Abstract
Previously, only one ribosome protection type of a tetracycline resistance gene, tetQ, had been identified in Bacteroides spp. During an investigation of anaerobic bacteria present in swine feces and manure storage pits, a tetracycline-resistant Bacteroides strain was isolated. Subsequent analysis showed that this new Bacteroides strain, Bacteroides sp. strain 139, did not contain tetQ but contained a previously unidentified tetracycline resistance gene. Sequence analysis showed that the tetracycline resistance gene from Bacteroides sp. strain 139 encoded a protein (designated Tet 36) that defines a new class of ribosome protection types of tetracycline resistance. Tet 36 has 60% amino acid identity over 640 aa to TetQ and between 31 and 49% amino acid identity to the nine other ribosome protection types of tetracycline resistance genes. The tet(36) region was not observed to transfer from Bacteroides sp. strain 139 to another Bacteroides sp. under laboratory conditions. Yet tet(36) was found in other genera of bacteria isolated from the same swine manure pits and from swine feces. Phylogenetic analysis of the tet(36)-containing isolates indicated that tet(36) was present not only in the Cytophaga-Flavobacter-Bacteroides group to which Bacteroides sp. strain 139 belongs but also in gram-positive genera and gram-negative proteobacteria, indicating that horizontal transfer of tet(36) is occurring between these divergent phylogenetic groups in the farm environment.
Due to their broad-spectrum activity and low toxicity, members of the tetracycline group of antibiotics, which includes tetracycline, chlortetracycline, minocycline, and doxycycline, have been used widely in human therapy. Tetracyclines have also been used in agriculture as growth promoters in farm animals and for prophylaxis in plant agriculture and in aquaculture. Use of tetracyclines to treat human infections has been associated with a significant rise in resistance to tetracyclines not only in human pathogens but also in human intestinal bacteria (6, 36).
Bacteroides species are not only among the numerically predominant genera of bacteria in the normal microfloras of the human colon but are also opportunistic human pathogens (12). In a recent survey of human clinical and intestinal isolates, Shoemaker et al. found that before 1970, 30% of human intestinal Bacteroides isolates were resistant to tetracycline whereas over 80% of the intestinal Bacteroides strains isolated in the 1990s were resistant to tetracycline (32). A similar rise in the incidence of tetracycline resistance was seen in both clinical and commensal Bacteroides isolates, indicating that the commensal bacterial species that make up the colonic microfloras are being affected by the use of antibiotics as much as the bacterial species causing infections (32).
Resistance to tetracycline among human clinical and intestinal Bacteroides isolates, whether from the pre-1970 period or the 1990s, was found to be due to a single tetracycline resistance gene, tetQ, which encodes a protein that protects ribosomes from tetracycline by a mechanism that is still not well understood (32). Results from the same study indicated that the spread of tetQ among human Bacteroides species was mediated by a type of conjugative transposon (CTn) exemplified by CTnERL and CTnDOT, two CTns that are virtually identical except for a 13-kb segment found in CTnDOT but not in CTnERL (32, 41, 42). CTns are DNA elements that are normally found integrated into the host chromosome, except during transfer, when they excise from the chromosome to form a circular transfer intermediate, a copy of which is transferred by conjugation to the recipient bacterium, where it integrates into the recipient chromosome (28).
Since tetracycline use in human medicine has been associated with, and probably caused, the increase in the incidence of tetracycline-resistant strains of human Bacteroides species, it might be expected that use of tetracycline in agriculture would also be associated with the spread of tetracycline resistance among bacteria in the intestines of farm animals (1, 2). Tetracycline resistance in Escherichia coli and other members of the proteobacteria has been documented, and this resistance is usually due to genes encoding efflux pumps rather than ribosome protection proteins (1, 22). There is little information about tetracycline resistance genes, however, in the numerically predominant populations such as the gram-positive anaerobes and Bacteroides. There have been a few reports of tetQ in isolates of ruminal Prevotella species, members of the Bacteroides phylogenetic group, but there have been no studies of tetracycline-resistant strains of Bacteroides or related genera in pigs.
A continuing problem in animal husbandry is the disposal of manure (8, 11, 18, 20, 39). Antibiotic resistance among bacteria in manure is of concern, because these organisms can leak into nearby groundwater. In a recent study of anaerobic bacteria from swine manure, numerous tetracycline-resistant isolates were found (10). In this report, we describe a new ribosome protection type of resistance gene that was found in one of the Bacteroides isolates. This same gene was also found in other genera of bacteria that are not members of the Bacteroides phylogenetic group.
Previously, 10 classes of ribosome protection types of tetracycline resistance proteins were known, including TetM, TetO, TetB(P), TetQ, TetS, TetT, TetW, OtrA, Tet, and Tet (32) (23, 24, 37). Members of the most recently discovered tetracycline resistance classes are now given a number rather than a letter designation. We show here that the manure pit isolate defines a new class of ribosome protection type tetracycline resistance protein that we have designated Tet36. Our findings demonstrate that new types of ribosome protection resistance genes remain to be found and that resistance genes found in the farm environment might differ in some cases from those discovered to date in human isolates.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Community isolates from 1996 and 1997 were obtained from students in the microbial diversity course at Woods Hole, Mass. (designations beginning with WH), while other strains are community (isolated in the 1960s) and clinical isolates obtained from various sources within the United States and isolated from before 1960 to the 1990s (32). The pure culture (PC) swine fecal isolates (PC111 and PC123B) and manure storage pit isolates (PC139, PC88, and PC128) were provided by the Fermentation Biotechnology Research Unit, National Center for Agricultural Utilization Research, Peoria, Ill. The strains are referred to by their numerical designations throughout this paper. The methods employed for growth of Bacteroides and E. coli strains, DNA manipulation, and conjugal transfer have been described elsewhere (29, 30; L. V. Holdeman and W. E. C. Moore, Anaerobe laboratory manual, 4th ed., Virginia Polytechnic Institute and State University, Blacksburg, Va., 1975). The methods employed for cultivation of swine fecal and manure storage pit isolates have been reported in detail elsewhere (10, 39, 40). Briefly, swine feces and waste pit samples were collected and suspended in anaerobic salts buffer and serial dilutions of the suspension were plated onto anaerobic complex medium containing tylosin or tetracycline and incubated in an anaerobic chamber at 37°C for up to 21 days. Colonies were streaked onto appropriate medium for further analyses. The antibiotic concentrations used were as follows: ampicillin, 100 μg/ml; cefoxitin, 20 μg/ml; erythromycin, 10 μg/ml; gentamicin, 200 μg/ml; and tetracycline, 3 μg/ml. Bacteroides spp. were initially isolated as gram-negative, nonmotile, obligate anaerobes that were aerotolerant and resistant to aminoglycosides.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant phenotype(s)a | Source and/or description and/or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5αMCR | RecA | Gibco-BRL |
| S17-1 | RecA Tpr Strr (ΩRP42-Tc::Mu-Km::Tn7a) | IncPα plasmid RP4 inserted into the S17-1 chromosome by bacteriophage Mu (35) |
| HB101 | RecA Strr | 5 |
| Bacteroides | ||
| BT4001 | Rifr | Spontaneous rifampin mutant of B. thetaiotaomicron 5482A (34) |
| BU1001 | Rifr | Spontaneous rifampin mutant of B. uniformis 0061 (31) |
| Bacteroides sp. strain 139 | Tpr Emr Tcr | Wild-type Bacteroides species isolated from swine pool (T. R. Whitehead, unpublished data) |
| 139ΩpGW142.2 | Tpr Emr Cefr | Bacteroides sp. strain 139 with suicide vector pGW142.2 inserted into tet(36) by homologous recombination (this study) |
| 139ΩpGW142.4 | Tpr Emr Cefr | Bacteroides sp. strain 139 with suicide vector pGW142.4 inserted into tet(36) by homologous recombination (this study) |
| Plasmids | ||
| pGWA34.2 | Apr (Cefr) | A 2.2-kb BamHI/EcoRI fragment from pFD351 (26) containing cefoxitin resistance gene cefA, cloned into the SspI site of pUC19oriTRK2; contains no Bacteroides replicon and so is a suicide vector in Bacteroides spp. (this study) |
| pGW47.17 | Kmr | A blunted 3.0-kb AatII/NarI fragment containing the replication and mobilization region from Bacteroides plasmid pBI143 was cloned into the SspI site of pK184 (19), a p15A replicon (this study) |
| pGWA48.3 | Kmr (Emr) | A blunted 1.3-kb PstI/SphI fragment containing the ermG resistance gene from CTn7853 (9) was cloned into the AflIII site of pGW47.17, generating a new Bacteroides-E. coli shuttle vector (this study) |
| pGW140.1 | Apr (Cefr) | A 1.1-kb PCR product containing an internal region of tet(36) amplified from Bacteroides sp. strain 139 by means of degenerate PCR primers DI and DII cloned into pGEMT (this study) |
| pGW141.1 | Apr (Cefr) | A 1.3-kb PCR product containing an internal region of tet(36) amplified from Bacteroides strain 139 by means of degenerate PCR primers DI and DII cloned into pGEMT (this study) |
| pGW142.2 | Apr (Cefr) | A 0.9-kb HincII internal tet(36) fragment from pGW140.1 cloned into the SmaI site of suicide vector pGWA34.2 in orientation A or B (this study) |
| pGW150.1 | Apr (Cefr) | A 3.6-kb SphI fragment rescued from Bacteroides sp. strain 139ΩpGW142.4 containing the carboxy-terminal end of tet(36) and downstream sequences (this study) |
| pGW151.1 | Apr (Cefr) | A 9.0-kb XbaI fragment rescued from Bacteroides sp. strain 139ΩpGW142.4 containing the carboxy-terminal end of tet(36) and downstream sequences (this study) |
| pGW152.1 | Apr (Cefr) | A 3.4-kb SstI fragment rescued from Bacteroides sp. strain 139ΩpGW142.2 containing the carboxy-terminal end of tet(36) and downstream sequences (this study) |
| pGW153.7 | Apr (Cefr) | A 3.8-kb XbaI fragment rescued from Bacteroides sp. strain 139ΩpGW142.2 containing the amino-terminal end of tet(36) and upstream sequences (this study) |
| pGW154.1 | Apr (Cefr) | A 3.0-kb SalI fragment rescued from Bacteroides sp. strain 139ΩpGW142.2 containing the amino-terminal end of tet(36) and upstream sequences (this study) |
| pGW155 | Apr, Tcs | A 2.4-kb PCR fragment amplified using primers 139F7 and 139R8, containing the entire tet(36) gene and 0.4-kb upstream of tet(36) cloned into pGEMT in orientation A or B (this study) |
| pGW156.2 | Kmr (Emr Tcr) | A 2.4-kb SstI/SphI fragment from pGW155A, containing the whole tet(36) gene and upstream regions cloned into Bacteroides shuttle vector pGWA48.3 (this study) |
Bacteroides phenotypes are shown in parentheses, and E. coli phenotypes are shown without parentheses. Resistances are indicated as follows: Ap, ampicillin; Cef, cefoxitin, Em, erythromycin; Km, kanamycin; Nal, nalidixic acid; Rif, rifampin; Str, streptomycin; Tp, trimethoprim; Tc, tetracycline.
PCR amplification using degenerate oligonucleotides.
Degenerate oligonucleotides were shown previously to specifically amplify ribosomal protection types of resistance genes from Streptococcus spp. (7). The sequences of the three degenerate oligonucleotides were as follows: for DI (4,096-fold degeneracy, forward primer), 5′-GAYACICCIGGICAYRTIGAYTT-3′; for DII (4,096-fold degeneracy, reverse primer), 5′-GCCCARWAIGGRTTIGGIGGIACYTC-3′; and for DIII (262,144-fold degeneracy, reverse primer), 5′-CKRAARTCIGCIGGIGTISWIRCIGG-3′. These degenerate primers were utilized to amplify an internal region of a tetracycline resistance gene from Bacteroides sp. strain 139. The reaction conditions utilized for PCR amplification with degenerate oligonucleotides were as follows: 5 min at 95°C, 2 min at 45°C, and then 35 cycles of 2 min at 72°C, 40 s at 92°C, and 40 s at 45°C, and a final extension of 10 min at 72°C. Taq polymerase (Gibco-BRL) was used for PCR amplification according to the manufacturer's instructions. Operon Technologies, Inc., synthesized the primers utilized in all experiments.
Dot blot and Southern blot analysis.
DNA dot blot analyses were performed using total DNA prepared from each of 311 community and human clinical Bacteroides isolates or from 48 swine feces and manure pit samples. For Southern blot and dot blot analysis, labeled DNA fragments were generated and detected using the Renaissance random primer fluorescein labeling kit and Renaissance nucleic acid chemiluminescence reagents, respectively, according to the manufacturer's instructions (Renaissance kit; Dupont NEN Life Sciences).
Plasmid rescue of sequences adjacent to tet(36).
A 0.94-kb HincII fragment containing an internal region of the tet(36) gene was purified from pGW140.1 and cloned into the SmaI site of the cefoxitin-resistant Bacteroides suicide vector pGWA34.2 (Table 1). The suicide vector, which contains a selectable marker that functions in Bacteroides, cannot replicate in Bacteroides, and so cefoxitin-resistant transconjugants result from homologous recombination between the suicide vector containing a fragment of the tet(36) gene and the chromosomal copy of the tet(36) gene. Transformants containing the insert in both possible orientations were selected, generating pGW142.2 and pGW142.4, respectively, which were transferred from E. coli strain S17-1 into Bacteroides sp. strain 139 by conjugation. Transconjugants were analyzed by Southern blotting to confirm that the tet(36) gene contained a single crossover disruption and to identify sites appropriate for the retrieval of the suicide vector and sequences upstream or downstream of tet(36) from the strain 139 chromosome. Genomic DNA containing the inserted suicide vector was digested with an appropriate restriction enzyme (XbaI, SalI, SphI, or SstI) and then ligated and transformed into E. coli strain DH5αMCR. Transformants were generated when the intact plasmid was rescued with contiguous chromosomal DNA from the host chromosome. This procedure was utilized to obtain sequences upstream of the tet(36) gene and repeated to obtain sequences downstream of the tet(36) gene.
Sequencing of tet(36) and contiguous upstream and downstream regions.
Sequencing of the tet(36) resistance gene and regions adjacent upstream and downstream of tet(36) and a 630-bp internal region of the tet(36) gene from strains 88, 123B, 128, and 111 was performed by the University of Illinois Biotechnology Genetic Engineering Facility with an Applied Biosystems model 373A (version 2.0.1A) dye terminator automated sequencer.
Nucleotide sequence accession numbers.
The 12-kb DNA sequence reported in this paper is available from the GenBank database under accession number AJ514254. 16S ribosomal DNA (rDNA) amplification of strains 123B, 128, 111, and 139 was performed using the method described by Weisburg et al. (38), and the resulting sequences have been submitted to the GenBank database under the accession numbers AJ514256, AJ514257, AJ514255, and AJ514258, respectively.
RESULTS
PCR amplification of an internal region of a ribosomal protection type of tetracycline resistance gene from Bacteroides sp. strain 139.
Bacteria resistant to tetracycline have been isolated from sewage material collected from swine feces and from swine manure pits (10, 39). 16S rDNA sequence analysis of one of these isolates, designated strain 139, indicated that it was most closely related to Bacteroides species, having 92% nucleotide (nt) identity to other Bacteroides 16S rRNA genes. The strain 139 isolate was also aminoglycoside resistant, gram negative, and an aerotolerant obligate anaerobe, all characteristics typical of Bacteroides species. So based on the 16S rDNA sequence, strain 139 has been classified as the type strain of a new Bacteroides species, Bacteroides sp. strain 139. In an effort to identify the tetracycline resistance gene responsible for the tetracycline resistance phenotype of strain 139, preliminary dot blot analyses were performed in which probes specific for other ribosome protection types of tetracycline resistance genes [tetQ, tetM, and tetB(P)] were utilized to identify any homologue present. These probes were chosen because CTn-associated tetQ is the only functional tetracycline resistance gene that had been isolated previously from Bacteroides spp. and tetM and tetB(P) are widely distributed in the microbial world on transferable elements, especially among gram-positive organisms. Probes specific for the tetQ, tetM, and tetB(P) genes did not hybridize to genomic DNA from strain 139 (data not shown), indicating that the resistance determinant present in strain 139 was not of these three classes and that a gene previously not found in Bacteroides species was responsible for the tetracycline resistance phenotype.
To determine whether the tetracycline resistance gene from strain 139 was another type of ribosome protection tetracycline resistance gene, we employed a PCR strategy in which degenerate primers DI, DII, and DIII were used to amplify a putative ribosome protection type of a tetracycline resistance gene from strain 139 (7). Primers DI and DII generated a 1.1-kb PCR product, while primers DI and DIII yielded a 1.3-kb product. The PCR products were cloned into pGEMT (Promega), generating pGW140.1 and pGW141.1, respectively (Table 1). In each of the two cases, the DNA segment amplified using the two degenerate primer sets generated a single product of the predicted size.
Preliminary sequence analysis indicated that at the nucleotide level, the cloned fragments had no significant identity to the DNA sequences available in the GenBank database. At the amino acid level, however, the cloned fragments encoded a protein with highest amino acid identity to a number of ribosome protection types of tetracycline resistance proteins, including TetQ (Table 2).
TABLE 2.
Sequence analysis of the tet(36) region of Bacteroides sp. strain 139
| Gene | Coordinates (5′-3′) | Size (aa) | % Identity (aa)a | Homolog (size of homolog or bp start) | Accession no. |
|---|---|---|---|---|---|
| rumB (N-term) | 378-0 | 126 | 67 (126) | Unfinished B. fragilis ATCC 25285 (strain Sanger 817; contig 98)b | NC003228 |
| 56 (123) | P. rettgeri RumB protein from R391 and V. cholerae RumB protein from SXT element (435 aa)c | AY090559, AY055428g | |||
| 34 (79) | B. thetaiotaomicron 5482 (ATCC 29148) (bp 5,809,737)d | AE015928 | |||
| orf2 | 982-560 | 140 | NAf | None | NA |
| rumA | 1586-1146 | 146 | 65 (141) | Unfinished B. fragilis ATCC 25285 (strain Sanger 817; contig 98)b | NC003228 |
| 44 (135) | P. rettgeri RumA protein from R391 and V. cholerae RumA protein from SXT element (149 aa) | AY090559, AY055428g | |||
| orf4 | 2207-1806 | 133 | 44 (85) | B. thetaiotaomicron 5482 (ATCC 29148) (bp 626,085)d | AE015928 |
| tet(36) | 2534-4456 | 640 | 60 (637) | B. thetaiotaomicron CTnDOT tetracycline resistance protein TetQ (641 aa) | X58717 |
| orf6 | 4768-5055 | 95 | NA | None | NA |
| orf7 | 5067-5369 | 100 | 56 (99) | Unfinished B. fragilis ATCC 25285 (strain Sanger 817; contig 12)b | NC003228 |
| orf8 | 5378-5599 | 73 | 46 (65) | Unfinished B. fragilis ATCC 25285 (strain Sanger 817; contig 12) | NC003228 |
| orf9 | 5601-6068 | 155 | 51 (159) | Unfinished B. fragilis ATCC 25285 (strain Sanger 817; contig 12)b | NC003228 |
| IntPCe | 6947-6369 | 192 | 34 (150) | Unfinished B. fragilis ATCC 25285 (strain Sanger 817; contig 96)b | NC003228 |
| 32 (167) | Pyrococcus horikoshii putative integrase (285 aa) | B71194 | |||
| 29 (176) | Staphylococcus aureus recombinase XerD (295) | AF173869 | |||
| ycgF | 7992-8645 | 217 | 66 (196) | Unfinished B. fragilis ATCC 25285 (strain Sanger 817; contig 63)b | NC003228 |
| 67 (196) | B. thetaiotaomicron 5482 (ATCC 29148) (bp 2,131,191)d | AE015928 | |||
| 32 (150) | B. subtilis hypothetical protein YcgF (209 aa) | Z99105 | |||
| 25 (117) | Brucella melitensis homoserine lactone efflux protein (249 aa) | AE009620 | |||
| orf12 | 8658-9206 | 182 | 58 (180) | Unfinished B. fragilis ATCC 25285 (strain Sanger 817; contig 63)b | NC003228 |
| 52 (167) | B. thetaiotaomicron 5482 (ATCC 29148) (bp 2,130,583)d | AE015928 | |||
| rmlD | 9209-10072 | 287 | 52 (286) | Unfinished B. fragilis ATCC 25285 (strain Sanger 817; contig 63)b | NC003228 |
| 46 (230) | B. thetaiotaomicron 5482 (ATCC 29148) (bp 2,130,043)d | AE015928 | |||
| 43 (266) | Serratia marcescens dTDP-l-rhamnose reductase (288 aa) | AF038816 | |||
| 39 (271) | C. perfringens dTDP-dehydrorhamnose reductase (294 aa) | AP003187 | |||
| prfC | 10131-11705 | 524 | 85 (524) | Unfinished B. fragilis ATCC 25285 (strain Sanger 817; contig 63)b | NC003228 |
| 85 (524) | B. thetaiotaomicron 5482 (ATCC 29148) (bp 2,129,189)%d; 75% nucleotide identity | AE015928 | |||
| 49 (520) | V. cholerae peptide chain RF-3 encoded by prfC (529 aa) | AE004152 |
No. of aa, the length over which the putative proteins (in amino acids) from the tet(36) region have identity to homologous proteins or genes.
B. fragilis strain ATCC 25685 unfinished chromosomal sequence; Sanger 817, accession no. NC003228.
In the V. cholerae SXT element the amino-terminal region of the rumB open reading frame is disrupted by a 17-kb insertion that contains multiple drug resistance genes (Fig. 2B).
B. thetaiotaomicron 5482 type strain recently sequenced. The locations of starts of the putative homologs are given (43).
The putative integrase gene, intPC, consisting primarily of the carboxy-terminal end of the protein, appears to be truncated and is approximately 93 aa shorter than the next most closely related known int gene from P. horikoshii.
NA, not applicable.
Paired accession numbers correspond to the respective proteins listed in the fifth column.
Isolation and analysis of the strain 139 tetracycline resistance gene and contiguous sequences.
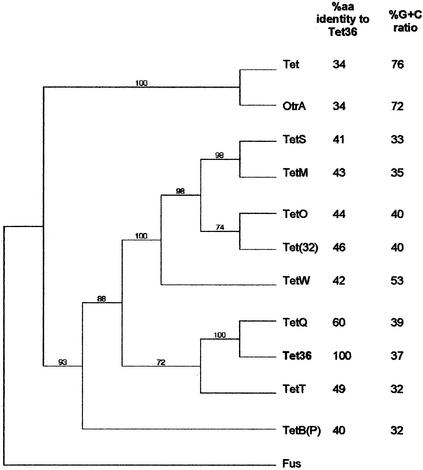
To isolate the sequences encoding the amino-terminal and carboxyl-terminal regions of the new tetracycline resistance gene and to demonstrate that it was responsible for the tetracycline resistance phenotype of strain 139, we employed a plasmid rescue strategy. An 11.8-kb region, comprising 2.5 kb upstream and 7.3 kb downstream of the tetracycline resistance gene, was cloned. The results of sequence analysis of this cloned segment are shown in Fig. 1 and Table 2. The putative tetracycline resistance gene had 6% nt identity and 60% amino acid identity with its closest relative, tetQ. Since the amino acid identity was less than 80%, the strain 139 tetracycline resistance gene represents a new class of a ribosome protection type of tetracycline resistance determinant. In accordance with recommendations for naming new tetracycline resistance genes, we obtained the designation tet(36) from the S. Levy group (23).
FIG. 1.
Phylogenetic relationship between Tet 36 and other ribosome protection types of tetracycline resistance proteins. The sequence of the Bacillus subtilis Fus protein for translation elongation factor EF-G was used as the outgroup to root the tree. The number at each node is the percentage of times that the tree configuration occurred in 10,000 bootstrap trials. The protein names and their organisms of origin and GenBank accession numbers are as follows: TetM, Enterococcus faecalis Tn916, GenBank accession no. U09422; TetS, Listeria monocytogenes BM4210 pIP811, Q48791; TetO, Streptococcus pneumoniae, P72533; TetW, Butyrivibrio fibrisolvens, AJ222769; Tet36, Bacteroides sp. strain 139, AJ514254; TetQ, B. thetaiotaomicron, X58717; TetT, S. pyogenes A498, L42544; TetB (P), Clostridium perfringens CW92, AE007656; OtrA, Streptomyces rimosus, S18572; Tet, S. coelicolor A3, CAC14348; Fus, B. subtilis, P80868; Tet(32), Clostridium sp. strain K10, AJ295238.
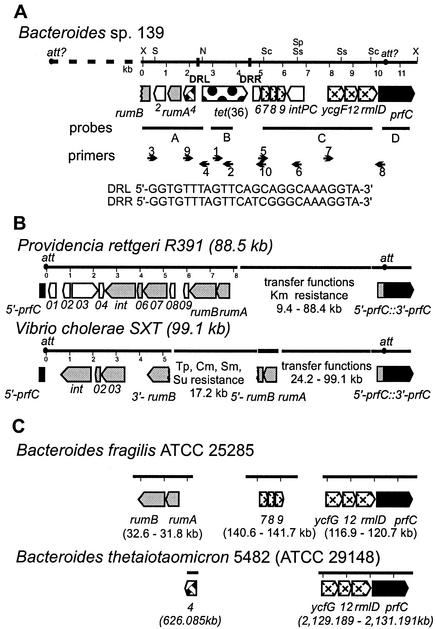
A schematic diagram of tet(36) and contiguous sequences is shown in Fig. 2A. Two direct repeats, DR1 and DR2, flanked the tet(36) gene. These repeats may have some role in the mobility of the tet(36) gene as a cassette. In addition to tet(36), 12 open reading frames (ORFs) were identified (Fig. 2A and Table 2). Upstream of tet(36), two putative ORFs were identified that encoded proteins that shared high amino acid identity (44 to 56%) with UV protection proteins RumA and RumB from proteobacterial CTn-like elements R391 (from Providencia rettgeri) and SXT (from Vibrio cholerae) (4, 15, 16) (Fig. 2B). Related genes are also present in the unfinished B. fragilis ATCC 25285 genome sequences, although the amino acid identity is only 65 to 67% (Fig. 2C and Table 2). There are no rumA or rumB homologues in the B. thetaiotaomicron 5482A (BT4000) genome sequence (43).
FIG. 2.
(A) Schematic diagram showing the organization of the tet(36) region from Bacteroides sp. strain 139. The numbers show the distance in kilobases from the leftmost XbaI site to the rightmost SphI site. Restriction sites shown are those of XbaI (X), SalI (S), NsiI (N), ScaI (Sc), SphI (Sp), and SstI (Ss). The major potential genes and their respective orientations are represented by arrows. Direct repeats flanking tet(36) are represented by filled boxes, and the sequences of the direct repeat upstream of tet(36) (DRL) and downstream of tet(36) (DRR) are shown. Probes used in Southern and dot blot analyses are represented by horizontal lines below the tet(36) region. Small arrows indicate the positions and directions of primers used for PCR analyses. (B) Schematic diagrams of related CTn-like elements R391 from P. rettgeri and SXT from V. cholerae are shown below the tet(36) region. (C) A schematic diagram of the organization and positions of genes from the B. fragilis type strain ATCC 25285 genome and the B. thetaiotaomicron type strain 5482 (ATCC 29148), which encode proteins homologous to those encoded by genes from the tet(36) region, is shown. ORFs are defined as follows: homologous amino acid sequences related to those present in both the SXT element and R391 are indicated by gray arrows, nonhomologous element sequences are indicated by unfilled arrows, the tet(36) gene sequence is indicated by a spotted arrow, homologous sequences not present in either the SXT element or R391 are indicated by hatched arrows, and prfC gene sequences are indicated by black arrows. orf4 (weave) is also found in B75482. Dashed lines indicate regions of the elements not drawn to scale. Antibiotic resistances are indicated as follows: kanamycin (Km), trimethoprim (Tp), chloramphenicol (Cm), streptomycin (Sm), and sulfonamides (Su).
Both the R391 and SXT elements have been shown to integrate site specifically into the host chromosome. In both cases the target site for integration is located in the 5′ end of prfC, a gene encoding peptide chain release factor 3 (15, 17). It is interesting that an ORF designated prfC (524 aa) was found 5 kb downstream of tet(36). This, together with the presence of genes encoding RumA and RumB homologues, suggests the possibility that the sequences surrounding tet(36) in strain 139 were derived from the acquisition of a CTn-like element related to the proteobacterial elements R391 and SXT. Other ORFs with matches to sequences were found, but the sequence similarities were generally fairly low (Table 2). The homologues detected in the B. fragilis and B. thetaiotaomicron chromosomal sequences and their arrangements relative to the tet(36) sequences of strain 139 are shown in Fig. 2C.
One way to test whether tet(36) is on a conjugal element is to test for conjugal transfer of the gene. Mating experiments were performed between a Bacteroides sp. strain 139 donor and BT4001 or BU1001 as recipient. No transfer was observed under conditions normally utilized for Bacteroides matings.
tet(36) confers tetracycline resistance in E. coli and in other Bacteroides strains in trans.
A 2.4-kb fragment that contained the potential promoter region and a complete copy of the tet(36) gene were initially cloned into the E. coli replicon pGEMT in both orientations, generating pGW155A and pGW155B. E. coli cells containing pGW155A were sensitive to tetracycline (10 μg/ml), while cells containing pGW155B were resistant to tetracycline (10 μg/ml), indicating that a promoter present in the pGEMT vector rather than the native promoter was driving the expression of the resistance gene in E. coli. Thus, Tet 36 is functional in E. coli when an appropriate promoter region is provided. To determine whether tet(36) was expressed in another species of Bacteroides, the 2.4-kb insert from pGW155A was inserted into the E. coli-Bacteroides shuttle vector pGWA48.3, generating pGW156.2. B. thetaiotaomicron BT4001 cells carrying pGW156.2, which is maintained at a copy number of 8 to 10 per cell, were resistant to 5 μg of tetracycline/ml, the same level as Bacteroides sp. strain 139 carrying a single copy of the tet(36) gene. The tet(36) gene appeared to be the only active tetracycline resistance gene present in strain 139, because a single-crossover disruption of tet(36), made by insertion of a suicide vector containing an internal fragment of tet(36) (pGW142.2 or pGW142.4), rendered the strain susceptible to tetracycline.
tet(36) was not present in human clinical and intestinal Bacteroides isolates but was found in diverse bacterial genera from swine manure.
Genomic DNAs from 311 human clinical and intestinal Bacteroides isolates, previously collected from various sources around the United States (32), were screened by dot blot analysis for the presence of tet(36) (probe B); however, none of the strains surveyed hybridized to the probe nor did they hybridize to probes A, C, or D containing sequences upstream and downstream of tet(36) (Fig. 2A).
To determine whether tet(36) and the region contiguous with tet(36) were present in other bacteria isolated from swine intestinal contents or manure pits, a PCR approach was utilized. Primers specific for tet(36) (primers 1 and 2) (Fig. 2A) were used to amplify a 630-bp internal fragment of tet(36) from 48 different strains that were resistant to tetracycline and/or tylosin. Four strains (123B, 128, 111, and 88) yielded a PCR product of the correct size that cross-hybridized to tet(36) in dot blot analyses. These fragments were cloned and sequenced and were shown to have 100% nucleotide identity to tet(36) from strain 139.
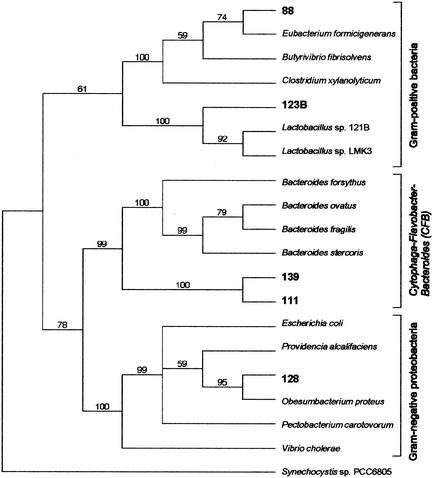
To determine the identity of these tet(36)-containing strains, a 16S rRNA gene from each was amplified and sequenced. Results are summarized in Fig. 3. The rDNA sequence of strain 123B had 98% nucleotide identity to rRNA genes from Lactobacillus sp. strain 121B and Lactobacillus sp. strain LMK3. The 16S rDNA sequence of strain 128 was most closely related to gram-negative proteobacteria (94% nucleotide identity to rDNA from Obesumbacterium proteus and 93% nucleotide identity to 16S rDNA from Pectobacterium carotovorum sp., Erwinia carotovora, and P. alcalifaciens). The 16S rDNA gene sequence of strain 88 had 94% nucleotide identity to a gene from Clostridium. Strain 111 is probably another strain of Bacteroides sp. strain 139, because its rDNA sequence was 99.9% identical to that of strain 139. Strain 111 was isolated directly from swine feces, whereas strain 139 was isolated from a manure pit. Also, it does not contain an erythromycin resistance gene, erm(35) (accession no. AF319779), that is present in, but is not adjacent to, tet(36) in strain 139 (T. R. Whitehead and M. A. Cotta, unpublished data).
FIG. 3.
Phylogenetic distribution of tet(36) in bacteria isolated from swine manure. The sequence of the Synechocystis sp. strain PCC6805 16S rRNA gene was used as the outgroup to root the tree. The number at each node is the percentage of times that the tree configuration occurred in 10,000 bootstrap trials. The rRNA gene sequences were obtained from the GenBank database as follows: Bacteroides sp. strain 139, GenBank accession no. AJ514258; Bacteroides sp. strain 111, AJ514255; B. ovatus, X83952; B. fragilis, X83943; B. stercoris, X83953; B. forsythus, X73962; E. coli, AF527827; P. alcalifaciens, AJ301684; unidentified proteobacterium strain 128, AJ514257; O. proteus, AJ233422; P. carotovorum, AF373182; V. cholerae, X74694; C. xylanolyticum, X76736; 88, AF445289; Eubacterium formicigenerans, L34619; B. fibrisolvens, AY029616; Lactobacillus sp. strain 121B, AF305930; Lactobacillus sp. strain 123B, AJ514257; Lactobacillus sp. strain LMK3, AJ251560; Synechocystis sp. strain, PCC6805, AB041938.
The presence of identical copies of tet(36) in phylogenetically diverse bacteria suggested that gene transfer has occurred recently and that a common mechanism of horizontal transfer might be involved in the acquisition of these genes. Consequently, genomic DNAs from strains 123B, 128, 88, and 111 were also analyzed by PCR and probed with sequences upstream and downstream of tet(36) to determine whether the sequences contiguous with tet(36) in Bacteroides sp. strain 139 were also contiguous with the tet(36) gene in strains 123B, 128, 88, and 111, as might be expected if tet(36) were located on the same type of DNA element in these other tet(36)-containing isolates. Only DNA from strains 88 and 111 cross-hybridized with sequences located upstream of tet(36) in Bacteroides sp. strain 139 (probe A) (Fig. 2A). PCR analysis (primers 3 and 4) (Fig. 2A) confirmed that as in Bacteroides sp. strain 139, these upstream sequences were also contiguous with tet(36) in strains 88 and 111 since they generated the same 2.1-kb product. Only DNA from strain 111 cross-hybridized to sequences located downstream of tet(36) (probes C and D) (Fig. 2A) in strain 139. Subsequent PCR analyses (primers 5 and 6, 7 and 8, and 9 and 10) (Fig. 2A) also indicated that the organization of sequences downstream of tet(36) was identical in Bacteroides sp. strains 139 and 111, since PCR fragments of the same size were amplified (data not shown).
DISCUSSION
Prior to this work, the only tetracycline resistance gene that has been demonstrated to be functional in Bacteroides spp. was tetQ, which encodes a ribosome protection type of tetracycline resistance and renders the host bacterium resistant to all of the tetracyclines used clinically (28). The tetQ gene has been found on conjugal elements in oral Prevotella and Porphyromonas spp., and transfer of tetQ between human Bacteroides isolates and an animal isolate of Prevotella ruminicola has been demonstrated in the laboratory. (3, 13, 14, 25, 33). The discovery of a new tetracycline resistance gene, tet(36), in a Bacteroides sp. isolated from swine manure pits is interesting ecologically. Bacteroides spp. comprise a small proportion of the microbial population in this environment, which consists primarily of gram-positive organisms with low G+C content (21, 27). A survey of 48 tetracycline- or tylosin-resistant bacteria isolated from the same swine manure source as Bacteroides sp. strain 139 led to the identification of four different isolates that contained tet(36), including two different gram-positive bacteria, a gram-negative proteobacterium, and an independent Bacteroides isolate. The sequences of the 16S rRNA genes of the new Bacteroides species defined currently by strains 139 and 111 are only 92% identical to the sequences of the closest human colonic species 16S rRNA. It is not uncommon for Bacteroides species isolated from animals to be so phylogenically distant from the human species (21). The characterized human colonic Bacteroides spp. are also quite divergent. For example, the 16S sequence of the B. uniformis type strain (AB050110) is only 92% identical to the 16S sequence of the B. thetaiotaomicron type strain (M58763).
Results of sequence comparisons of tet(36) genes from each of the five phylogenetically diverse isolates (Fig. 3) indicated that this gene might have been transferred among these isolates. Like other bacteria, Bacteroides spp. harbor a variety of transmissible elements that are involved in the transfer of antibiotic resistance genes, including plasmids, nonmobilizable and mobilizable transposons, and CTns. Although Bacteroides sp. strain 139 was recently discovered to contain a single cryptic plasmid, p139EF (GenBank accession no. AF448250), tet(36) is not carried on this plasmid (T. R. Whitehead et al., unpublished). Thus, if tet(36) is on a transmissible element, the element may be an integrated element such as a CTn. However, we were not able to demonstrate transfer of tet(36) under laboratory conditions. The Bacteroides CTnDOT-type CTns require tetracycline stimulation to trigger horizontal transfer. Accordingly, if tet(36) is carried on a CTn or other integrated element, it might be necessary to induce transfer with an as-yet-unknown inducer. There was no detectable transfer of the tet(36) even when the donor cells were grown in medium containing either tetracycline or erythromycin. Another possibility is that if the element carrying tet(36) is related to the SXT/R391 elements, it may, like them, require sequence identity between the end of the element and the 5′ end of the prfC in the recipient. The BT4001 recipient used in the transfer studies had only 75% nucleotide identity to the prfC in strain 139. Thus, transfer may only be detected if the recipient is more isogenic to strain 139.
The finding that genes in the tet(36) region of Bacteroides sp. strain 139 were also present in the unfinished B. fragilis genome sequence (Fig. 2C and Table 2) raises the possibility that sequences contiguous with tet(36) in Bacteroides sp. strain 139 were not acquired with the tet(36) gene and that the tet(36) gene came in on some other element and subsequently integrated in this region of the Bacteroides chromosome. It is notable, however, that in addition to Bacteroides sp. strains 139 and 111, gram-positive Clostridium sp. strain 88 also contained rumA and rumB upstream of tet(36), homologues that were similar enough to cross-hybridize under high-stringency conditions. This is significant because the rumA and rumB alleles from B. fragilis and Bacteroides sp. strain 139 have insignificant nucleotide identity and B. thetaiotaomicron lacks homologues to these genes (43). This observation raises yet another possibility, which is that the tet(36) genes present in Clostridium sp. strain 88 and Bacteroides sp. strains 139 and 111 are carried on the same transmissible element, which has since undergone deletions or other rearrangements downstream of the tet(36) gene. Whatever the mechanism of transfer, it appears that tet(36) is moving between species of bacteria found in the porcine intestine. If so, it is interesting that this tet gene has not yet been found in any of the human colonic species so far tested.
Acknowledgments
This work was supported by grant AI22383 from the National Institutes of Health.
We thank Jorge Frias-Lopez for assistance with assembly of dendrograms.
REFERENCES
- 1.Aminov, R. I., J. C. Chee-Sanford, N. Garrigues, B. Teferedegne, I. J. Krapac, B. A. White, and R. I. Mackie. 2002. Development, validation, and application of PCR primers for detection of tetracycline efflux genes of gram-negative bacteria. Appl. Environ. Microbiol. 68:1786-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aminov, R. I., N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrés, M. T., W. O. Chung, M. C. Roberts, and J. F. Fierro. 1998. Antimicrobial susceptibilities of Porphyromonas gingivalis, Prevotella intermedia, and Prevotella nigrescens spp. isolated in Spain. Antimicrob. Agents Chemother. 42:3022-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184:4259-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer, H. B., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification system of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 6.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clermont, D., O. Chesneau, G. De Cespédès, and T. Horaud. 1997. New tetracycline resistance determinants coding for ribosomal protection in streptococci and nucleotide sequence of tet(T) isolated from Streptococcus pyogenes A498. Antimicrob. Agents Chemother. 41:112-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole, D. J., V. R. Hill, F. J. Humenik, and M. D. Sobsey. 1999. Health, safety, and environmental concerns of farm animal waste. Occup. Med. 14:423-448. [PubMed] [Google Scholar]
- 9.Cooper, A. J., N. B. Shoemaker, and A. A. Salyers. 1996. The erythromycin resistance gene from the Bacteroides conjugal transposon Tcr Emr 7853 is nearly identical to ermG from Bacillus sphaericus. Antimicrob. Agents Chemother. 40:506-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotta, M. A., T. R. Whitehead, and R. L. Zeltwanger. Isolation, characterization, and comparison of bacteria from swine feces and manure storage pits. Environ. Microbiol., in press. [DOI] [PubMed]
- 11.Esiobu, N., L. Armenta, and J. Ike. 2002. Antibiotic resistance in soil and water environments. Int. J. Environ. Health Res. 12:133-144. [DOI] [PubMed] [Google Scholar]
- 12.Finegold, S. M., and W. L. George. 1989. Anaerobic infections in humans. Academic Press, San Diego, Calif.
- 13.Flint, H. J., A. M. Thomson, and J. Bisset. 1988. Plasmid-associated transfer of tetracycline resistance in Bacteroides ruminicola. Appl. Environ. Microbiol. 54:855-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guiney, D. G., and P. Hasegawa. 1992. Transfer of conjugal elements in oral black-pigmented Bacteroides (Prevotella) spp. involves DNA rearrangements. J. Bacteriol. 174:4853-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hochhut, B., J. W. Beaber, R. Woodgate, and M. K. Waldor. 2001. Formation of chromosomal tandem arrays of the SXT element and R391, two conjugative chromosomally integrating elements that share an attachment site. J. Bacteriol. 183:1124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hochhut, B., J. Marrero, and M. K. Waldor. 2000. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J. Bacteriol. 182:2043-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hochhut, B., and M. K. Waldor. 1999. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol. Microbiol. 32:99-110. [DOI] [PubMed] [Google Scholar]
- 18.Jensen, L. B., S. Baloda, M. Boye, and F. M. Aarestrup. 2001. Antimicrobial resistance among Pseudomonas spp. and the Bacillus cereus group isolated from Danish agricultural soil. Environ. Int. 26:581-587. [DOI] [PubMed] [Google Scholar]
- 19.Jobling, M. G., and R. K. Holmes. 1990. Construction of vectors with the p15a replicon, kanamycin resistance, inducible lacZ alpha and pUC18 or pUC19 multiple cloning sites. Nucleic Acids Res. 18:5315-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhne, M., D. Ihnen, G. Moller, and O. Agthe. 2000. Stability of tetracycline in water and liquid manure. J. Vet. Med. A Physiol. Pathol. Clin. Med. 47:379-384. [DOI] [PubMed] [Google Scholar]
- 21.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Moller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy, S. B. 1978. Emergence of antibiotic-resistant bacteria in the intestinal flora of farm inhabitants. J. Infect. Dis. 137:689-690. [PubMed] [Google Scholar]
- 23.Levy, S. B., L. M. McMurry, T. M. Barbosa, V. Burdett, P. Courvalin, W. Hillen, M. C. Roberts, J. I. Rood, and D. E. Taylor. 1999. Nomenclature for new tetracycline resistance determinants. Antimicrob. Agents Chemother. 43:1523-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melville, S. E., C. S. Gerrard, and J. M. Blackwell. 1999. Multiple causes of size variation in the diploid megabase chromosomes of African tyrpanosomes. Chromosome Res. 7:191-203. [DOI] [PubMed] [Google Scholar]
- 25.Okamoto, M., K. Takano, and N. Maeda. 2001. Distribution of the tetracycline resistance determinant tetQ gene in oral isolates of black-pigmented anaerobes in Japan. Oral Microbiol. Immunol. 16:224-228. [DOI] [PubMed] [Google Scholar]
- 26.Parker, A. C., and C. J. Smith. 1993. Genetic and biochemical analysis of a novel Ambler class A β-lactamase responsible for cefoxitin resistance in Bacteroides species. Antimicrob. Agents Chemother. 37:1028-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pryde, S. E., A. J. Richardson, C. S. Stewart, and H. J. Flint. 1999. Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig. Appl. Environ. Microbiol. 65:5372-5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salyers, A. A., N. B. Shoemaker, A. M. Stevens, and L. Y. Li. 1995. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59:579-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 30.Shoemaker, N. B., C. Getty, E. P. Guthrie, and A. A. Salyers. 1986. Regions in Bacteroides plasmids pBFTM10 and pB8-51 that allow Escherichia coli-Bacteroides shuttle vectors to be mobilized by IncP plasmids and by a conjugative Bacteroides tetracycline resistance element. J. Bacteriol. 166:959-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shoemaker, N. B., E. P. Guthrie, A. A. Salyers, and J. F. Gardner. 1985. Evidence that the clindamycin-erythromycin resistance gene of Bacteroides plasmid pBF4 is on a transposable element. J. Bacteriol. 162:626-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shoemaker, N. B., H. Vlamakis, K. Hayes, and A. A. Salyers. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shoemaker, N. B., G. R. Wang, and A. A. Salyers. 1992. Evidence for natural transfer of a tetracycline resistance gene between bacteria from the human colon and bacteria from the bovine rumen. App. Environ. Microbiol. 58:1313-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shoemaker, N. B., G. R. Wang, A. M. Stevens, and A. A. Salyers. 1993. Excision, transfer, and integration of NBU1, a mobilizable site-selective insertion element. J. Bacteriol. 175:6578-6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon, R., U. Prkseifer, and A. Puhler. 1983. A broad-host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 36.Speer, B. S., N. B. Shoemaker, and A. A. Salyers. 1992. Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin. Microbiol. Rev. 5:387-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor, D. E., and A. Chau. 1996. Tetracycline resistance mediated by ribosomal protection. Antimicrob. Agents Chemother. 40:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitehead, T. R., and M. A. Cotta. 2001. Characterisation and comparison of microbial populations in swine faeces and manure storage pits by 16S rDNA gene sequence analyses. Anaerobe 7:181-187. [Google Scholar]
- 40.Whitehead, T. R., and M. A. Cotta. 2001. Sequence analyses of a broad host-range plasmid containing ermT from a tylosin-resistant Lactobacillus sp. isolated from swine feces. Curr. Microbiol. 43:17-20. [DOI] [PubMed] [Google Scholar]
- 41.Whittle, G., B. D. Hund, N. B. Shoemaker, and A. A. Salyers. 2001. Characterization of the 13-kilobase ermF region of the Bacteroides conjugative transposon CTnDOT. Appl. Environ. Microbiol. 67:3488-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whittle, G., N. B. Shoemaker, and A. A. Salyers. 2002. The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell. Mol. Life Sci. 59:2044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]