD and tested in reporter gene assays. However, NIX1 inhibits receptor-mediated gene expression in an AF2-D-dependent manner presumably by interfering with coactivator binding. By docking to the holo-LBD of nuclear receptors, NIX1 might displace coactivators and result in suppression of the transcriptional activity of liganded nuclear receptors. Therefore, NIX1 might fall into the same category of coregulatory proteins such as RIP140 (30), which is described to suppress the activation of certain agonist-bound hormone receptors (31, 32). Nuclear receptors and their associated proteins, each capable of being activated by ligands and other signals, constitute interacting networks that produce complex patterns of gene activation and repression. Because of its distinct neuronal expression pattern, NIX1 might act as an additional but special player, in regulating brain-specific gene expression by nuclear receptors. However, additional biochemical and genetic experiments have to be addressed to investigate how NIX1 impinges on particular nuclear receptor functions in the nervous system. Importantly, NIX1 displays features that differ from nuclear receptor coactivators. NIX1 binds nuclear receptors in their holo conformation, but down-regulates their activation function.
Acknowledgments
We thank Birgit Egle for performing the in situ hybridization stainings; Richard A. Heyman for providing ligands; Dino Moras, Jean-Marie Wurtz, Jean-Paul Renaud, and Anke Steinmetz for fruitful discussion; and the members of the Schüle lab for support and criticism. This work was supported in part by a grant from the Deutsche Forschungsgemeinschaft (DFG Schu 688/4-2) (to R.S.).
Abbreviations
- ERα
estradiol receptor α (NR3A1)
- GST
glutathione S-transferase
- NIX1
neuronal-interacting factor X1
- RARα
retinoic acid receptor α (NR1B1)
- RORβ
retinoid-related orphan receptor β (NR1F2)
- RXRα
retinoid X receptor α (NR2B1)
- TRβ
thyroid hormone receptor β (NR1A2)
- DBD
DNA-binding domain
- LBD
ligand-binding domain
- AF2-D
activation function 2
- EMSA
electrophoretic mobility-shift assay
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ278170).
References
- 1.Mangelsdorf D J, Evans R M. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 2.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Nature (London) 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 3.Brzozowski A M, Pike A C W, Dauter Z, Hubbard R E, Bonn T, Engström O, Öhman L, Greene G L, Gustafsson J A, Carlquist M. Nature (London) 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 4.Renaud J P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Nature (London) 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 5.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. Nature (London) 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 6.Greiner E F, Kirfel J, Greschik H, Dörflinger U, Becker P, Mercep A, Schüle R. Proc Natl Acad Sci USA. 1996;93:10105–10110. doi: 10.1073/pnas.93.19.10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu L, Glass C K, Rosenfeld M G. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 8.Chen J D, Evans R M. Nature (London) 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 9.Hörlein A J, Näär A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Söderström M, Glass C K, Rosenfeld M G. Nature (London) 1995;377:397–403. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 10.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 11.Nomura T, Khan M M, Kaul S C, Dong H D, Wadhwa R, Colmenares C, Kohno I, Ishii S. Genes Dev. 1999;13:412–423. doi: 10.1101/gad.13.4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman S C, Rose D W, Glass C K, Rosenfeld M G. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 13.Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 14.Hong H, Kohli K, Garabedian M J, Stallcup M R. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voegel J J, Heine M J, Zechel C, Chambon P, Gronemeyer H. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 16.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 17.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 18.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. Nature (London) 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 19.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Nature (London) 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 20.Pazin M J, Kadonaga J T. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 21.Struhl K. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 22.Puigserver P, Wu Z, Park C W, Graves R, Wright M, Spiegelman B M. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 23.Xu J, Qiu Y, DeMayo F J, Tsai S Y, Tsai M J, O'Malley B W. Science. 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- 24.Nuclear Receptors Committee. Cell. 1999;97:161–162. [Google Scholar]
- 25.Pfitzner E, Becker P, Rolke A, Schüle R. Proc Natl Acad Sci USA. 1995;92:12265–12269. doi: 10.1073/pnas.92.26.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaeren-Wiemers N, Gerfin-Moser A. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 27.Heery D M, Kalkhoven E, Hoare S, Parker M G. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 28.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, et al. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller J M, Isele U, Metzger E, Rempel A, Moser M, Pscherer A, Breyer T, Holubarsch C, Buettner R, Schüle R. EMBO J. 2000;19:359–369. doi: 10.1093/emboj/19.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavaillès V, Dauvois S, L'Horset F, Lopez G, Hoare S, Kushner P J, Parker M G. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee C H, Wei L N. J Biol Chem. 1999;274:31320–31326. doi: 10.1074/jbc.274.44.31320. [DOI] [PubMed] [Google Scholar]
- 32.Treuter E, Albrektsen T, Johansson L, Leers J, Gustafsson J A. Mol Endocrinol. 1998;12:864–881. doi: 10.1210/mend.12.6.0123. [DOI] [PubMed] [Google Scholar]