Abstract
A plasmid coding for the nisin two-component regulatory proteins, NisK and NisR, was constructed; in this plasmid a gfp gene (encoding the green fluorescent protein) was placed under control of the nisin-inducible nisF promoter. The plasmid was transformed into non-nisin-producing Lactococcus lactis strain MG1614. The new strain could sense extracellular nisin and transduce it to green fluorescent protein fluorescence. The amount of fluorescence was dependent on the nisin concentration, and it could be measured easily. By using this strain, an assay for quantification of nisin was developed. With this method it was possible to measure as little as 2.5 ng of pure nisin per ml in culture supernatant, 45 ng of nisin per ml in milk, 0.9 μg of nisin in cheese, and 1 μg of nisin per ml in salad dressings.
The type A lantibiotic nisin (16), produced by some Lactococcus lactis strains, is a small antimicrobial peptide that inhibits the growth of a wide range of gram-positive bacteria, such as Bacillus, Clostridium, Listeria, and Staphylococcus species. It is nontoxic to humans (19) and is broadly used as a food preservative (E234) (6) in more than 50 countries, including the United States, countries in the European Union, and the People's Republic of China. So far, two natural nisin variants, nisin A (4) and nisin Z (15, 25), which differ in a single amino acid residue, have been described. National laws concerning the presence and maximum levels of nisin in different food products vary greatly (1). Therefore, there is a demand by national food authorities for better methods to identify and quantify nisin reliably in different food matrices. The inhibitory effect of nisin on a given test organism (9) is the basis for most quantification methods that have been developed so far. The sensitivity and accuracy of the agar diffusion method (13, 35) are affected by several parameters (38). Moreover, due to the better diffusion properties of nisin Z, it produces larger inhibition zones than equal amounts of nisin A produce (8). Immunological tests have also been described. Falahee et al. (12) described an enzyme-linked immunosorbent assay (ELISA) for nisin A in cheese based on sheep polyclonal antibodies. This method is considerably more sensitive than the agar diffusion assay, but it is not totally reliable due to cross-reactivity with the lantibiotic subtilin (11). Suárez et al. developed competitive direct ELISAs for nisin with polyclonal (31) and monoclonal (32) antibodies from mice. Bouksaim et al. (2) described an immunodot detection method in which rabbit antiserum against nisin Z in milk and whey is used. The same authors (3) also introduced an ELISA method in which affinity-purified polyclonal rabbit antibodies are used for quantification of nisin Z in complex media and milk. The flow injection immunoassay described by Nandakumar et al. (26) has an advantage over the other nisin quantification methods because it allows workers to monitor the concentration of nisin in a fermentation broth online. Dadoudi et al. (5) developed the first immunoassay capable of distinguishing between nisin A and nisin Z. They described a competitive enzyme immunoassay based on mice monoclonal antibodies that recognize only nisin Z. The method could be used to measure nisin Z contents in culture supernatant, milk, and whey. In conclusion, no relevant progress in detection limits and sample matrices has been made in the field of nisin quantification by immunological methods since these methods were first introduced.
The proteins responsible for nisin biosynthesis, regulation, and producer self-immunity are encoded by genes arranged in two inducible operons, nisA/ZBTCIPRK and nisFEG (24) The transmembrane histidine kinase NisK and the response regulator NisR form a two-component regulation system (10, 20, 36), in which NisK autophosphorylates at a specific histidine residue after exposure to extracellular nisin and subsequently transfers the phosphate moiety to NisR. The phosphorylated NisR binds to two regulated promoters in the nisin gene cluster (i.e., the nisA/Z and nisF promoters), thereby activating transcription of the structural gene nisA and the downstream genes nisBTCIPRK from the nisA/Z promoter and transcription of the nisFEG genes from the nisF promoter (7, 22, 28, 29) (Fig. 1). Currently, the most sensitive quantification method that has been described is based on the autoinducibility of the nisin promoter PnisF and bioluminescence derived from bacterial luciferase genes fused to the nisin promoter. This method, described by Wahlstöm and Saris (37), detects nisin concentrations of 0.0125 ng/ml in water and culture supernatants and 0.075 ng/ml in milk (final assay mixture concentrations). The advantage of the luciferase assay over all the methods described above is its ability to quantify nisin in undiluted milk samples without any sample pretreatment. However, in this assay there are problems with addition of the substrate for luciferase (n-decyl-aldehyde) following the 3 h of nisin induction: due to the chemical nature of luciferase bioluminescence, the indicator cells in all samples must be in the same energetic state, which requires stringent timing for substrate addition and subsequent measurement of luminescence, thus limiting the number of samples that can be processed at the same time. Therefore, we developed the assay described in this paper by using a gfp gene as a reporter instead of luciferase.
FIG. 1.
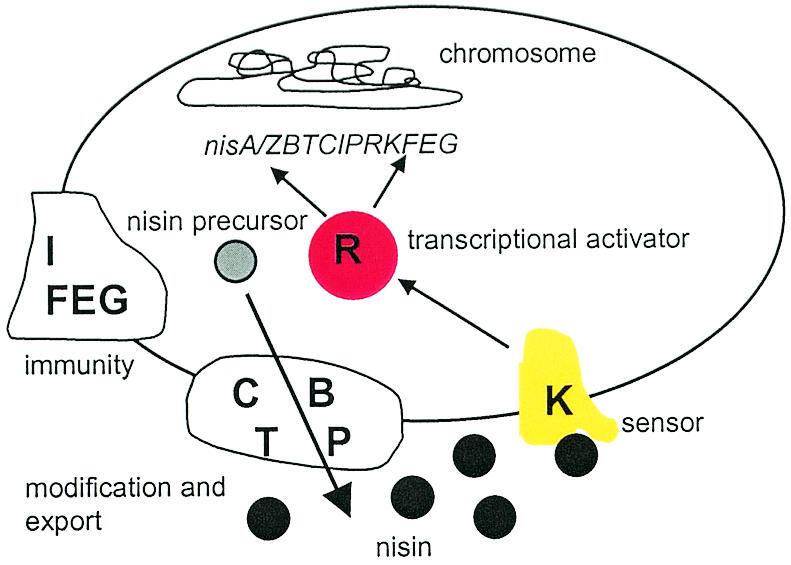
Autoregulation of nisin biosynthesis and immunity in nisin-producing L. lactis strains. The nisin precursor is posttranslationally modified and secreted by the biosynthetic NisBCTP machinery, and the producer strain is protected from the antimicrobial activity of nisin by the immunity proteins NisIFEG. When confronted with nisin, the transmembrane histidine kinase NisK autophosphorylates and subsequently transfers the phosphate to NisR. The phosphorylated NisR binds to the nisA and nisF promoters, thereby activating transcription of the genes downstream from the promoters.
MATERIALS AND METHODS
Construction of the indicator strain.
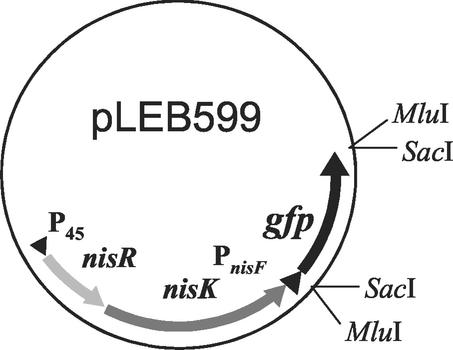
A gene fragment containing a ribosome-binding site and having a red-shifted P11 mutation of the gfp gene (17) was excised from plasmid pKPSPgfp (30) as a SacI fragment. This fragment was inserted with the aid of the MluI-SacI linkers 5′-CGCGTGGGCCCGGGTCTAGAGCT-3′ and 5′-CTAGACCCGGGCCCA-3′ into MluI-restricted plasmid pLEB338, a derivative of plasmid pLEB124 (27) containing intact nisRK genes and the nisF promoter from plasmid pLEB189 (20) originally cloned from the nisin Z-producing strain L. lactis subsp. lactis N8 (15). In the resulting plasmid, pLEB599, the gfp gene was placed under control of the PnisF promoter (Fig. 2). This construct was electroporated (18) into the non-nisin-producing strain L. lactis MG1614 (14) and plated at 30°C on M17 (33) plates containing 0.5% (wt/vol) glucose (M17G) or 0.5% (wt/vol) sucrose and erythromycin (5 μg/ml). The resulting indicator strain was designated LAC240.
FIG. 2.
Schematic representation of the essential parts of plasmid pLEB599.
Nisin fluorescence assay.
The indicator strain LAC240 was grown overnight in M17G supplemented with erythromycin (5 μg/ml). Then 0.2 volume of glycerol (87%) was added, and the cells were stored at −20°C. Prior to nisin detection these precultured cells were diluted 1:100 in M17G supplemented with erythromycin (5 μg/ml), and Tween 80 was added to a final concentration of 0.1% (wt/vol) in order to prevent adsorption of nisin to the polypropylene tube and tip surfaces (21). Nisin standards (Sigma) were prepared in 0.1% Tween 80 dissolved in distilled water acidified to pH 2.5 with HCl (referred to below as 0.1% Tween 80). Nisin was added to the diluted indicator bacteria so that the concentration of nisin in the culture medium ranged from 2.5 to 125 ng/ml. The bacterial suspensions were then divided into 225-μl aliquots in a microtiter plate, in which the cells were grown overnight at 30°C without aeration. The next day 175 μl of each supernatant was removed, and the cells were frozen for 30 min at −20°C and thawed at room temperature before detection of fluorescence. Fluorescence was expressed in relative fluorescence units (RFU) as determined with a Fluoroscan Ascent 374 fluorometer connected to a computer by using Ascent software (version 1.2; Labsystems, Helsinki, Finland); the excitation and emission filters were 485- and 538-nm filters, respectively. Growth was measured by determining the optical density at 600 nm with an UltrospecII spectrophotometer (Pharmacia LKB).
Detection of nisin in cheese, milk, and salad dressings.
Amounts of processed cheese (20% fat; Valio Ltd., Helsinki, Finland) ranging from 15 to 300 mg were dissolved in 1 ml of 0.1% Tween 80 and incubated for 10 min at 80°C. Milk (1.5% fat; Valio Ltd.) was serially diluted into 0.1% Tween 80. Various amounts of nisin and 210 μl of each cheese and milk solution were added to 945 μl of a 1:100 dilution of LAC240 in M17G containing erythromycin (5 μg/ml) and 0.1% (wt/vol) Tween 80. These preparations were divided into 225-μl aliquots on a microplate; otherwise the samples were treated as described above.
One gram of Thousand Island (Saarioisten säilyke Ltd., Huittinen, Finland) or French (Los Toros Ltd., Pirkkala, Finland) salad dressing was spiked with 1 to 5 μg of nisin. The volumes were adjusted to 40 ml with 0.1% Tween 80, and 300-μl portions of these extracts were combined with 1,200 μl of a 1:100 dilution of LAC240 in M17G containing erythromycin (5 μg/ml) and 0.1% (wt/vol) Tween 80. Otherwise the samples were processed as described above.
RESULTS
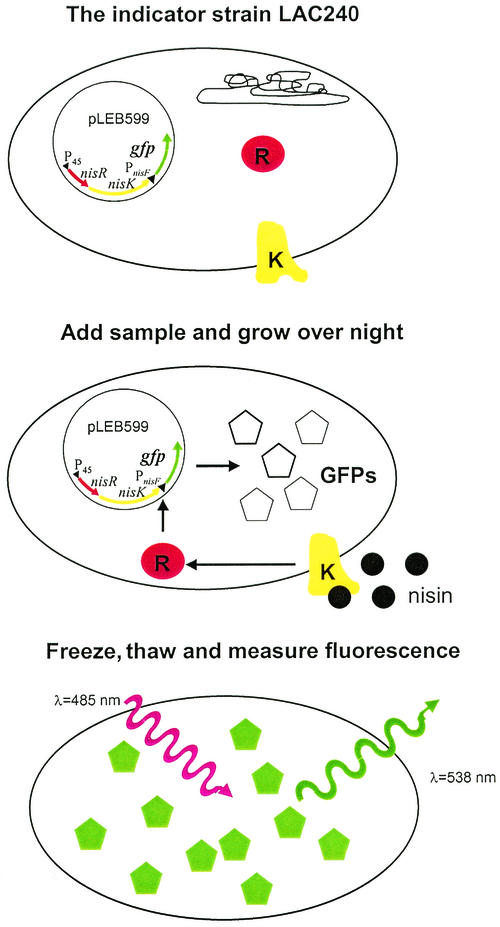
First, indicator strain LAC240, which is capable of producing green fluorescent protein (GFP) molecules upon nisin induction, was constructed. Fluorophore formation leading to photoactivity is assumed to require at least 4 h after translation in the case of a P11 GFP mutant (17). Also, the effects of possible inhibitory agents in sample matrices on the growth rate of the indicator strain had to be excluded. Therefore, overnight incubation was used as the induction conditions in all experiments. The maturation of newly synthesized GFP molecules is also a temperature-dependent process; maturation is faster at low temperatures and is almost halted at temperatures of 30°C or more (23). Nisin-induced LAC240 cells had to be kept at 4°C for several hours before any fluorescence from within the intact cells could be detected (results not shown). Freezing the cells for 30 min at −20°C and thawing them at room temperature prior to analysis shortened this unwanted step. The growth medium M17G (Oxoid), which is a red-brown color, absorbs light at the same wavelengths as GFP. Therefore, 175 μl of the supernatant above the cell pellets was removed before fluorescence was measured. Thus, the GFP-based nisin bioassay used in this study consisted of the following steps: addition of a sample to LAC240 cells, overnight incubation, removal of 175 μl of the supernatant, freezing and thawing, and measurement of fluorescence (Fig. 3).
FIG. 3.
Protocol for the GFP fluorescence assay for nisin with the indicator strain L. lactis LAC240. A LAC240 inoculum is supplemented with a sample and grown overnight at 30°C on a microplate. Prior to measurement the supernatant is removed, and the cells are frozen and thawed. The fluorescence is expressed in RFU as determined with a fluorometer; the excitation light is red (λ = 485 nm), and the emission light is green (λ = 538 nm).
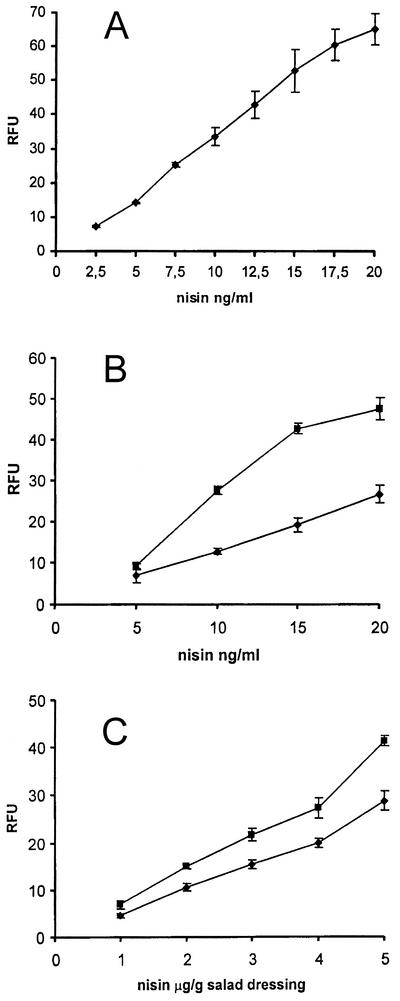
When this protocol was used, nisin concentrations ranging from 2.5 to 20 ng/ml, (final assay mixture concentrations) could be detected quantitatively and reliably directly in the culture medium (Fig. 4A). At nisin concentrations greater than 20 ng/ml, the fluorescence signal started to decrease, and the signal reached the background level when the concentration of nisin was approximately 60 ng/ml. The standard error increased at concentrations greater than 20 ng/ml, probably reflecting the inhibitory effect of nisin on the indicator strain. This assumption was supported by the visually detectable decrease in the sizes of the bacterial pellets grown with nisin concentrations greater than 25 ng/ml compared to the pellet sizes for bacteria grown with lower concentrations of nisin.
FIG. 4.
Standard curves for nisin concentrations in 0.1% Tween 80 (A) and in milk (▪) and cheese (♦) (B), expressed as final assay concentrations. The data are the means and standard deviations (error bars) of four parallel experiments. (C) Standard curves for Thousand Island (▪) and French (♦) salad dressings spiked with nisin, expressed as concentrations of nisin in the dressings. The data are the means andstandard deviations (error bars) of six parallel experiments. The background fluorescence values for samples not supplemented with nisin (26.2 ± 0.95 RFU for a pure solution, 56.9 ± 1.13 RFU for milk, 23.6 ± 0.76 RFU for cheese, 22.7 ± 0.61 RFU for Thousand Island dressing, and 21.6 ± 1.85 RFU for French dressing) were subtracted from the fluorescence values obtained with the samples spiked with nisin.
The usefulness of the fluorescence assay for detecting nisin in food samples was tested with cheese, milk, and salad dressings. It was found that cheese concentrations of 50 mg/ml or less in 0.1% Tween 80 were best for the assay protocol. When the concentration of cheese was 50 mg/ml, the concentration of nisin that was quantitatively detectable ranged from 10 to 20 ng/ml. For concentrations of cheese of 25 mg/ml or less the detection range was a little broader (5 to 20 ng of nisin per ml) (Fig. 4B). When milk was diluted 1:2 or more in 0.1% Tween 80, a linear dose-response relationship in the fluorescence signal was observed with nisin concentrations of 5 to 20 ng/ml (Fig. 4B). Two types of salad dressing were used; the Thousand Island dressing was a representative of mayonnaise-based dressings, and the French dressing was a representative of vegetable oil- and vinegar-based salad dressings. Nisin is added to salad dressings at levels of 1.25 to 5 μg/g (34), and by using the GFP assay it was possible to measure concentrations of nisin ranging from 1 to 5 μg/g of salad dressing (Fig. 4C).
DISCUSSION
In most previous papers concerning nisin quantification the authors emphasized the lowest detectable amount of nisin, which was given as the final assay concentration. However, this value seldom describes the most important numerical value giving true limits to the usefulness of the method in question, namely, the lowest detectable concentration of nisin in a food sample. Indeed, in some studies food material was spiked with amounts of nisin far from the linear dose-response range of the assay described, and the authors did not reveal the solvent into which the food extract was diluted prior to measurement (3, 5). Because of the sensitivity of immunological methods to interfering substances in a sample matrix, this kind of reporting makes it impossible for a reader to decide whether the assay in question is useful for his or her application.
At present, the most widely used quantification assay for nisin, the agar diffusion method, which was developed by Tramer and Fowler in 1964, is more sensitive than most of the immunological methods; the detection limit for nisin in a sandwich spread in this test was 100 ng/g (35). The major drawback of this assay, however, is its inability to differentiate nisin from other inhibitory substances (13). The ELISA test described by Falahee et al. suffers from the same problem and gives false-positive results when samples contain subtilin (11), the lantibiotic structurally most similar to nisin. The NisRK signal transduction system utilized in the GFP assay is not activated by subtilin (37). Also, the detection limit for nisin in cheese (250 ng/ml, corresponding to 1.25 μg/g of cheese) is lower than the detection limit of the agar diffusion assay (12). The monoclonal antibodies against nisin produced in mice recognized 50 ng of nisin per g of cheese, which is less than the 900 ng/g detected by the GFP test. However, reliable quantification of nisin by this competitive direct ELISA is impossible due to cross-reactivity of nisin Z and nisin A; nisin Z gives a signal that is approximately three times stronger than the signal given by nisin A (32). In contrast, the NisRK pathway has been demonstrated to produce approximately the same response in the bioluminescence assay for both nisin variants (37). The immunodot method described by Bouksaim et al. (2) could sense a nisin concentration of 155 ng/ml in milk and whey, whereas the detection limit for nisin in milk was 45 ng/ml with the GFP assay. The ELISA test described by the same authors based on rabbit polyclonal antibodies against nisin Z had a true detection limit for nisin in milk of 5 μg/ml due to a high dilution factor prior to the ELISA (3). The first nisin Z-specific immunoassay has a detection limit of 78 ng/ml for nisin Z in pure solution (5); in comparison, the same value for the GFP test is 2.5 ng/ml. The usefulness of this method (5) with food samples remains enigmatic, since the authors did not reveal the solvent used when dilutions were prepared from milk, whey, and culture supernatant. The luciferase assay developed by Wahlström and Saris (37) is the most sensitive quantification method for nisin so far, requiring a nisin concentration of only 1 ng/ml in milk. However, the indicator cells have to be in the same energetic state (more specifically, in the early log phase) in order to obtain the highest signal-to-background level when the luciferase substrate is added. This means that the growth of each sample and known standard should be measured if the effect of possible interfering substances in the sample matrices is to be excluded. Also, even if every sample is treated in this way and the substrate is added at exactly the same point in growth to each individual sample, the subsequent 3-h period before luminescence is measured must be timed independently for each sample. Clearly, these demands seriously affect the number of samples that can be processed at the same time and also make automation of the method difficult. In contrast, in the GFP test the amount of fluorescence is based on the amount of GFP molecules; therefore, a lag phase in growth is an adequate time for measurement of fluorescence. Furthermore, since GFP fluorescence is induced solely by photoactivation, no substrate addition is required. The simplicity of the GFP test allows analysis of hundreds of samples at the same time to detect 45 ng of nisin per ml in milk, 0.9 μg of nisin per g in cheese, and 1 μg of nisin per g in salad dressings, compared to the levels of more than 1 μg/ml or 1 μg/g used in food manufacturing (9, 34), and makes it possible to analyze nisin in different kind of food matrices. Therefore, the GFP assay could be widely used in the food industry, as well as in basic research.
REFERENCES
- 1.Anonymous. 1986. International acceptance of nisin as a food additive. Issue 1/ 86. Applin & Barrett Ltd., Trowbridge, Wiltshire, England.
- 2.Bouksaim, M., I. Fliss, J. Meghrous, R. Simard, and C. Lacroix. 1998. Immunodot detection of nisin in milk and whey using enhanced chemiluminescence. J. Appl. Bacteriol. 84:176-184. [DOI] [PubMed] [Google Scholar]
- 3.Bouksaim, M., C. Lacroix, R. Bazin, and R. E. Simard. 1999. Production and utilization of polyclonal antibodies against nisin in an ELISA and for immuno-location of nisin in producing and sensitive bacterial strains. J. Appl. Microbiol. 87:500-510. [DOI] [PubMed] [Google Scholar]
- 4.Buchman, G. W., S. Banerjee, and J. N. Hansen. 1988. Structure, expression, and evolution of a gene encoding the precursor of nisin, a small protein antibiotic. J. Biol. Chem. 263:16260-16266. [PubMed] [Google Scholar]
- 5.Dadoudi, L., C. Turcotte, C. Lacroix, and I. Fliss. 2001. Production and characterization of anti-nisin Z monoclonal antibodies; suitability for distinguishing active from inactive forms through a competitive enzyme immunoassay. Appl. Microbiol. Biotechnol. 56:114-119. [DOI] [PubMed] [Google Scholar]
- 6.Delves-Broughton, J., P. Blackburn, R. J. Evans, and J. Hugenholz. 1996. Applications of the bacteriocin, nisin. Antonie Leeuwenhoek 69:193-202. [DOI] [PubMed] [Google Scholar]
- 7.de Ruyter, P. G. G.A., O. P. Kuipers, M. M. Beerthuyzen, I. Van Alenboerrigter, and W. M. de Vos. 1996. Functional analysis of promoters in nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Voss, W. M., J. W. M. Mulders, R. J. Siezen, J. Hugenholtz, and O. P. Kuipers. 1993. Properties of nisin Z and distribution of its gene, nisZ, in Lactococcuslactis. Appl. Environ. Microbiol. 59:213-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Vuyst, L., and E. J. Vandamme. 1994. Nisin, a lantibiotic produced by Lactococcus lactis subsp. lactis: properties, biosynthesis, fermentation and applications, p. 152-199. In L. de Vuyst and E. J. Vandamme (ed.), Bacteriocins of lactic acid bacteria. Chapman & Hall, The Alden Press, Oxford, United Kingdom.
- 10.Engelke, G., Z. Gutowski-Eckel, P. Kiesau, K. Siegers, M. Hammelmann, and K.-D. Entian. 1994. Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Appl. Environ. Microbiol. 60:814-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falahee, M. B., and M. R. Adams. 1992. Cross-reactivity of bacteriocins from lactic acid bacteria and lantibiotics in a nisin bioassay and ELISA. Lett. Appl. Microbiol. 15:214-216. [DOI] [PubMed] [Google Scholar]
- 12.Falahee, M. B., M. R. Adams, J. W. Dale, and B. A. Morris. 1990. An enzyme immunoassay for nisin. Int. J. Food Sci. Technol. 25:590-595. [Google Scholar]
- 13.Fowler, G. G., B. Jarvis, and J. Tramer. 1975. The assay of nisin in foods. Soc. Appl. Bacteriol. Tech. Ser. 8:91-105. [Google Scholar]
- 14.Gasson, M. J. 1983. Plasmid components of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graeffe, T., H. Rintala, L. Paulin, and P. Saris. 1991. A natural nisin variant, p. 260-268. In G. Jung and H.-G. Sahl (ed.), Nisin and novel lantibiotics. ESCOM Science Publishers B. V., Leiden, The Nedherlands.
- 16.Gross, E., and J. L. Morell. 1971. The structure of nisin. J. Am. Chem. Soc. 93:4634-4635. [DOI] [PubMed] [Google Scholar]
- 17.Heim, R., D. C. Prasher, and R. Y. Tsien. 1994. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. USA 91:12501-12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurst, A. 1981. Nisin. Adv. Appl. Microbiol. 27:85-123. [Google Scholar]
- 20.Immonen, T., S. Ye, R. Ra, L. Paulin, and P. E. J. Saris. 1995. The codon usage of the nisZ operon in Lactococcus lactis N8 suggests a non-lactococcal origin of the conjugative nisin-sucrose transposon. DNA Sequence 5:203-218. [DOI] [PubMed] [Google Scholar]
- 21.Joosten, H. M. L. J., and M. Nuñes. 1995. Adsorption of nisin and enterocin 4 to polypropylene and glass surfaces and its prevention by Tween 80. Lett. Appl. Microbiol. 21:389-392. [Google Scholar]
- 22.Kuipers, O. P., M. M. Beerthuyzen, P. G. G. A. de Ruyter, E. J. Luesink, and W. M. de Vos. 1995. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 270:27299-27304. [DOI] [PubMed] [Google Scholar]
- 23.Lim, C. R., Y. Kimata, M. Oka, K. Nomaguchi, and K. Kohno. 1995. Thermosensitivity of green fluorescent protein fluorescence utilized to reveal novel nuclear-like compartments in a mutant nucleoporin NSP1. J. Biochem. 118:13-17. [DOI] [PubMed] [Google Scholar]
- 24.McAuliffe, O., R. P. Ross, and C. Hill. 2001. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 25:285-308. [DOI] [PubMed] [Google Scholar]
- 25.Mulders, J. W. M., I. J. Boerrigter, H. S. Rollema, R. J. Siezen, and W. M. de Vos. 1991. Identification and characterization of the lantibiotic nisin Z, a natural nisin variant. Eur. J. Biochem. 201:581-584. [DOI] [PubMed] [Google Scholar]
- 26.Nandakumar, R., M. P. Nandakumar, and B. Mattiasson. 2000. Quantification of nisin in flow-injection immunoassay systems. Biosens. Bioelectron. 15:241-247. [DOI] [PubMed] [Google Scholar]
- 27.Qiao, M., T. Immonen, O. Koponen, and P. E. J. Saris. 1995. The cellular location and effect on nisin immunity of the NisI protein from Lactococcus lactis N8 expressed in Escherichia coli and L. lactis. FEMS Microbiol. Lett. 131:75-80. [DOI] [PubMed] [Google Scholar]
- 28.Qiao, M., S. Ye, O. Koponen, R. Ra, M. Usabiaga, T. Immonen, and P. E. J. Saris. 1996. Regulation of the nisin operons in Lactococcus lactis N8. J. Appl. Bacteriol. 80:626-634. [DOI] [PubMed] [Google Scholar]
- 29.Ra, S. R., M. Qiao, T. Immonen, I. Pujana, and P. E. J. Saris. 1996. Genes responsible for nisin synthesis, regulation and immunity form a regulon of two operons and are induced by nisin in Lactococcus lactis N8. Microbiology 142:1281-1288. [DOI] [PubMed] [Google Scholar]
- 30.Scott, K. P., D. K. Mercer, L. A. Glover, and H. J. Flint. 1998. The green fluorescence protein as a visible marker for lactic acid bacteria in complex ecosystems. FEMS Microbiol. Ecol. 26:219-230. [Google Scholar]
- 31.Suárez, A. M., J. M. Rodríguez, P. E. Hernández, and J. I. Azcona-Olivera. 1996. Generation of polyclonal antibodies against nisin; immunization strategies and immunoassay development. Appl. Environ. Microbiol. 62:2117-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suárez, A. M., J. M. Rodríguez, P. Morales, P. E. Hernándes, and J. I. Azcona-Olivera. 1996. Development of monoclonal antibodies to the lantibiotic nisin A. J. Agric. Food Chem. 44:2936-2940. [Google Scholar]
- 33.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas, L. V., M. R. Clarkson, and J. Delves-Broughton. 2000. Nisin, p. 463-524. In A. S. Naidu (ed.), Natural food antimicrobial systems. CRC Press, Boca Raton, Fla.
- 35.Tramer, J., and J. J. Fowler. 1964. Estimation of nisin in foods. J. Sci. Food Agric. 15:522-528. [Google Scholar]
- 36.van der Meer, J. R., J. Polman, M. M. Beerthuyzen, R. J. Siezen, O. P. Kuipers, and W. M. de Vos. 1993. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J. Bacteriol. 175:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wahlstöm, G., and P. E. J. Saris. 1999. Nisin bioassay based on bioluminescence. Appl. Environ. Microbiol. 65:3742-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolf, C. E., and W. R. Gibbons. 1996. Improved method for quantification of the bacteriocin nisin. J. Appl. Bacteriol. 80:453-457. [DOI] [PubMed] [Google Scholar]