Abstract
Endophytic filamentous actinobacteria were isolated from surface-sterilized roots of wheat plants. Endophytic colonization of germinating wheat seed was examined using one of these endophytes, Streptomyces sp. strain EN27, tagged with the egfp gene. Endophytic colonization was observed from a very early stage of plant development with colonization of the embryo, endosperm, and emerging radicle.
In order to find effective biocontrol agents for fungal plant pathogens of cereal crops, we isolated actinobacterial strains from surface-sterilized roots of healthy wheat plants (3; J. T. Coombs and C. M. M. Franco, submitted for publication). There have been reports of actinobacteria, other than Frankia spp., inhabiting the tissues of healthy plants (5, 15, 16). However, spatial colonization of the root tissue of cereals by actinobacteria has not been reported, and evidence of their distribution as endophytes is required to prove endorhizosphere competence. A representative endophytic strain, Streptomyces sp. strain EN27, was tagged with green fluorescent protein (GFP) to study early colonization events after it was applied to wheat (Triticum aestivum L.) seeds. Expression of GFP has been used to visualize a number of unicellular bacteria-plant interactions (6, 9, 10, 14, 21), but this is the first report on a filamentous actinobacterial endophyte. For actinobacteria, gfp expression has been optimized (4, 20) and coupled to a constitutively expressed promoter, ermEp (17).
Streptomyces sp. strain EN27, identified by 16S ribosomal DNA (rDNA) sequence analysis to be closely related to Streptomyces caviscabies, was isolated from surface-sterilized wheat root tissue (3; Coombs and Franco, submitted). Control experiments to validate the sterilization procedure were done by subjecting five individual actinobacterial endophytes and two endophytic pseudomonads, at 107 to 109 CFU per ml or per g of seed, to the sterilization protocol. They were tested as coatings on wheat seeds and as suspensions. Simple washing steps with sterile water do not easily remove the microbial coatings, but the sterilization protocol was effective in removing all surface-adhering microorganisms.
Streptomyces sp. strain EN27 was selected as a model organism with which to investigate the colonization of germinating seeds of the wheat host due to its wide distribution among wheat plants in the field and its ability to promote plant growth and control a number of root-infective phytopathogenic fungi (Coombs and Franco, submitted; J. T. Coombs, P. P. Michelsen, and C. M. M. Franco, submitted for publication). Streptomyces sp. strain EN27 was transformed with egfp by using an 8.0-kb vector, pIJ8641 (20), containing the egfp gene downstream of a strong constitutive ErmE promoter, an apramycin-resistant marker (aac(3)IV), an oriT/RK2 region, and a lambda phage chromosomal integration sequence (IntC31). Competent Escherichia coli S17.1 transformed with pIJ8641 DNA was used for intergeneric recombination with Streptomyces sp. strain EN27 by the intergeneric recombination protocol of Flett et al. (8), which is a modification of the method of Mazodier et al. (13), as described in Practical Streptomyces Genetics (12).
Approximately 100 T. aestivum L. cv. Excalibur seeds were surface sterilized by a 6-min wash in 3.125% NaOCl, followed by three double-volume rinses in sterile water. A spore suspension of the egfp-tagged actinobacterium, Streptomyces sp. strain EN27(pIJ8641), was added to half the seeds, while 1.5 ml of sterile water was added to the other half, to act as a control. The seeds were placed on a mannitol-soy flour (MS) medium plate and allowed to germinate. Five inoculated seeds were planted aseptically, in duplicate, in twice-autoclaved sterile sandy-loam soil placed in sterile 500-ml screw-cap flasks to a depth of 7 cm. The flasks were watered with sterile water and incubated in a plant growth chamber with a 16-h-light, 8-h-dark cycle at 25°C.
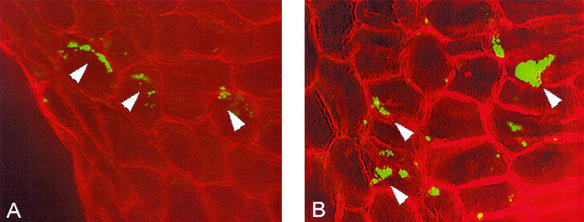
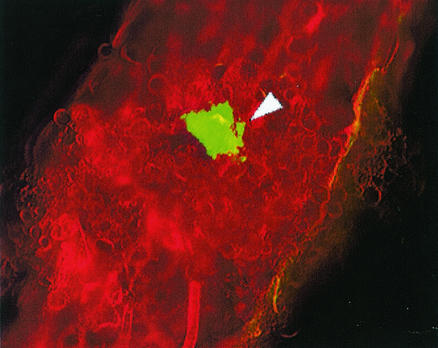
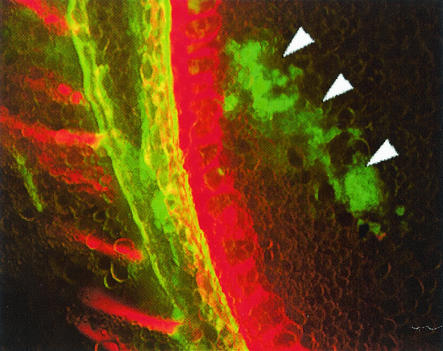
The egfp-tagged Streptomyces sp. strain EN27 showed strong GFP expression when visualized under a Nikon laser scanning confocal microscope, in comparison to control wild-type Streptomyces sp. strain EN27, which displayed no fluorescence (micrographs not shown). The stability of the integrated vector was observed for at least 7 successive progeny over a 7-week period; subcultures from spores obtained from agar plates with no selective pressure were resistant to 50 μg of apramycin per ml and expressed GFP. Germinating seeds coated with Streptomyces sp. strain EN27 and untreated control seeds were harvested every 24 h and were cut into 60- to 80-μm-thick sections by using a Leitz Wetzlar microtome with a freezing stage attachment. Sections were examined under an Olympus BX-50 microscope using a mercury vapor lamp. The GFP-expressing streptomycete was visualized with a Chroma 31001 filter block with excitation at 465 to 495 nm and emission at 515 to 555 nm. The structure of the plant tissue was visualized by using the autofluorescence of the tissue itself, with an Olympus U-MNUA filter set that gave an excitation wavelength of 360 to 370 nm and an emission wavelength of 420 to 460 nm. After 24 h the presence of the strain was detected only in the embryo and around the break in the seed husk where the embryo emerges from the seed. No fluorescence was observed on the outer seed husk, indicating that these cells were nonviable and no longer expressing GFP, or, more likely, that these cells were washed away when the seeds were immersed in the freezing step during sectioning. Figure 1 shows the actinobacteria inhabiting the embryo tissue. The image generated from the green-light detection of the tagged Streptomyces was digitally colored green, and the image generated from the blue-light detection of the plant tissue was digitally colored red. The GFP-expressing strain did not show any fluorescence under blue-light detection in any of the sections. The images were then overlaid by using Confocal Assistant, version 4.0. It was observed that the actinobacteria grew preferentially in close proximity to the plant cell walls. It is possible that this is intercellular growth and that the microscope stage was slightly moved (down and to the right in the image) between the two image captures, as the growth of the actinobacteria appears to mimic the shape of the plant cell wall in many places. After 3 days, GFP-expressing microcolonies of the actinobacteria were seen more frequently in the embryo tissue of the seed than at 24 h, indicating that the actinobacteria were actively growing in the plant tissue. Actinobacterial microcolonies were also detected in the emerging radicle (young root) of the embryo (Fig. 2). After 3 days, actinobacterial growth was observed in the endosperm of the wheat seed, which was not observed at 24 h (Fig. 3).
FIG. 1.
Microcolonies of Streptomyces sp. strain EN27 expressing enhanced GFP (EGFP) in wheat embryo tissue (plumule) at 24 h (A) and 3 days (B) after germination of coated seed. The image represents the merge of two micrographs of EGFP detection (shown in green) and plant autofluorescence (shown in red). Magnification for all images, ×200. Arrowheads indicate EGFP-expressing Streptomyces sp. strain EN27.
FIG. 2.
Enhanced GFP (EGFP)-expressing Streptomyces sp. strain EN27 microcolony in the emerging radicle. Magnification for all images, ×200. The arrowhead indicates EGFP-expressing Streptomyces sp. strain EN27.
FIG. 3.
Microcolonies of Streptomyces sp. strain EN27 expressing enhanced GFP (EGFP) in the endosperm after 3 days. Magnification for all images, ×200. Arrowheads indicate EGFP-expressing Streptomyces sp. strain EN27.
These observations show that the endophytic actinobacterium was able to associate with its host at a very early stage in the development of the plant. Actinobacterial microcolonies were observed in the wheat embryo 24 h after infection. Direct infection across the pericarp seems unlikely, as no early infection of the endosperm was seen. However, the endophytic actinobacteria may be carried initially into the embryo through the break in the seed husk and then into the endosperm as the seed hydrates from the embryo end. This hypothesis would explain the lack of endosperm infection at 24 h, followed by infection of the endosperm at 3 days. The actinobacteria appear to form microcolonies intracellularly in the plant tissue and were generally found in close proximity to the cell walls of the plant tissue. It is possible that the actinobacteria are within the wall of the plant cell but have not penetrated the plant cell membrane, like the arbuscles of arbuscular mycorrhizal fungi (11).
This visualization technique can be employed to determine the colonization response of endophytic actinobacteria to environmental conditions, as was observed with Frankia (1) or plants grown in hydrocarbon-contaminated sites (18). In the same way, it would be interesting to find any correlation between the pathogen inoculum level and the level of colonization of wheat by endophytes with the ability to control the pathogen.
The level of autofluorescence in the plant tissue may represent one of the biggest challenges for the visualization of endophytic organisms by using fluorescent proteins. Under blue- and green-light excitation, the autofluorescence of the plant tissue was particularly bright and several subcellular plant structural features could also be observed (data not shown). The GFP-expressing actinobacteria were detected by using green emission, together with the absence of red emission (whereas most plant structures exhibited fluorescence across a range of wavelengths). The use of uninfected control plants was also necessary to distinguish those signals from the endophytic actinobacteria and to gauge the autofluorescence of normal plant structures. This problem may be overcome by the use of specific fluorescent proteins with emission wavelengths (19) that coincide with low levels of autofluorescence in plant tissue. It may also be possible to transform the endophytic actinobacteria with multiple copies of the egfp gene in order to increase the fluorescence intensity, as demonstrated with Pseudomonas spp. (21). To be able to readily visualize the narrow actinobacterial hyphae in the plant tissue, one could employ an electron microscopy method as demonstrated by Berg and McDowell (2) for the visualization of Frankia in Casuarina nodules. In studying colonization by an actinobacterial inoculant, the limitation of this method is that it would not be possible to guarantee that that the observed actinobacterial hyphae originated from the inoculant. This problem could be overcome by detection of the organism through an electron microscopy-based technique that remained specific for the particular strain, such as immunogold labeling (7). Immunogold labeling combined with scanning electron microscopy has a resolving power of better than 1 nm and uses the highly specific binding characteristics of an antibody, which should enable visualization of the actinobacterial hyphae.
Acknowledgments
This work was supported by the Australian Grains Research and Development Corporation (GRDC).
We thank Johan Kers of Agri-Food Canada for the plasmids and Michelle Lewis of the Flinders Medical Centre for technical assistance.
REFERENCES
- 1.Benoit, L. F., and A. M. Berry. 1990. Actinorhizal plants in forestry, landscapes and revegetation. In D. D. Baker, J. D. Tjepkema, and C. R. Schwintzer (ed.), The biology of Frankia and actinorhizal plants. Academic Press, San Diego, Calif.
- 2.Berg, R. H., and L. McDowell. 1987. Cytochemistry of the wall of infected cells in Casuarina actinorhizae. Can. J. Bot. 66:2038-2047. [Google Scholar]
- 3.Coombs, J. T., C. M. M. Franco, and R. Loria. 2003. Complete sequencing and analysis of pEN2701, a novel 13-kb plasmid from an endophytic Streptomyces sp. Plasmid 49:86-92. [DOI] [PubMed]
- 4.Cormack, B. P., R. H. Valdiva, and S. Falkow. 1996. FACS optimised mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 5.de Araujo, J. M., A. C. da Silva, and J. L. Azevedo. 2000. Isolation of endophytic actinomycetes from roots and leaves of maize (Zea mays L.). Braz. Arch. Biol. Technol. 43:447-451. [Google Scholar]
- 6.Egener, T., T. Hurek, and B. Reinhold-Hurek. 1999. Endophytic expression of nif genes of Azoarcus sp. strain BH72 in rice roots. Mol. Plant-Microbe Interact. 12:813-819. [DOI] [PubMed] [Google Scholar]
- 7.Faulk, W., and G. Taylor. 1971. An immunocolloid method for the electron microscope. Immunochemistry 8:1081-1083. [DOI] [PubMed] [Google Scholar]
- 8.Flett, F., V. Mersinas, and C. P. Smith. 1997. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 155:223-229. [DOI] [PubMed] [Google Scholar]
- 9.Gage, D. J., T. Bobo, and S. R. Long. 1996. Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa). J. Bacteriol. 178:7159-7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hallmann, J., A. Quadt-Hallmann, W. G. Miller, R. A. Sikora, and S. E. Lindow. 2000. Endophytic colonisation of plants by the biocontrol agent Rhizobium etli G12 in relation to Meloidogyne incognita infection. Phytopathology 91:415-422. [DOI] [PubMed] [Google Scholar]
- 11.Harrison, M. J. 1998. Development of the arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 1:360-365. [DOI] [PubMed] [Google Scholar]
- 12.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood (ed.). 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 13.Mazodier, P., R. Petter, and C. Thompson. 1989. Intergeneric conjugation between Escherichia coli and Streptomyces spp. J. Bacteriol. 171:3583-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Normander, B., N. B. Hendriksen, and O. Nybroe. 1999. Green fluorescent protein-marked Pseudomonas fluorescens: localization, viability, and activity in the natural barley rhizosphere. Appl. Environ. Microbiol. 65:4646-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okazaki, T., K. Takahashi, M. Kizuka, and R. Enokita. 1995. Studies on actinomycetes isolated from plant leaves. Annu. Rev. Sankyo Res. Lab. 47:97-106. [Google Scholar]
- 16.Sardi, P., M. Saracchi, S. Quaroni, B. Petrolini, G. E. Borgonovi, and S. Merli. 1992. Isolation of endophytic Streptomyces strains from surface-sterilized roots. Appl. Environ. Microbiol. 58:2691-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitt-John, T., and J. W. Engels. 1992. Promoter constructions for efficient secretion expression in Streptomyces lividans. Appl. Microbiol. Biotechnol. 36:493-498. [DOI] [PubMed] [Google Scholar]
- 18.Siciliano, S. D., N. Fortin, A. Mihoc, G. Wisse, S. Labelle, D. Beaumier, D. Oulette, R. Roy, L. G. Whyte, M. K. Banks, P. Schwab, K. Lee, and C. W. Greer. 2001. Selection of specific endophytic bacterial genotypes by plants in response to soil contamination. Appl. Environ. Microbiol. 67:2469-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stuurman, N., C. P. Bras, H. R. M. Schlaman, and A. H. M. Wijfjes. 2000. Use of green fluorescent protein color variants expressed on stable broad host range vectors to visualise rhizobia interacting with plants. Mol. Plant-Microbe Interact. 13:1163-1169. [DOI] [PubMed] [Google Scholar]
- 20.Sun, J., G. H. Kelemen, J. M. Fernandez-Albos, and M. J. Bibb. 1999. Green fluorescent protein as a reporter for spatial and temporal expression in Streptomyces coelicolor A3(2). Microbiology 145:2221-2227. [DOI] [PubMed] [Google Scholar]
- 21.Tombolini, R., D. J. van der Gaag, B. Gerhardson, and J. K. Jansson. 1999. Colonization pattern of the biocontrol strain Pseudomonas chlororaphis MA 342 on barley seeds visualized by using green fluorescent protein. Appl. Environ. Microbiol. 65:3674-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]