Abstract
The CCAAT displacement protein/cut homologue (CDP/cut) is a divergent homeodomain protein that is highly conserved through evolution and has properties of a potent transcriptional repressor. CDP/cut contains three conserved cut-repeat domains and a conserved homeobox, each involved in directing binding specificity to unique nucleotide sequence elements. Furthermore, CDP/cut may play a role as a structural component of chromatin through its direct interaction with nucleosomal DNA and association with nuclear matrix attachment regions. CDP/cut is cell-cycle regulated through interactions with Rb, p107, specific kinases and phosphatases directing the transcriptional activity of CDP/cut on such genes encoding p21WAF1,CIP1, c-myc, thymidine kinase, and histones. Our previous studies indicate that CDP/cut is associated with histone deacetylase activity and is associated with a corepressor complex through interactions with histone deacetylases. Here, we report the interaction of CDP/cut with CBP and p300/CREB-binding protein-associated factor (PCAF) along with the modification of CDP/cut by the histone acetyltransferase PCAF. Acetylation of CDP/cut by PCAF is directed at conserved lysine residues near the homeodomain region and regulates CDP/cut function. These observations are consistent with the ability of CDP/cut to regulate genes as a transcriptional repressor, suggesting acetylation as a mechanism that regulates CDP/cut function.
It is generally accepted that histone acetylation is associated with transcriptionally competent chromatin (1). Consistent with that observation is the close relationship of hypoacetylated histones with transcriptionally inert or silent chromatin (2). Despite this perception, it is clear that the random or general increase in core histone acetylation is not necessarily associated with widespread gene activation in chromatin (3). Thus, it is implied that acetylation and deacetylation of core histones by histone acetyltransferases (HATs) and histone deacetylases (HDACs) is a tightly regulated process in chromatin that plays a direct role in gene-specific transcription (4). However, the notion that the acetylation of substrates other than histones by protein acetyltransferase activities, encoded by coactivators of transcription such as p300/CREB-binding protein (CBP) (5–7), has been only a recent revelation, suggesting protein acetyltransferases also participate in transcription through targeting specific protein substrates directly involved in the transcriptional apparatus (8–10). The observation that the coactivator complexes of p300/CBP can associate with a diverse and growing number of DNA-binding transcription factors (11) implicates the intrinsic acetyltransferase activity expressed by p300/CBP (12) as a potential enzymatic function directed at specific proteins associated with p300/CBP.
The CCAAT displacement protein/cut homologue (CDP/cut) repressor is a 180- to 190-kDa polypeptide (13, 14) closely related to the cut protein of Drosophila (15), which determines cell fate in Drosophila (16, 17). CDP/cut was first described as a CCAAT box-binding protein bound with the sea urchin sperm histone promoter (18). Since then, CDP/cut has been shown to regulate many diverse cellular and viral genes. Such genes include gp91phox, c-myc, thymidine kinase (TK), c-mos, TGFβ-type II receptor, CFTR, cdk inhibitor p21WAF1/CIP1, and histone genes in a variety of cell types (14, 19–24). CDP/cut is also involved in the regulation of gene transcription through nuclear matrix attachment regions and postulates a role for CDP/cut as an architectural nuclear protein (25, 26). Sequence analysis of CDP/cut cDNAs reveals that it is a homeobox protein that contains three highly conserved DNA-binding domains (DBDs) designated as cut repeats (27, 28). In addition to the DNA-binding motifs, CDP/cut encodes two closely linked transcriptional repression domains located within the distal C-terminal region of CDP/cut (19, 29).
Recently, we have demonstrated that CDP/cut functions to repress gene transcription through the recruitment of HDAC activity and interaction with HDAC1 both in vivo and in vitro (19). Because it is believed that CDP/cut functions as an architectural protein in chromatin, recent studies have affirmed this notion by demonstrating direct interactions between CDP/cut and nucleosomes associated with the histone H4 promoter (30). Despite the prevailing model for CDP/cut as a potent transcriptional repressor, studies suggest the presence of CDP/cut within a transcriptional activating complex(es) (31). However, little is known about the regulation of CDP/cut function and the role of CDP/cut in a process to modify chromatin. In light of recent observations, we tested whether CDP/cut is a target of HAT activity. We show that CDP/cut interacts with HAT coactivators CBP and PCAF. Furthermore, we demonstrate that the potential for CDP/cut to bind DNA and repress transcription is regulated through acetylation of specific lysine residues near the homeodomain (HD) of CDP/cut. Together, these data support the finding that transcriptional repressors are targets for HAT activity and could be regulated by acetylation.
Materials and Methods
In Vivo Immunoprecipitations and Immunoblot Analysis.
Approximately 2 × 106 HeLa cells were cotransfected with the plasmids for CBP (pRSV-HA/CBP) (32) and CDP/cut (pMT2-CDP) (13) by Lipofectamine-Plus transfection reagent according to manufacturer's instructions (Life Technologies, Grand Island, NY). All procedures were performed at 4°C. After 48 h, transfected cells were washed with PBS, scraped, prepared as lysate, and immunoprecipitated as described (19) with either anti hemagglutinin (anti-HA) antibody, anti-CDP/cut antiserum (19), or normal preimmune serum from guinea pig and mouse. Immunoprecipitated material was separated by SDS/PAGE. Immunoblotting onto poly(vinylidene)fluoride membrane was performed as previously described (19). The membrane was incubated further with either a mouse mAb against the HA-tag of CBP or with polyclonal anti-CDP antiserum. The bound antibodies were detected with peroxidase-conjugated secondary antibody as either goat anti-mouse IgG antibody (Sigma) or goat anti-guinea pig IgG1 antibody (Sigma) and detected by ECL (Amersham Pharmacia Biotech) according to the manufacturer's recommendations. In vivo acetylation of CDP/cut was determined from extracts of HeLa cells transfected with pMT2-CDP immunoprecipitated with anti-CDP/cut or preimmune serum and goat anti-guinea pig IgG agarose (Sigma). Immunoprecipitates were separated by SDS/PAGE, followed by transfer onto nitrocellulose and immunoblotting with rabbit polyclonal antiacetylated lysine antibody (New England Biolabs).
In Vitro Protein–Protein Interaction Assays.
FLAG-tagged CBP fusion proteins were constructed by the PCR and expressed in pcDNA 3.1 (Invitrogen). Fragments of CBP were generated by PCR by using specific primer sets corresponding to the N terminus through amino acid residue 786 and amino acid residue 1621 through the C terminus of mouse CBP. The [35S]-labeled human CDP/cut polypeptide was synthesized in vitro by use of the pGEM 7Zf(+) plasmid in the TNT system (Promega) according to the manufacturer's directions. The [35S]-labeled CDP/cut protein was then incubated with the corresponding FLAG-tagged CBP protein fragment in 250 μl of HEMG binding buffer as described (19). The protein complexes were then immunoprecipitated with anti-FLAG (M2) antibodies conjugated to agarose (Sigma) in HEMG buffer and eluted with FLAG peptide (Sigma). Immunoprecipitates were washed four times in 1 ml of HEMG buffer in the absence of protease inhibitors and BSA. Immunoprecipitates were resolved by SDS/PAGE and visualized by autoradiography.
In Vitro Protein Acetyltransferase Assays.
Protein acetyltransferase reactions were performed as previously described (33). CDP/cut was synthesized by using the TNT system (Promega) in the absence of radioisotope or in the presence of [14C]leucine (50mCi/mmol, New England Nuclear). For the bacterial expression plasmid glutathione S-transferase (GST)-CDP (C3-HD), expression and purification of CDP/cut fragments were previously described (19). For acetyltransferase reactions, approximately 1 μg of in vitro translated product or GST-CDP (C3-HD) and 100 ng of immunoprecipitated FLAG-tagged proteins of CBP (wild type), PCAF (wild type), PCAF (Δ579–608) were incubated with [14C]acetyl-CoA (55mCi/mmol, New England Nuclear) for 1 h at 30°C. After acetyltransferase reactions, GST-CDP (C3-HD) protein was immunoprecipitated with monoclonal anti-GST antibody (Sigma), washed extensively, and the incorporation of [14C]- or [3H]acetyl-CoA was determined by filter-binding assays. Equivalent amounts (50 ng) of input FLAG-tagged PCAF (wild type), PCAF (Δ579–608), or CBP (wild type) were determined in parallel from cell lysates labeled with [35S]methionine (NEN Life Sciences). For electrophoretic assays, the entire reaction mixture was loaded onto a 10% SDS/PAGE and subjected to electrophoresis. Gels containing [14C]-labeled proteins were stained and fixed with 10% glacial acetic acid and 40% methanol for 1 h, soaked in Amplify (Amersham Pharmacia) for 40 min, and autoradiographed. For the preparation of unlabeled acetylated GST-CDP (C3-HD) for electrophoretic mobility-shift assay (EMSA), [14C]acetyl-CoA was replaced with unlabeled acetyl-CoA (Sigma) in a standard acetyltransferase reaction.
EMSAs.
EMSA was carried out essentially as described (19). Sequences of the complementary double-stranded oligonucleotide contained the CDP/cut binding sequences for the human TK promoter, as human TK promoter (TK2C) was previously described (23). The DNA-binding reactions were performed with 200 ng of affinity-purified GST-CDP (C3-HD). For the peptide acetyltransferase competition assays, synthetic peptides were introduced (2 ng = 100-fold molar excess) at the same time as the GST-CDP/cut fusion protein, before the addition of [32P]-labeled TK2C double-stranded oligonucleotide.
GAL4 DBD Fusions with CDP/cut and Transactivation Assays.
To localize repression domains within the CDP/cut protein, various segments of human CDP/cut cDNA were inserted to the GAL4 DBD of plasmid pM commercial reagent (CLONTECH) (34). The cDNA fragments of the human CDP/cut cDNA were amplified with a SalI restriction site linker by using the EXpand system (Roche Diagnostics), and corresponding PCR fragments were directly cloned into the EXpand vector II (Roche). The cDNA fragments were excised with SalI and were then transferred into the SalI site of the GAL4 DNA-binding vector, pM (CLONTECH). Construction of the pG4-CDPΔ1186–1256 plasmid from the parental GAL4-CDP/cut fusion expression plasmid pG4-CDP (wild type) was created by excision with Mro 1 (Roche). After digestion with Mro 1, backbone plasmid pG4-CDP (wild type) DNA was recovered by gel purification and religated with a 964-bp Mro1/XbaI fragment of the CDP/cut cDNA. In plasmid pG4-CDP1256-term, the 964-bp Mro1/XbaI fragment from pG4-CDP (wild type) was blunt end inserted in frame with the GAL4 DBD into pM. The target plasmid GAL4InrCAT was used (described previously) (35). For cotransfections, human kidney (293) cells were grown in DMEM supplemented with 10% FBS (HyClone). Cells were transfected and collected between 24–48 h after transfection, and chloramphenicol acetyltransferase (CAT) assays were performed as described (19) with 10 μg DNA per 10-cm plate; total amounts of DNA were normalized by using the parental (pM) GAL4 expression vector and the GAL4InrCAT target plasmid. In vivo acetylation of GAL4 fusions with CDP/cut, G4-CDPΔ1186–1256, and G4-CDP (wild type) from cotransfected 293 cells was determined with an anti-GAL4 mAb (CLONTECH) and antiacetyllysine antibody by immunoprecipitation and immunoblot analysis by using the same procedure as described above.
Results and Discussion
Interaction of CDP/cut with HATs, CBP, and PCAF.
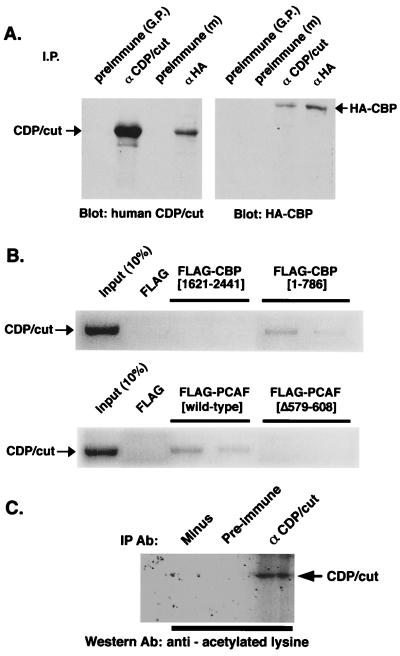
Because transcriptional repression by CDP/cut is associated with HDAC activity, and the function of CDP/cut is suppressed by both HAT activity and the HDAC inhibitor, TSA (19), we suspected that CDP/cut interacts with CBP. Cells were cotransfected with constructs that direct the overexpression of CDP/cut (pMT2-CDP) and the HA-tagged CBP (pRSV-HA/CBP). Immunoprecipitation of specific CDP/cut and CBP complexes was followed by immunoblot analysis of CDP/cut. Fig. 1A Left shows that CDP/cut is immunoprecipitated with a HA-tagged (HA-) CBP complex in extracts from cotransfected HeLa cells. A corresponding immunoblot of HA-CBP was performed after immunoprecipitation of both CDP/cut and HA-CBP. Results in Fig. 1A Right indicate that CBP as well is immunoprecipitated with CDP/cut. We conclude that CBP interacts with CDP/cut in vivo. To determine whether CDP/cut interacts with either of the prevalent protein interaction domains of CBP (11), an in vitro “pull-down” assay was performed with domains of CBP expressed as FLAG-tagged fusion proteins. FLAG-tagged proteins were constructed and expressed as fusions with CBP between the residues (1–786) or (1621–2441) and correspond to the N and C termini of CBP, respectively. CDP/cut was synthesized in vitro with [35S]methionine (Fig. 1B Upper). To demonstrate whether the interaction of CDP/cut with CBP was specific to either the N or C termini of CBP, [35S]-labeled CDP/cut was mixed with FLAG-tagged protein fragments of CBP, immunoprecipitated with anti-FLAG (M2) antibody, resolved by SDS/PAGE, and autoradiographed. Results of experiments shown in Fig. 1B indicate that CBP interacts with CDP/cut through the N terminus of CBP in vitro. In a similar experiment, CDP/cut was again synthesized and radiolabeled. In vitro-translated CDP/cut were mixed with FLAG-tagged PCAF or FLAG-tagged PCAF with a deletion in the HAT domain (Δ579–608). Results show that PCAF interaction in vitro with CDP/cut requires the HAT domain of PCAF.
Figure 1.
Interaction of CBP and PCAF with CDP/cut. (A) Immunoprecipitation/Western blot analysis of transfected HA-tagged CBP and CDP/cut. HeLa cells were cotransfected with expression vectors for CBP and CDP/cut. Immunoprecipitations with antibody or antiserum directed to either the HA-tag (αHA) or CDP/cut, respectively, from the lysates of transfected cells were recovered and subjected to immunoblotting by antisera indicated under the blot. Arrows indicate the predicted size of the detectable immunoblotted protein. (B) [35S]-labeled CDP/cut was synthesized in vitro and mixed with purified FLAG-tagged CBP, corresponding to the residues of CBP as indicated, immunoprecipitated with anti-FLAG M2 antibody (Sigma), electrophoresed, and detected by autoradiography. (Lower) [35S]-labeled CDP/cut was synthesized in vitro and mixed with purified FLAG-tagged PCAF (wild type) or PCAF (Δ579–608) and visualized as in Upper. (C) Acetylation of CDP/cut in vivo. Lysates from HeLa cells, transfected with pMT2-CDP, were immunoprecipitated with anti-CDP/cut antiserum or preimmune serum as a control. Immunoprecipitates were analyzed by immunoblot analysis by using a polyclonal antiacetylated lysine antibody.
Because CDP/cut can interact with CBP and PCAF, we tested whether CDP/cut is acetylated in vivo. Antibodies raised against acetylated lysine residues from histone H4 have been shown to have the ability to recognize acetylated nonhistone proteins in vivo (10, 36) and were used in an experiment shown in Fig. 1C. HeLa cells, previously transfected with constructs that direct expression of CDP/cut (pMT2-CDP), were used as a source of CDP/cut and immunoprecipitated with anti-CDP/cut antibody or preimmune serum. The antiacetylated lysine antibody was then used in an immunoblot. Fig. 1C shows that the specifically immunoprecipitated CDP/cut protein is recognized by the antiacetylated lysine antibody.
HAT Activity of PCAF Inhibits DNA Binding by CDP/cut.
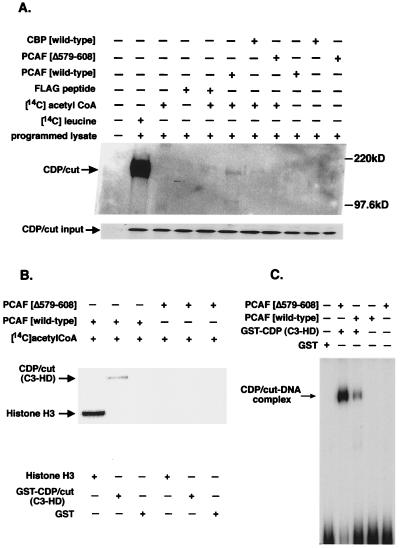
To investigate whether the interaction of CDP/cut with CBP results in the specific acetylation of CDP/cut, protein acetyltransferase assays were performed by using immunopurified preparations of recombinant HAT proteins, CBP and PCAF, expressed as HA- and FLAG-tagged fusion protein, respectively. Again, CDP/cut was synthesized in vitro and subjected to acetyltransferase reactions in the presence of [14C]acetyl-CoA with HA- or FLAG-tagged recombinant proteins of CBP (wild type), PCAF (wild type), or mutant PCAF (Δ576–608), respectively. Reactions of CDP/cut were analyzed by SDS/PAGE. To ensure the synthesis of the correct CDP/cut protein, a translated product of CDP/cut was labeled with [14C]leucine and immunoprecipitated (Fig. 2A, second lane from left). A corresponding immunoblot of in vitro-translated CDP/cut was used to normalize the amount of CDP/cut used as substrate for acetyltransferase reactions in vitro shown in Fig. 2A Lower. Results show in Fig. 2A (sixth lane from the left) that acetyltransferase activity of PCAF on CDP/cut is detectable after exposure of the gel by autoradiography. However, a comparative study shows that acetyltransferase activity from CBP is not detectable on the CDP/cut protein (Fig. 2A). Consistent with the results that PCAF-HAT activity is responsible for the acetylation of CDP/cut, the same assay was conducted with the PCAF mutant lacking HAT activity (Δ579–608) and fails to show any detectable acetyltransferase activity on CDP/cut. Therefore, our result shows that HAT activity expressed by PCAF preferentially acetylates CDP/cut, whereas CBP HAT activity does not produce a readily detectable signal in this regard. Because the third cut repeat (C3) and HD of CDP/cut are highly conserved and are important for DNA-binding specificity (37), we tested whether this DNA-binding region of CDP/cut is subjected to acetylation by PCAF in vitro. Results shown in Fig. 2B indicate that the regions encoding the C3 and HD of CDP/cut and histone H3 are readily acetylated by recombinant PCAF when compared with the mutant PCAF (Δ579–608) lacking HAT activity. To demonstrate that acetylation of the C3 and HD region of CDP/cut affects DNA binding to a cognate sequence, the fusion protein of CDP/cut (C3-HD) was prepared and tested in a gel-shift experiment after treatment with PCAF and acetyl-CoA (Fig. 2C). Results in Fig. 2C show that the acetylation in vitro of the GST-CDP (C3-HD) fusion protein by PCAF inhibit CDP/cut binding to the TK2C sequence (23).
Figure 2.
CDP/cut is a target of acetylation by the HAT PCAF. (A) In vitro acetyltransferase reactions were used on in vitro-synthesized and unlabeled CDP/cut. Equivalent molar amounts of CBP (wild type), PCAF (wild type), and PCAF (Δ579–608) were immunoprecipitated from cells expressing HA-tagged CBP (wild type), FLAG-tagged PCAF (wild type), and FLAG-tagged PCAF (Δ579–608) and used in an in vitro reaction along with [14C]acetyl-CoA where indicated. CDP/cut was synthesized with [14C]leucine in vitro and immunoprecipitated as a control. Immunoprecipitated products were resolved on a SDS/PAGE gel and autoradiographed. Lower corresponds to an immunoblot of the in vitro-translated product of CDP/cut as the input amount for protein acetyltransferase reactions in vitro. (B) Acetylation of C3 and HD of CDP/cut. GST fusion protein of CDP/cut (C3-HD) was affinity purified and subjected to acetylation reactions with the same FLAG-tagged reagents described above. Acetylated product of the GST fusion with CDP/cut was separated by SDS/PAGE and autoradiographed. Free histone H3 served as a positive control for the acetyltransferase activity of PCAF. Gels were stained with Coomassie stain for estimation of protein levels before autoradiography (not shown). (C) Acetylation of CDP/cut disrupts DNA binding to the TK promoter. EMSA of the acetylated product of GST-CDP/cut (C3-HD) after in vitro acetyltransferase reactions as described above with the exception that reactions were performed with unlabeled acetyl-CoA.
Regulation of CDP/cut Repression by PCAF Occurs Near the HD of CDP/cut and Corresponds with the Acetylation of CDP/cut in Vivo.
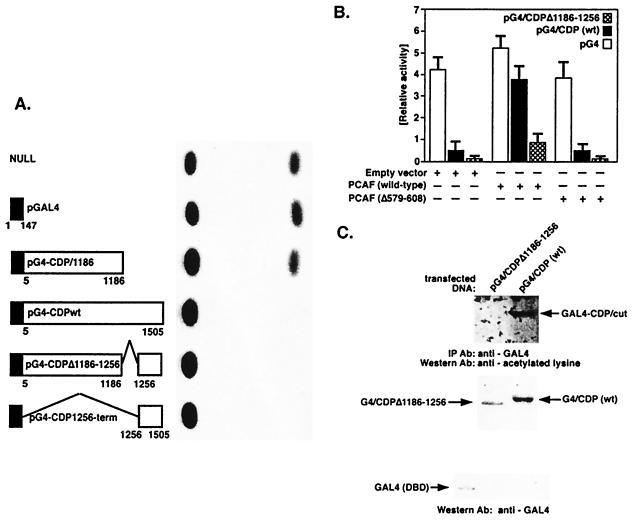
Because repression by CDP/cut is expressed through the C-terminal domain (29, 38) and repression correlates with the ability of CDP/cut to interact with the HDAC HDAC1 (19), we examined whether this phenotype affects the ability of CDP/cut to repress transcription. In light of previous studies documenting the repression potential of CDP/cut localized within the C-terminal domain (19, 29), we tested whether several deletions of CDP/cut would affect repression of a basal promoter directed by tandem GAL4 upstream activating sequences (UAS). Several CDP/cut cDNA fragments were fused to GAL4 and tested in a transactivation assay (Fig. 3A). Results show the ability of the CDP/cut to repress transcription of a UAS-directed promoter (Fig. 3 A and B). Deletion of the C-terminal region from CDP/cut fails to repress transcription. This result is consistent with previous studies (29). However, results in Fig. 3A show that full-length CDP/cut was less effective at repression when compared with the fusion of the C-terminal region to the DBD of GAL4 alone. Surprisingly, a deletion created between C3 and HD of CDP/cut (pG4-CDPΔ1186–1256) corresponded to a moderate increase in repression by CDP/cut by more than 3-fold. Because CDP/cut can repress gene transcription by two distinct (or separate) mechanisms (29), and CDP/cut function is altered by HAT activity (19), we tested whether PCAF was capable of regulating repression by CDP/cut independent of specific DNA binding by CDP/cut. The deletion of residues between 1186 and 1256 increased the potential for CDP/cut repression; further delineation of the CDP/cut amino acid sequence indicates high homology to the amino terminal of histone H4 between the C3 and HD of CDP/cut. In an attempt to determine whether this region of CDP/cut was potentially regulated by PCAF, cotransfection studies were performed. Fig. 3B shows a GAL4 transactivation assay performed with the CDP/cut fragments fused to the GAL4 DBD. Fig. 3B demonstrates that PCAF HAT activity can modulate repression directed by CDP/cut. However, a deletion to CDP/cut between C3 and HD fusion of CDP/cut with GAL4, shown as pG4-CDPΔ1186–1256, is unaffected by PCAF cotransfection (Fig. 3B). To confirm that fragments of CDP/cut fused to GAL4 are substrates for acetylation, in vivo immunoprecipitation with anti-GAL4-specific mAb was performed. The immunoprecipitates of the two GAL4 fusion proteins of CDP/cut, pG4-CDPΔ1186–2256 and pG4-CDP (wild type) were then characterized by immunoblot analysis with antiacetylated lysine antibody (Fig. 3C). Results show (Fig. 3C Upper) that the GAL4 fusion protein, corresponding to the wild-type CDP/cut protein, pG4-CDP (wild type), is readily detected by antiacetylated lysine-specific antibody. However, the same GAL4 (DBD) protein fragment fused with CDP/cut but lacking residues between 1186 and 1256 (pG4-CDPΔ1186–1256) is not recognized by the antiacetylated lysine-specific antibody (Fig. 3C). Therefore, acetylation of CDP/cut in vivo by PCAF corresponds to the inhibition of CDP/cut-mediated repression by PCAF in this cotransfection study.
Figure 3.
CDP/cut is regulated by PCAF HAT activity through sequences between the C3 and HD and corresponds to the acetylation of CDP/cut in vivo. (A) Schematic diagram depicts various deletion mutants of the CDP/cut cDNA fused in frame to the DBD of GAL4. The deletion mutants of CDP/cut cDNA were cloned into parental plasmid pM (CLONTECH) containing the DBD of GAL4. The human CDP/cut cDNA fragments, fused to GAL4, were tested in a transactivation assay and monitored for the repression of the cotransfected target CAT reporter gene, GAL4InrCAT, as shown in the photo (Right). The NULL (A Upper) sample corresponds to the background expression of the reporter gene construct GAL4InrCAT. (B) Comparison of the relative activities of the individual CDP/cut cDNAs shown (A) fused to the GAL4 DBD is shown in the presence (or absence) of the cotransfected PCAF HAT activity. Results compare the ability of specific CDP/cut deletions to repress activity of the GAL4InrCAT target. Full-length wild-type CDP/cut [pG4-CDP (wild type)] and deletion mutant of CDP/cut (pG4-CDPΔ1186–1256) are compared with the background level of CAT activity rendered in the presence of the parental GAL4 expression vector shown as pGAL4 (n = 10). (C) To analyze the in vivo acetylation of GAL4 derived fusion proteins of CDP/cut, G4-CDP (wild type) and G4/CDPΔ1186–1256, 8 μg of PCAF (wild type) plasmid was cotransfected with 5 μg of either pG4-CDP (wild type) or pG4/CDPΔ1186–1256 into 293 cells. Acetylation of the CDP/cut fusions with the DBD of the GAL4 protein was determined by immunoprecipitation of equivalent amounts of G4/CDP (wild type) and G4/CDPΔ1186–1256 protein with a mouse monoclonal anti-GAL4 antibody and precipitated with goat anti-mouse IgG agarose. The immunoprecipitates, separated by SDS/PAGE, were immunoblotted with antiacetylated lysine antibodies (Upper). A corresponding immunoblot (Lower) by using an anti-GAL4 mAb was performed with the identical cell lysates [as shown (Upper)] from 293 cells, cotransfected with PCAF and either the parental GAL4 vector pM, pG4-CDPΔ1186–1256, or pG4-CDP (wild type).
Acetylation of CDP/cut at Conserved Lysine Residues Alters DNA Binding.
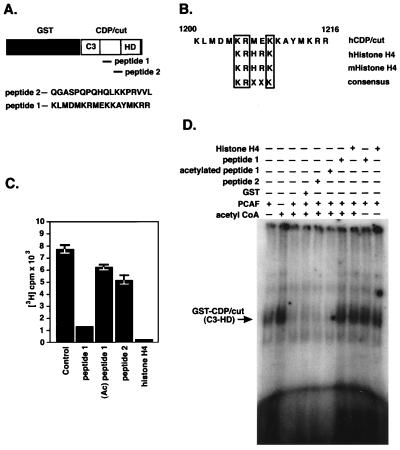
Because acetylation of CDP/cut by PCAF had been determined (Fig. 2), evidence that specific lysine residues act as substrates for PCAF HAT activity was still hypothetical, and a prospective consensus was identified by comparing sequences prototypic of HAT substrates through sequence database analysis. The consensus KRXXK was identified in a span of 59 amino acid residues between the C3 and HD of CDP/cut and conforms to the acetylation of other nonhistone proteins (8, 10, 36). The consensus of residues was conserved throughout other orthologues of CDP/cut (39, 40) and suggests a consensus typical of histone H4 (41), corresponding to the lysine at position 16 of both mouse and human (Fig. 4B). To determine whether the residues between C3 and HD are substrates for PCAF HAT activity, a GST fusion protein encoding the C3 HD of CDP/cut was expressed and affinity purified as a substrate for PCAF. After the protein acetyltransferase reactions with FLAG-tagged PCAF, GST-CDP/cut was immunoprecipitated with anti-GST antibody, and acetylation status was quantified by filter-binding assays. To determine the specificity of acetyltransferase activity in vitro, two peptides were synthesized and used to determine substrate specificity of PCAF acetyltransferase activity on CDP/cut by competition analysis of the peptides as competing substrates for PCAF (Fig. 4A). Results of acetylation assays performed in vitro on the recombinant GST-CDP/cut substrate show that PCAF acetylates the fraction of CDP/cut encoding the C3 through the HD. Furthermore, we were able to determine that peptide 1 and histone H4 were both capable of competing for acetyltransferase activity expressed by PCAF, whereas peptide 2 or the preacetylated peptide 1 failed to compete for acetyltransferase activity by PCAF. Therefore, we conclude that the analogous peptide sequence in CDP/cut is the bona fide substrate for PCAF.
Figure 4.
Localization of PCAF acetyltransferase activity on CDP/cut. (A) A schematic diagram depicts the location of two synthetic peptides used in the competition assay relative to the structure of CDP/cut, between C3 and HD in a GST fusion protein. The primary sequence of residues for the competing peptides is shown (Lower). (B) Consensus site of residues within the sequence derived from peptide 1. Sequence comparison of residues located within the histone-like consensus of CDP/cut and sequences of the histone H4 from man and mouse. (C) Peptide competition for acetyltransferase activity was performed with CDP/cut and PCAF. Synthetic peptides shown in A were used in an in vitro protein acetyltransferase assay as competing substrates with immunoaffinity-purified FLAG-tagged PCAF corresponding to the wild-type PCAF in the presence of [3H]acetyl-CoA. Synthetic nonacetylated peptides 1 and 2, preacetylated peptide 1 [100 mol/mol of GST-CDP (C3-HD)] or a synthetic peptide corresponding to the 24 amino terminal of mouse histone H4 were introduced into acetyltransferase reaction as substrates before the addition of GST-CDP (C3-HD). The GST-CDP (C3-HD) fusion protein was introduced into the acetyltransferase reactions and quenched with unlabeled acetyl-CoA after 10 min. Incorporation of [3H]acetyl-CoA into GST-CDP (C3-HD) was determined by filter-binding assays. (D) EMSA of acetylated GST-CDP (C3-HD). Acetyltransferase assay was performed as described in C with the exception that the acetyltransferase reactions with GST-CDP (C3-HD) were conducted with unlabeled acetyl-CoA. After the termination of the acetyltransferase reactions conducted in vitro, GST-CDP (C3-HD) was affinity purified with glutathione–agarose and eluted with 10 mM glutathione. Affinity-purified GST-CDP (C3-HD) products of the acetyltransferase reactions were then added to DNA-binding reactions by using the [32P]-labeled TK2C oligonucleotide, and reactions were loaded onto 4% polyacrylamide gels for EMSA.
To determine the effects of acetylation on DNA binding within the region encoding the C3 and HD of CDP/cut, EMSA was performed with GST fused to the C3 and HD of CDP/cut. Shown in Fig. 4D is the result of an EMSA by using the oligonucleotide (TK2C), homologous to the human TK promoter sequence (23). Fusion proteins were subjected to acetyltransferase reactions with FLAG-tagged PCAF, acetyl-CoA, and competing peptides followed by EMSA. Our results show (Fig. 4D) that PCAF dramatically reduces the ability of CDP/cut to bind the TK promoter element through the C3 repeat and HD. The DNA-binding potential of CDP/cut is preserved in the presence of the two competing acetyltransferase substrates, peptide 1 and a synthetic peptide corresponding to histone H4. Based on results of this experiment, we conclude that acetylation of CDP/cut within the consensus analogous to peptide 1 serves as a negative regulatory motif in CDP/cut.
In conclusion, we have demonstrated that CDP/cut interacts with CBP, but acetylation of CDP/cut occurs primarily with PCAF HAT activity. Acetylation of CDP/cut by PCAF affects both DNA binding and transcriptional repression by CDP/cut. Our studies indicate that DNA binding and transcriptional repression are distinct properties encoded separately in CDP/cut. These results could be reconciled by the fact that two independent mechanisms of repression expressed by CDP/cut, active repression and competition for binding-site occupancy, are independent of each other (29). This is consistent with the notion that activity of CDP/cut may be expressed through acetylation as one of many mechanisms regulating CDP/cut function. Acetylation of CDP/cut, like that of nucleosomal core histones, likely influences the ability of CDP/cut to associate with nucleosomal DNA and thus recruit HDAC activity to specific nucleosomes (19, 24). This process, in turn, may represent a mechanism to direct the position of nucleosomes and may contribute to either repression or activation of gene transcription. These results provide insight on how signaling pathways expressed through protein acetyltransferases such as PCAF may determine the function of transcriptional repressors and ability to regulate target genes.
Acknowledgments
We thank R. H. Goodman (Vollum Institute, University of Oregon, Portland, OR) for providing plasmid pRSV-HA/CBP, P. Nakatani (National Institute of Child Health and Human Development, National Institutes of Health) for FLAG-tagged PCAF expression plasmids, T. Kouzarides (Wellcome/CRC Institute, University of Cambridge) for plasmid GAL4InrCAT, E. Neufeld (Children's Hospital, Harvard University) for pMT2-CDP plasmid DNA and CDP/cut antiserum, and E. Stefayano (University of Athens, Athens, Greece) for synthetic acetylated peptides. We also thank J. J. Bieker and F. Ramirez for helpful discussions and for critically reviewing this manuscript. This work was supported by an award from the National Institutes of Health of the United States Public Health Service. M.J.W. is supported by the Ruth Larkey Foundation for Children.
Abbreviations
- HAT
histone acetyltransferase
- CDP/cut
CCAAT-displacement protein/cut homologue
- p300/CBP
p300/CREB-binding protein
- PCAF
p300/CBP-associated factor
- HDAC
histone deacetylase
- GST
glutathione S-transferase
- DBD
DNA binding domain
- EMSA
electrophoretic mobility-shift assay
- CAT
chloramphenicol acetyltransferase
- HA
hemagglutinin
- TK2C
human thymidine kinase promoter
- HD
homeodomain
- C3
third cut repeat
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.130028697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.130028697
References
- 1.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 2.Kadosh D, Struhl K. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 3.Pazin M J, Kadonaga J T. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 4.Kadosh D, Struhl K. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 6.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 7.Blanco J C, Minucci S, Lu J, Yang X J, Walker K K, Chen H, Evans R M, Nakatani Y, Ozato K. Genes Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu W, Roeder R G. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 9.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 10.Zhang W, Bieker J J. Proc Natl Acad Sci USA. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldman P S, Tran V K, Goodman R H. Recent Prog Horm Res. 1997;52:103–119. [PubMed] [Google Scholar]
- 12.Bannister A J, Kouzarides T. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 13.Neufeld E J, Skalnik D G, Lievens P M, Orkin S H. Nat Genet. 1992;1:50–55. doi: 10.1038/ng0492-50. [DOI] [PubMed] [Google Scholar]
- 14.Skalnik D G, Strauss E C, Orkin S H. J Biol Chem. 1991;266:16736–16744. [PubMed] [Google Scholar]
- 15.Blochlinger K, Jan L Y, Jan Y N. Development (Cambridge, UK) 1993;117:441–450. doi: 10.1242/dev.117.2.441. [DOI] [PubMed] [Google Scholar]
- 16.Blochlinger K, Bodmer R, Jan L Y, Jan Y N. Genes Dev. 1990;4:1322–1331. doi: 10.1101/gad.4.8.1322. [DOI] [PubMed] [Google Scholar]
- 17.Jack J, Dorsett D, Delotto Y, Liu S. Development (Cambridge, UK) 1991;113:735–747. doi: 10.1242/dev.113.3.735. [DOI] [PubMed] [Google Scholar]
- 18.Barberis A, Superti-Furga G, Busslinger M. Cell. 1987;50:347–359. doi: 10.1016/0092-8674(87)90489-2. [DOI] [PubMed] [Google Scholar]
- 19.Li S, Moy L, Pittman N, Shue G, Aufiero B, Neufeld E J, LeLeiko N S, Walsh M J. J Biol Chem. 1999;274:7803–7815. doi: 10.1074/jbc.274.12.7803. [DOI] [PubMed] [Google Scholar]
- 20.Coqueret O, Berube G, Nepveu A. EMBO J. 1998;17:4680–4694. doi: 10.1093/emboj/17.16.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dufort D, Nepveu A. Mol Cell Biol. 1994;14:4251–4257. doi: 10.1128/mcb.14.6.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.el-Hodiri H M, Perry M. Mol Cell Biol. 1995;15:3587–3596. doi: 10.1128/mcb.15.7.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim E C, Lau J S, Rawlings S, Lee A S. Cell Growth Differ. 1997;8:1329–1338. [PubMed] [Google Scholar]
- 24.Wang Z, Goldstein A, Zong R T, Lin D, Neufeld E J, Scheuermann R H, Tucker P W. Mol Cell Biol. 1999;19:284–295. doi: 10.1128/mcb.19.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banan M, Rojas I C, Lee W H, King H L, Harriss J V, Kobayashi R, Webb C F, Gottlieb P D. J Biol Chem. 1997;272:18440–18452. doi: 10.1074/jbc.272.29.18440. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Barnett A, Neufeld E J, Dudley J P. Mol Cell Biol. 1999;19:4918–4926. doi: 10.1128/mcb.19.7.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harada R, Dufort D, Denis-Larose C, Nepveu A. J Biol Chem. 1994;269:2062–2067. [PubMed] [Google Scholar]
- 28.Aufiero B, Neufeld E J, Orkin S H. Proc Natl Acad Sci USA. 1994;91:7757–7761. doi: 10.1073/pnas.91.16.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mailly F, Berube G, Harada R, Mao P L, Phillips S, Nepveu A. Mol Cell Biol. 1996;16:5346–5357. doi: 10.1128/mcb.16.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Last T J, van Wijnen A J, de Ridder M C, Stein G S, Stein J L. Mol Biol Rep. 1999;26:185–194. doi: 10.1023/a:1007058123699. [DOI] [PubMed] [Google Scholar]
- 31.Yoon S O, Chikaraishi D M. J Biol Chem. 1994;269:18453–18462. [PubMed] [Google Scholar]
- 32.Kwok R P, Laurance M E, Lundblad J R, Goldman P S, Shih H, Connor L M, Marriott S J, Goodman R H. Nature (London) 1996;380:642–646. doi: 10.1038/380642a0. [DOI] [PubMed] [Google Scholar]
- 33.Herrera J E, Bergel M, Yang X J, Nakatani Y, Bustin M. J Biol Chem. 1997;272:27253–27258. doi: 10.1074/jbc.272.43.27253. [DOI] [PubMed] [Google Scholar]
- 34.Sadowski I, Ptashne M. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Metz R, Bannister A J, Sutherland J A, Hagemeier C, O'Rourke E C, Cook A, Bravo R, Kouzarides T. Mol Cell Biol. 1994;14:6021–6029. doi: 10.1128/mcb.14.9.6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Nature (London) 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 37.Harada R, Berube G, Tamplin O J, Denis-Larose C, Nepveu A. Mol Cell Biol. 1995;15:129–140. doi: 10.1128/mcb.15.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ai W, Toussaint E, Roman A. J Virol. 1999;73:4220–4229. doi: 10.1128/jvi.73.5.4220-4229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andres V, Chiara M D, Mahdavi V. Genes Dev. 1994;8:245–257. doi: 10.1101/gad.8.2.245. [DOI] [PubMed] [Google Scholar]
- 40.Ludlow C, Choy R, Blochlinger K. Dev Biol. 1996;178:149–159. doi: 10.1006/dbio.1996.0205. [DOI] [PubMed] [Google Scholar]
- 41.Schiltz R L, Mizzen C A, Vassilev A, Cook R G, Allis C D, Nakatani Y. J Biol Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]