Abstract
The efficiency of enterobacterial disinfection is dependent largely on enterobacterial community physiology. However, the relationship between enterobacterial community physiology and wastewater processing is unclear. The purpose of this study was to investigate this relationship. The influence of wastewater treatment processes on enterobacterial community physiology was examined at the single-cell level by using culture-independent methods. Intracellular concentrations of two conserved proteins, the growth-related protein Fis and the stationary-phase protein Dps, were analyzed by epifluoresence microscopy of uncultivated cells by using enterobacterial group-specific polyclonal fluorochrome-coupled antibodies. Enterobacterial single-cell community protein profiles were distinct for different types of biological treatment. The differences were not apparent when bulk methods of protein analysis were used. Trickling filter wastewater yielded Fis-enriched communities compared to the communities in submerged aeration basin wastewater. Community differences in Fis and Dps contents were used to predict disinfection efficiency. Disinfection of community samples by heat exposure combined with cultivation in selective media confirmed that enterobacterial communities exhibited significant differences in sensitivity to disinfection. These findings provide strategies that can be used to increase treatment plant performance, reduce the enterobacterial content in municipal wastewater, and minimize the release of disinfection by-products into receiving water.
The growing demand for water resources is increasing interest in reclamation of municipal wastewater for potable and nonpotable applications (30, 53). Primary wastewater treatment removes solid materials. In secondary treatment a range of strategies collectively based on biological treatment, including activated sludge and biofilms, is used to reduce the organic carbon content and thereby reduce the biological oxygen demand (37). Despite their differences, secondary treatment strategies overcome variations in microbial community structure, water chemistry, and substrate composition to effect water purification.
Wastewater quality is judged by the content of a subgroup of the Enterobacteriaceae termed fecal enterobacteria (fecal coliforms) that provide a biological indicator of fecal pollution (10). In most forms of tertiary wastewater treatment chemical disinfection with chlorine or chloramines is used to inactivate resident microbes. When the receiving water is classified as recreational, residual chlorine must be neutralized by sulfur dioxide treatment (46). The Environmental Protection Agency also mandates that discharged municipal effluent can contain no more than 4,000 fecal coliforms per liter to reduce the risk of fecal contamination in the receiving water. Many treatment plants, however, achieve acceptable microbial contents at the expense of releasing considerable amounts of disinfection by-products (5). Chlorination generates trihalomethane and other by-products with mutagenic and carcinogenic properties (47). Wastewater reuse must therefore balance removing pathogenic microbes with minimizing formation and release of disinfection by-products.
In laboratory experiments the efficiency of disinfection of pure microbial cultures and of mixed-taxon microcosms is dependent upon a range of physiologic characteristics. These include the microbial growth state (25, 31), the availability of nutrients (18, 43), the induction of starvation proteins (4, 19, 24, 25, 36, 43), and alterations in membrane structure (23, 25). Despite the availability of information, the use of culture-based microbiological methods imposes limits on a complete understanding of the relationship between the treatment process and enterobacterial disinfection in environmental samples.
Culture-independent methods provide a direct approach for obtaining information about endogenous microbes in environmental samples. In situ hybridization for detection of individual taxa (32) was initially performed with radiolabeled oligonucleotides (12), and the procedure was quickly refined to incorporate fluorescently labeled oligonucleotide probes. Detection of individual cells and identification of specific taxa in activated sludge also were accomplished by using immunofluorescence-based techniques (13, 29, 48). However, such approaches have been replaced by nucleic acid-based strategies applied to activated sludge (6, 7, 8, 11, 21, 41, 49, 50, 51) and biofilm (22, 35, 44) processes. The studies have focused primarily on the occurrence and distribution of a variety of taxa, such as Gordonia, Acinetobacter, and Nitrosococcus, to understand how these processes influence microbial community composition (15, 52). Although such studies provide important information concerning the taxonomic compositions of communities, the relationship between community physiology and water processing remains obscure (33). Because wastewater release depends on enterobacterial content, particularly the content of feces-associated taxa, it is important to clarify how the treatment process and facility management influence these organisms. To address this question, we developed a culture-independent single-cell method to determine enterobacterial physiological status based on the growth-state-specific protein content (40).
In Escherichia coli, Fis is an 11-kDa DNA binding protein (14, 16) which plays a critical role in coordinating rRNA synthesis with growth (27). Fis is present in replicating cells, and its abundance is directly correlated with the growth rate (3, 45). Fis abundance in a cell varies more than 500-fold between the extremes of rapid growth and the stationary phase, and Fis is conserved in the Enterobacteriaceae (2, 40). Therefore, we considered Fis abundance a positive indicator of cells in a balanced growth (2, 40). Dps is a highly conserved 19-kDa DNA binding protein (1, 20) that is important in stationary-phase stress physiology (1, 20, 39). Dps abundance is inversely correlated with the growth rate, and the cellular concentration of Dps varies more than 100-fold between the extremes of the stationary phase and rapid growth (1, 20, 28, 39). Dps abundance was therefore used as a positive indicator of cells that had entered the stationary phase (2, 40). In the present investigation, the impact of primary and secondary wastewater treatment on enterobacterial community physiology was determined by measuring single-cell levels of Fis and Dps with previously characterized enterobacterium-specific fluorochrome-conjugated polyclonal antibodies (40). Protein profiling was then combined with direct measurement of disinfection efficiency. The results provide new process strategies for improving enterobacterial disinfection in municipal wastewater.
MATERIALS AND METHODS
Cloning, purification, and production of antibodies for Fis and Dps.
The fis gene was amplified by PCR by using 5′ TTGAATTCATGTTCGAACAACGCG 3′ (forward primer) and 5′ TTCTTAAGAGCATTTAGCTAACC 3′ (reverse primer) from E. coli strain PBL500 (39). The resulting PCR product was cloned into the NcoI and XhoI sites of pET28b (Novagen), creating plasmid pPB916, placing fis under control of the T7lac promoter, and adding a hexahistidine N-terminal fusion peptide. For induction of the Fis protein we employed E. coli strain BL21 DE3 (Novagen). Fis was purified by nickel affinity chromatography by using a His∗Bind Quick column (Novagen) as described by the manufacturer. Preparation and purification of recombinant Dps were performed as described previously (39, 40). Rabbit anti-Fis and anti-Dps antibodies were prepared as described previously (39, 40). An E. coli fis mutant strain (PB918) was reconstructed by homologous recombination by using a phage-encoded mutant allele of fis as described previously (40). Recombinants devoid of integrated phage were verified by PCR after successive cycles of purification for single colonies on Luria-Bertani agar plates containing 0.5% (wt/vol) deoxycholic acid and then Luria-Bertani agar plates containing tetracycline (25 μg/ml).
Antibody probes.
Rabbit sera containing anti-Fis and anti-Dps antibodies were processed with acetone powders derived from homologous mutant strains PB918 and PB664 and then fractionated by affinity chromatography with protein A-Sepharose as described previously (17, 40). Anti-Fis antibodies were coupled to Alexa Fluor 488 (Molecular Probes), and anti-Dps antibodies were coupled to Alexa Fluor 568 (Molecular Probes). Coupling procedures and the extent of antibody labeling were determined as described by the manufacturer. The optimal coupling ratios were 4 to 9 mol of dye per mol of Fis antibodies and 2 to 6 mol of dye per mol of Dps antibodies. The conjugated antibodies were stored at 4°C in light-impermeable containers and were stable for several months. The degree of labeling was determined periodically to ensure that probe quality was maintained. Sample aliquots were used for long-term storage at −20°C to minimize freeze-thaw cycles and light exposure.
Oligonucleotide probe.
The 16S rRNA probe ENT specific for the Enterobacteriaceae described previously (26) (sequence, 5′-CATGAATCACAAAGTGGTAAGCGCC-3′) was purchased prelabeled with flourescein by the manufacturer (Gibco-BRL). Single-cell 16S rRNA analysis with the fluorescein-labeled oligonucleotide was performed as previously described (12). The fixed cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) (0.1 μg/ml) as described previously (34).
Wastewater and sample analysis.
Wastewater was obtained from a local municipal facility that serves a population of 200,000 individuals and treats 18 million gallons of water daily. Grab samples were recovered in 125-ml sterile bottles and processed immediately for protein profiling or placed on ice for transport to the laboratory. Total carbon levels were determined by the carbon combustion method (Leco Corp.). Dissolved oxygen concentrations were determined by using a dissolved oxygen meter (Sper Scientific) as described previously (10). Grab samples were analyzed by plating on selective media, including Endo agar (Becton Dickinson) and A-1 media (Difco).
Fluorescence microscopy and sample preparation.
Grab samples were fixed by addition of paraformaldehyde to a final concentration of 4% (vol/vol) by using a freshly prepared 40% (wt/vol) stock solution prepared in phosphate-buffered saline (PBS) (10 mM NaH2PO4, 150 mM NaCl; pH 7.2) and maintained at room temperature protected from light in brown glass bottles. Before fixation, samples were filtered with a 10-μm-pore-size filter paper disk (diameter, 5.5 cm; Fisher) to remove autofluorescent particulate material, fixed with paraformaldehyde, and shaken gently for 2 h at 25°C. Cells were recovered by filtration on 0.2-μm-pore-size membrane filters (Millipore), resuspended in PBS containing 4% (vol/vol) paraformaldehyde, and incubated at 4°C for 1 h with mixing. The cells were then resuspended in 4°C PBS by centrifugation for 5 min at 3,000 × g. Washed cells were then resuspended in PBS-100% ethanol (1:1, vol/vol) and stored at −20°C until they were used. Gelatin-subbed slides were prepared by dipping new slides into an aqueous solution containing 0.1% (wt/vol) type A gelatin (approximately 175 bloom; Sigma) and 0.01% (wt/vol) chromium potassium sulfate dodecahydrate [CrK(SO4)2 · 12H2O] and then dried for 10 min at 25°C. Fixed cells were applied to treated slides and air dried at 37°C, and then they were dehydrated by successive rinses in 50, 80, and 98% ethanol solutions. Cell permeabilization was accomplished by a lysozyme-EDTA treatment (54) with lysozyme (5 mg/ml) in 100 mM Tris-HCl-50 mM EDTA (pH 8.0). Rinsed and dried slides were then simultaneously treated with two fluorochrome-coupled antibody probes in the dark. Antibody probes in probing buffer (4% [wt/vol] bovine serum albumin, 150 mM NaCl, 100 mM Tris-HCl) were applied to the permeabilized cells on a slide and incubated for 2 h in a humidified chamber at 25°C. After incubation, the slides were washed in a buffer containing 150 mM NaCl, 100 mM Tris-HCl (pH 7.5), and 0.1% (wt/vol) sodium dodecyl sulfate for 20 min, washed in water for another 20 min, and air dried at 37°C. Fluorochrome bleaching was minimized by application of an aqueous solution of phenylenediamine (1 mg/ml) to probed cells, followed by sealing of the coverslip with clear nail polish. Fluorescence emission from specifically bound fluorochrome-labeled antibody probes was detected with an epifluorescence microscope (Olympus AX 70) fitted with an LEI-750 charge-coupled device camera (DE1 750 Optronic) and light filter sets for fluorescein and rhodamine (Olympus). Images were captured, cells were counted, and fluorescence was quantified by using the Image-1 image analysis software (version 4.0; Universal Imaging). The relationship of fluorescence to the number of cells or to different amounts of Fis or Dps has been described previously (40), and the experiments included mixing experiments in which known numbers of cells having or lacking one of the target proteins (Fis or Dps) were analyzed by fluorescence and bright-field microscopy.
Image analysis.
Single-cell fluorescence intensity was determined by converting wavelength-specific cell brightness into a grey scale pixel value. The total pixel values ranged from 0 to 255 per cell. A midrange pixel value of 100 to 200 per cell was selected for data analysis. The midrange fluorescence data set excluded very faint and very bright fluorescent cells. At least some portion of the weakly fluorescent cells resulted from nonspecific probe binding that reflected insufficient target blocking as a compromise strategy to avoid signal quenching. Also excluded were rare unusually bright cells that fluoresced strongly in both emission ranges. For image analysis we employed automatic fluorescence measurements. The reliability of the automatic mode was validated by comparing automatic fluorescence measurements to manual measurements for identical samples containing a total of 500 cells. Noncellular fluorescent debris and particles were excluded from the analysis by using standard shapes, spheres and rods, for recognition of stationary and growing enterobacteria, respectively. Single-cell fluorescence was converted to a percentage of the maximum fluorescence observed for all cells in each sample for each of the two target proteins. Fluorescent cells were then divided into groups based on 10% incremental relative fluorescence values. The percentage of cells in each incremental group was then determined by determining the fraction of the total number of cells in all incremental groups. The average of the percentages for each incremental group for two replicate water samples was then plotted. The experimental data points included drop lines and a range of data point ball sizes. Ball sizes were proportional to the abundance of the fluorescent cell groups, and the following five ball sizes were used: largest, 100 to 20%; large, 20 to 10%; medium, 10 to 5%; small, 5 to 1.0%; and smallest, 1.0% to undetectable. Since most fluorescent cells fell within the 20 to 1.0% range, a nonlinear division was selected for graph construction.
Heat killing.
To assess the thermal tolerance of enterobacterial community samples, grab samples were collected in 125-ml sterile bottles, placed on ice, and then transported to the laboratory. The time between collection and processing in the laboratory was less than 1 h. To determine the impact of chilling on the community physiology in wastewater samples, E. coli K-12 cells (106 CFU/ml) were exposed to ice for 60 min and subjected to heat killing at 51°C. The percentages of killed cells before and after exposure to ice were 99.70 and 99.85%, respectively. Since there was no significant difference between the percentages of killed cells, sample chilling appeared to have little impact on subsequent viability determinations. Prior to heating, samples were filtered by using a 10-μm-pore-size filter paper disk (diameter, 12.5 cm; VWR) to remove large particulate material, which recovered more than 99% of the cells. Two-milliliter portions of the filtered samples were subjected to heating at 51°C in a prewarmed heating block for different periods of time. Heated samples were then diluted in sterile distilled water, spread onto duplicate Endo agar (Becton Dickinson) plates, and incubated overnight at 37°C. The surviving population was counted, and the percentage of the unheated population was calculated.
Statistical analysis.
The statistical significance of differences in enterobacterial physiology was investigated for different wastewater processing steps during the 5 weeks of successive sampling. Since there were two variables, week and treatment process, that could affect the dependent variable (growth state physiology), a two-way mixed-design analysis of variance (ANOVA) was used for data analysis. For the clusters of cells with fluorescent Fis and Dps grouped in 10% increments based on individual fluorescence, the fluorescent cell types with 80, 90, and 100% of the total cellular fluorescence were included for statistical analysis. The cells were selected because they exhibited maximum expression of growth-state-specific proteins and thus correlated most closely with the extremes of growth and the stationary phase. Since each week's sample data had been normalized previously to 100%, a portion of the data was included for statistical analysis to compare the variables based on the mean data. The SAS program (version 8.0) was used for ANOVA. Correlation analysis and a Student's t test were performed by using MS Excel.
RESULTS
Single-cell enterobacterial protein profiling during wastewater treatment.
The main components of the wastewater treatment plant and the locations of sampling sites are indicated in Fig. 1. Incoming wastewater (influent wastewater) was aerated and diverted into three primary clarifiers after an aerated grit removal process. The three primary clarifiers received approximately equal volumes of wastewater, and the wastewater from these clarifiers had completed primary treatment. Secondary treatment (biological treatment and clarification) followed this initial process and involved the use of trickling filters or activated sludge by surface aeration or submerged aeration methods. Each type of biological treatment was distinguished by the mode and extent of aeration. Wastewater from each biological treatment process was then individually clarified and pooled for tertiary treatment (chlorination). Residual chlorine was neutralized prior to entry into the receiving water.
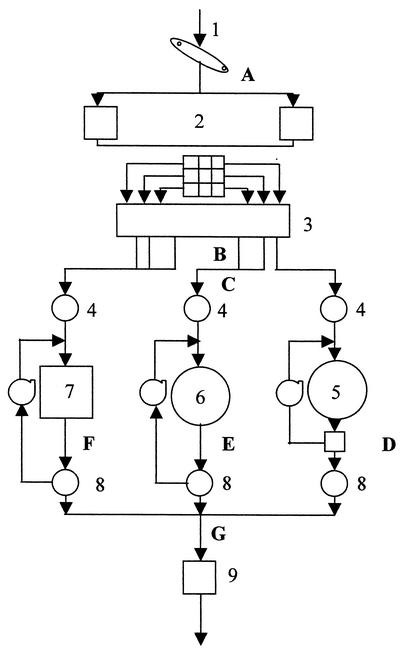
FIG. 1.
Diagram of the wastewater treatment plant. 1, plant influent; 2, pump station; 3, aerated grit removal; 4, primary clarifier; 5, trickling filter; 6, submerged aeration basin; 7, surface aeration basin; 8, secondary clarifier; 9, chlorine contact basin. The sampling locations were influent (A), aerated grit removal (B), primary clarifier (C), trickling filter (D), submerged aeration basin (E), surface aeration basin (F), and pooled secondary clarifier (G). Samples were collected from the outlet at each location.
The specificity and suitability of immunofluorescence techniques for the environmental samples were tested to assess detection of organisms other than enterobacteria present in the wastewater. Fixed cells from each sampling location were probed first with ENT, an oligodeoxynucleotide probe specific for the 16S rRNA of members of the Enterobacteriaceae, and counterstained with DAPI. Fixed cells also were probed with both anti-Fis or anti-Dps antibodies and then counterstained with DAPI. There was no statistically significant difference between the percentages of cells detected with the ENT probe and the percentages of cells detected with the anti-Fis and anti-Dps probes (Table 1). This indicated that the antibody probes were specific for enterobacteria and did not detect other organisms.
TABLE 1.
Specificity of antibody probesa
| Sampling location | No. of ENT-positive cells per DAPI- positive cell | No. of Dps- or Fis-positive cells per DAPI-positive cell |
|---|---|---|
| Influent | 31.77 ± 19.70 | 27.97 ± 15.46 |
| Aerated grit removal | 25.65 ± 17.70 | 23.33 ± 19.53 |
| Primary clarifier | 38.50 ± 22.38 | 36.93 ± 20.83 |
| Trickling filter | 9.94 ± 3.91 | 12.70 ± 2.63 |
| Submerged aeration basin | 8.22 ± 3.22 | 10.20 ± 3.88 |
| Surface aeration basin | 5.92 ± 2.64 | 5.77 ± 3.73 |
| Pooled secondary clarifier | 9.84 ± 4.72 | 7.53 ± 3.65 |
Values are means ± standard deviations.
Single-cell levels of Fis and Dps were determined over a 5-week sampling period for uncultivated enterobacterial cells present in wastewater treatment samples. Samples were collected from the outlet of a treatment process, and cells were fixed immediately after sample collection to prevent variation in the Fis and Dps contents during sample transport. Protein profiles were determined by simultaneous application of enterobacterium-specific fluorochrome-labeled antibody probes. A representative micrograph showing the appearance of these probed cells is shown in Fig. 2. Fis and Dps levels were determined by measuring the fluorescence intensities of the corresponding fluorochrome-labeled antibodies for approximately 1,000 cells per time point over a 5-week period at selected sampling locations (Fig. 3). All samples contained cells with either Dps or Fis (Fig. 3), but the relative proportions varied considerably. In addition, the relative levels of fluorescence of cells within samples varied widely for both proteins at all sampling locations. In wastewater treatment plant influent samples, there were few cells with an abundance of Fis. However, more cells of this type appeared following primary treatment (Fig. 3A to C). The trickling filter process resulted in an increase in the number of cells with an abundance of Fis (Fig. 3D) compared to the number of such cells in primary clarifier wastewater (Fig. 3C). Cells containing large amounts of Fis also were rare in samples after the submerged aeration process (Fig. 3E). In most samples there were few cells with large amounts of Dps. However, the levels observed varied significantly between locations. In most cases, the cellular levels of Dps were low. Cells with relatively high levels of Dps were most evident in primary clarifier wastewater (Fig. 3C) and in submerged aeration basin wastewater (Fig. 3E).
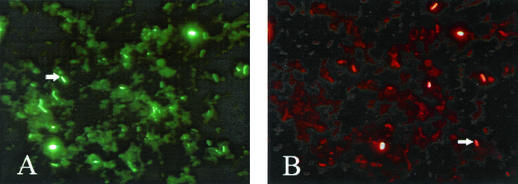
FIG. 2.
Growth state of enterobacteria in wastewater samples: fluorescence micrographs showing wastewater samples probed with anti-Fis antibody (A) and anti-Dps antibody (B). The arrows in panels A and B indicate cells with significant levels of Fis and Dps, respectively.
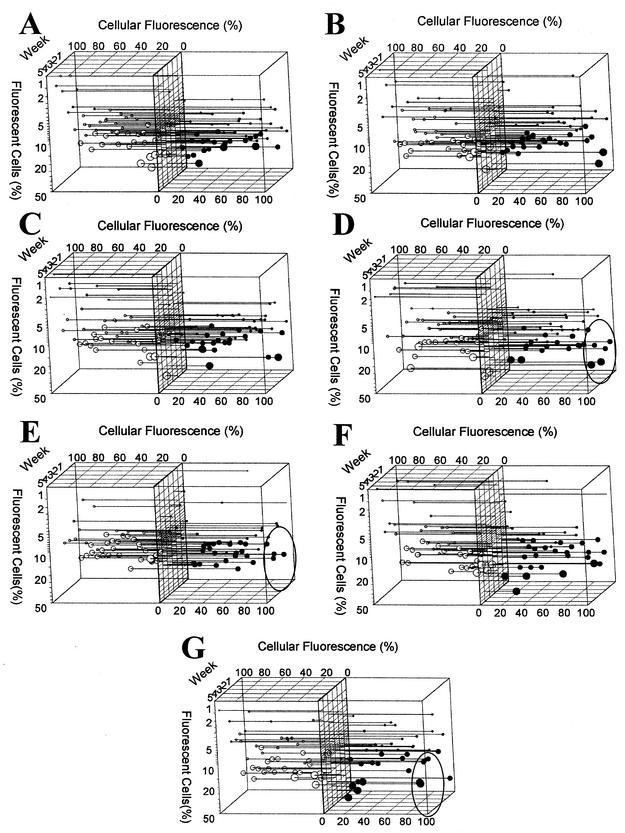
FIG. 3.
Single-cell protein profiles of the wastewater enterobacterial communities. (A) Influent; (B) aerated grit removal; (C) primary clarifier; (D) trickling filter; (E) submerged aeration basin; (F) surface aeration basin; (G) pooled secondary clarifier. The cellular contents of Dps (○) and Fis (•) are expressed as percentages of the cellular fluorescence of the brightest cells in each sample (y axis). The amounts of cells exhibiting particular levels of fluorescence are expressed as percentages of the fluorescent cell type (x axis). The time series represents 5 weeks of samples (z axis). The circle size is proportional to the abundance of the fluorescent cell group. The following five sizes are shown: largest, 100 to 20%; large, 20 to 10%; medium, 10 to 5%; small, 5 to 1.0%; smallest, 1.0% to undetectable. Cells are grouped into clusters based on 10% increments of individual cellular fluorescence. Experimental data points are plotted with drop lines with a range of data point ball sizes. Two graphs (Fis and Dps) were merged after 90o rotation for each sampling location. Significant differences in Fis abundance are indicated by ovals in panels D, E, and G. Samples were obtained weekly in the winter beginning in January 2002.
Secondary wastewater from the biological treatment processes was pooled before chlorination (Fig. 1). Protein profiles of this pooled material revealed an abundance of cells with high levels of Fis (Fig. 3G). These cells could have resulted from the mixing of the distinct communities created during biological treatment. Such cells were present in trickling filter wastewater and absent in submerged aeration basin wastewater.
Average values for Fis and Dps cell contents also were determined for each of the treatment plant sampling locations (Table 2). These values were determined by adding all of the fluorescence values for Fis or Dps and normalizing the sum to the total number of cells examined. The resulting data reflected enterobacterial community averages. The values for sampling locations, however, were not statistically significantly different.
TABLE 2.
Community average levels of Fis and Dps
| Samples | Fis cellular fluorescencea | Dps cellular fluorescencea |
|---|---|---|
| Influent | 51.33 ± 6.42 | 35.90 ± 6.82 |
| Aerated grit removal | 52.55 ± 8.68 | 37.18 ± 6.34 |
| Primary clarifier | 52.55 ± 9.87 | 36.06 ± 7.22 |
| Trickling filter | 55.85 ± 14.36 | 36.91 ± 6.31 |
| Submerged aeration basin | 50.22 ± 6.72 | 40.30 ± 4.35 |
| Surface aeration basin | 51.51 ± 9.82 | 34.01 ± 8.80 |
| Pooled secondary clarifier | 47.84 ± 14.36 | 40.51 ± 6.31 |
The values indicated (means ± standard deviations) represent the total number of pixels for X number of cells averaged over 10 samples per location.
Efficiency of disinfection of community samples by heat inactivation.
The protein profile study revealed the presence of an enterobacterial community in submerged aeration basin wastewater containing the smallest number of cells with an abundance of Fis. In contrast, the enterobacterial community present in the trickling filter wastewater contained the largest number of cells with an abundance of Fis. In general, the pattern for the number of cells with an abundance of Dps was the inverse of the pattern observed with Fis. However, the range of variation between samples was more constrained. To test the hypothesis that the protein profiles were indicative of growing and nongrowing enterobacterial communities, differences in the efficiency of disinfection were examined by using samples from the trickling filter (the location where the Fis cellular fluorescence was highest) and the submerged aeration basin (the location where the Fis cellular fluorescence was lowest). New samples obtained from the treatment plant were placed on ice and transported to the laboratory. Reconstruction experiments demonstrated that short periods of chilling did not alter the survival kinetics. Attempts to use chlorination were confounded by the presence of variable amounts of total carbon in the wastewater samples that titrated added chlorine and interfered with cell killing. Since the general pattern for growth state and resistance to chlorination also applied to killing by heat exposure (9, 38, 39), heating was used as an alternative means of disinfection.
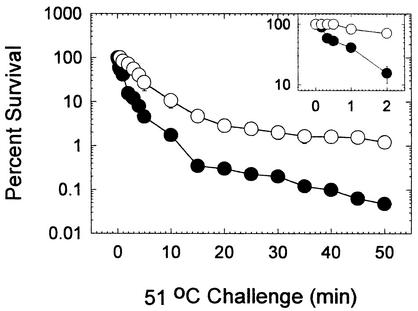
The ambient temperature varied considerably in different seasons in the region occupied by the target wastewater treatment facility. Therefore, analysis of the additional samples also addressed the temporal stability of enterobacterial community protein profiles. Replicate samples of cells from trickling filter and submerged aeration wastewater were subjected either to protein profiling or thermal killing. These samples were obtained approximately 6 months after those described above were obtained (Fig. 3), and they were obtained during the summer months. This study again revealed the presence of both Fis-containing and Dps-containing cells in the water from the two processes, and the distributions resembled the distributions obtained in the experiment described above (Fig. 4). The number of cells with high levels of Fis was large in trickling filter wastewater and small in submerged aeration wastewater. The inverse pattern for Dps was also detected, as observed previously. In thermal killing studies we employed a heat treatment regimen consisting of exposure to 51°C for 50 min. Viability was assessed by plating samples at various intervals onto Endo agar plates to detect the enterobacterial segment of the community (Fig. 5). The thermal tolerance of enterobacteria derived from the submerged aeration wastewater was significantly different from the thermal tolerance of enterobacteria derived from the trickling filter wastewater. Enterobacteria in the trickling filter wastewater were highly sensitive to heat killing, and significant reductions in population sizes were apparent after 10 s of exposure. In contrast, no alteration in the population size was apparent for enterobacteria from the submerged aeration wastewater even after 30 s of exposure (Fig. 5, inset). Most of the enterobacteria in the trickling filter wastewater were killed after 2 min of exposure, compared with only one-half of the enterobacteria from the submerged aeration basin wastewater. More than 1% of enterobacteria from the submerged aeration wastewater were heat resistant even after 50 min of exposure. Similarly, less than 0.01% of enterobacteria in the trickling filter wastewater were resistant to heat killing.
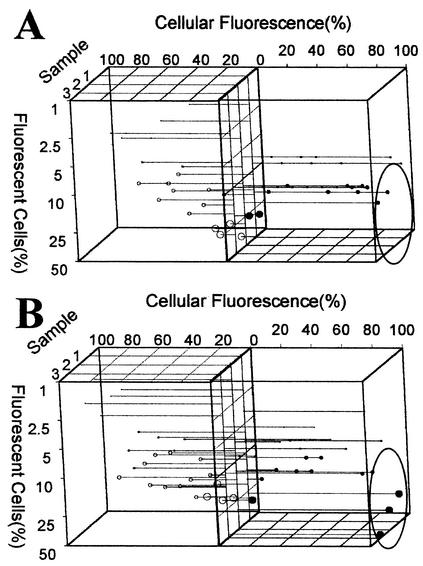
FIG. 4.
Influence of seasons on single-cell protein profiles. (A) Submerged aeration basin; (B) trickling filter. Samples were obtained weekly in the summer beginning in July 2002. The axes, data representation, and sample times are the same as those described in the legend to Fig. 3. Significant differences in the abundance of Fis-containing cells are indicated by ovals.
FIG. 5.
Enterobacterial thermal tolerance after biological treatment. The number of surviving cells was normalized to the number of viable cells in an untreated but otherwise identical sample. The values are the averages for three duplicate samples, and the error bars indicate the standard errors. The inset shows the percentage of survival for 2 min after the initial exposure. ○, submerged aeration basin samples; •, trickling filter samples.
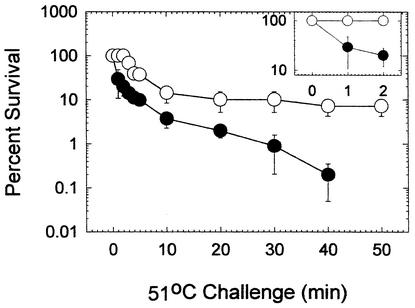
These data reflected the physiological response of the total enterobacterial community to thermal killing because they were obtained by using Endo agar as the plating medium. Since wastewater release into receiving water is regulated by the content of fecal enterobacteria, additional efforts were made to examine this subset of the enterobacterial group. The heat resistance of fecal enterobacteria derived from the wastewater after both forms of biological treatment was examined by using the most-probable-number method of analysis and A-1 medium (42). The sensitivity to heat killing when this medium was used resembled that observed with Endo agar. Fecal enterobacteria derived from trickling filter wastewater were more sensitive to killing than fecal enterobacteria derived from submerged aeration basin wastewater (Fig. 6). After 2 min of exposure, there was no reduction in the number of fecal enterobacteria in water from the submerged aeration process, but after 1 min of exposure approximately 70% of the organisms from the trickling filter water were killed. While there were no surviving fecal enterobacteria after 50 min of exposure in trickling filter wastewater, a significant fraction remained culturable after 50 min of exposure in submerged aeration basin wastewater. These two studies confirmed that the physiology of both total enterobacteria and fecal enterobacteria in submerged aeration basin wastewater was significantly different from the physiology of enterobacteria in trickling filter wastewater.
FIG. 6.
Fecal enterobacterial thermal tolerance after biological treatment. The number of surviving cells was normalized to the number of viable cells in an untreated but otherwise identical sample. The values are the averages for three duplicate samples, and the error bars indicate the standard errors. The inset shows the percentage of survival during the initial 2-min treatment period. ○, submerged aeration basin samples; •, trickling filter samples.
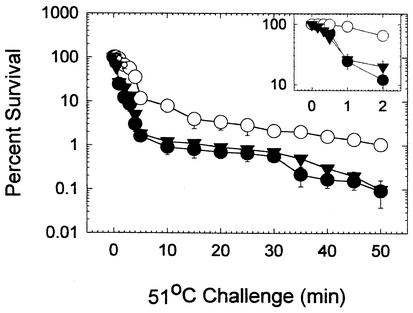
To test whether the thermal killing profiles observed for the enterobacterial communities from biological treatment wastewater remained apparent after secondary clarification and before pooling and chlorine disinfection, additional samples were subjected to thermal killing. Survival again was measured after plating on a selective medium (Fig. 7). A distinctive pattern was observed, in which the thermal resistance was significantly greater in wastewater from the submerged aeration basin clarifier than in wastewater from either the trickling filter or surface aeration basin secondary clarifier. The dissolved oxygen and total carbon contents were studied by using three duplicate samples collected simultaneously from the individual and pooled secondary clarifier wastewaters (Table 3). Both the dissolved oxygen and total carbon contents were found to be highest in the trickling filter secondary clarifier. The submerged aeration secondary clarifier wastewater also differed from the trickling filter secondary clarifier wastewater (P = 0.002) and the surface aeration basin secondary clarifier wastewater (P = 0.05), while the dissolved oxygen content was lowest in samples from the submerged aeration basin secondary clarifier. However, the oxygen levels in the three secondary clarifier wastewaters did not differ significantly from those in the final pooled secondary clarifier wastewater. There was no statistically significant difference among the total carbon levels in these secondary clarifier wastewaters. The variation in sample replicates, however, precluded assessment of the significance of these parameters for establishing or maintaining enterobacterial physiological status during wastewater treatment.
FIG. 7.
Enterobacterial thermal tolerance after secondary clarification. The number of surviving cells was normalized to the number of viable cells in an untreated but otherwise identical sample. The values are the averages for three duplicate samples, and the error bars indicate the standard errors. The inset shows the percentage of survival for 2 min after the initial exposure. ○, submerged aeration basin secondary clarifier samples; •, trickling filter secondary clarifier samples; ▾, surface aeration secondary clarifier samples.
TABLE 3.
Dissolved oxygen contents and total carbon levels in secondary clarifiers
| Secondary clarifier location | Dissolved oxygen concn (mg/liter)a | Total carbon level (%)b |
|---|---|---|
| Trickling filter | 6.41 ± 0.637 | 0.050 ± 0.020 |
| Submerged aeration basin | 5.03 ± 0.518 | 0.040 ± 0.020 |
| Surface aeration basin | 5.60 ± 0.478 | 0.038 ± 0.018 |
| Pooled secondary clarifier | 5.71 ± 0.686 | 0.036 ± 0.017 |
Means ± standard deviations of five samples.
Means ± standard deviations of three samples.
DISCUSSION
Our results reveal that there are distinct enterobacterial communities with characteristic protein profiles that reflect discrete phases of wastewater treatment. Protein profiles were used to predict physiological parameters, including sensitivity to disinfection. Single-cell analysis provided information that was essential for physiological predictions since bulk measurements of Fis and Dps contents that reflected community averages were nearly identical for different sample locations and thus failed to provide information that could be used to predict physiological differences between communities. Single-cell analysis of cellular Fis and Dps contents allowed detection of predictive trends in subsegments of the community that were otherwise obscured by invariant community components.
Two-way ANOVA revealed that there was a significant difference (P = 0. 0001) in the number of cells with fluorescent Fis between weekly samples, but the difference between locations was not significant. The data for the combined variables (week and location), however, did reveal that there was a significant difference (P = 0.0039). Consequently, to understand the difference among the numbers of cells with fluorescent Fis in the processes, a contrast study was performed for the processes, especially the three different types of biological processing. The contrast study revealed that the number of cells with fluorescent Fis in the trickling filter wastewater differed significantly from the number in the submerged aeration basin wastewater (P = 0.0345) and the number in the surface aeration basin wastewater (P = 0.0362) at the 95% confidence level. Similarly, the cell types with fluorescent Dps present in the five weekly samples were examined by ANOVA. There were significant differences among the cells with fluorescent Dps when different sample weeks were compared, but the difference between locations was not significant. The combined interaction of week and location was also not significant. A contrast study of the processes, especially the three different types of biological processing, revealed that there was not a significant difference between locations for Dps.
If a treatment process favors exponential-phase cells (cells with fluorescent Fis) at one location, there must be a corresponding reduction in the abundance of stationary-phase cells (cells with fluorescent Dps). To confirm the existence of treatment selection in the trickling filter and the submerged aeration basin, a correlation study was performed by using cells with fluorescent Fis and Dps present in the trickling filter and submerged aeration basin wastewaters. The mean values for the cells with fluorescent Fis or Dps (averages of the 5-week sample set) were used for the correlation study. In this study we found that there was a significant negative correlation (correlation coefficient, −760) between cells with fluorescent Fis and cells with fluorescent Dps at a 99% confidence level in trickling filter wastewater. There was not a significant correlation between cells with fluorescent Fis and cells with fluorescent Dps in submerged aeration basin wastewater. The existence of the opposite trend for the types of exponential-phase and stationary-phase cells in the trickling filter wastewater indicated that physiological selection during the trickling filter process promoted the growth of enterobacteria.
The main finding of this study was that enterobacteria and fecal enterobacteria in trickling filter wastewater are much easier to kill than enterobacteria in submerged aeration basin wastewater. Protein profile and disinfection analyses conducted for later stages of the treatment process suggested that the distinctive enterobacterial communities were stable and contributed significantly to the community that underwent tertiary treatment (chlorination). Consequently, inefficient tertiary treatment may result from treatment processes that produce disinfection-resistant enterobacterial communities. In addition, improved tertiary treatment may be obtained by directed manipulation of enterobacterial physiology, which has the potential for reducing chlorine use and minimizing release of disinfection by-products into receiving water.
Cells with an abundance of Dps occurred rarely in our studies. This suggests that the enterobacterial communities were metabolically active and capable of proliferating during wastewater treatment. However, the occurrence of cells with variable (although small) amounts of Dps indicated that these cells were in different physiological states in the early stages of the stationary phase. Even though oxygen and nutrients are generally available during wastewater treatment, the presence of Dps indicated that the bacteria were unable to grow well. This condition could reflect the presence of growth-inhibiting compounds or physiological stress.
In influent samples, only a few (<1%) of the cells exhibited high levels of Fis. However, a higher percentage of such cells appeared during subsequent treatment steps. This indicates that there was a metabolic shift or growth transition precipitated by the treatment process. Such differences were particularly apparent when modes of biological treatment were compared. The differences prompted the use of heat killing to measure the efficiency of disinfection. Since the enterobacterial community present in the submerged aeration basin wastewater was notably more resistant that the enterobacterial community present in trickling filter wastewater, the subfraction comprising fecal enterobacteria was examined. The efficiencies of disinfection of this subfraction exhibited similar process-dependent differences, providing results that are relevant to wastewater tertiary treatment and subsequent water release. Prolonged exposure to heat resulted in the presence of a small percentage of enterobacterial cells that exhibited extreme resistance to killing. Such cells are likely to have entered the stationary phase accumulating high levels of Dps and exhibiting concomitant resistance to killing. These findings suggest that most cells in the wastewater enterobacterial community occur in a growing state and are not starving.
It is important to note that the methods employed in this study to examine uncultivated cell physiology failed to distinguish between enterobacterial taxa. Much of the variation observed could reflect species-specific physiological differences. Methods that combine obtaining growth state information with determining taxonomic identity are currently being developed. Together with current strategies, such methods may increase treatment plant performance, reduce wastewater enterobacterial content, and minimize the release of disinfection by-products into receiving water.
Acknowledgments
The cooperation of wastewater treatment plant personnel is gratefully acknowledged. We thank R. Morita and A. C. Matin for their insight and emphasis on studies concerning microbial physiology. We also thank Kent Eskridge and Anil K. Jayaprakash, Department of Biometry/Statistics, University of Nebraska, for performing the statistical analysis with SAS.
This research was supported in part by grant 00-HHE-1 from the Water Environment Research Foundation, by Environmental Protection Agency assistance agreement CR827345-01-0, and by grant LB2106 from the state of Nebraska.
REFERENCES
- 1.Almiron, M., A. J. Link, D. Furlong, and R. Kolter. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6:2646-2654. [DOI] [PubMed] [Google Scholar]
- 2.Azam, T. A., A. Iwata, A. Nishimura, S. Ueda, and A. Ishihama. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181:6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball, C. A., R. Osuna, K. C. Ferguson, and R. C. Johnson. 1992. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J. Bacteriol. 174:8043-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg, J. D., A. Matin, and P. V. Roberts. 1982. Effects of antecedent growth conditions on sensitivity of Escherichia coli to chlorine dioxide. Appl. Environ. Microbiol. 44:814-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chauret, P. C., C. Z. Radziminski, M. Lepuil, R. Creason, and R. C. Andrews. 2001. Chlorine dioxide inactivation of Cryptosporidium parvum oocysts and bacterial spore indicators. Appl. Environ. Microbiol. 67:2993-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De los Reyes, F. L., W. Ritter, and L. Raskin. 1997. Group-specific small-subunit rRNA hybridization probes to characterize filamentous foaming in activated sludge systems. Appl. Environ. Microbiol. 63:1107-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De los Reyes, M. F., F. de los Reyes, L. Hernandez, and L. Rakin. 1998. Quantification of Gordonia amarae strains in foaming activated sludge and anaerobic digester systems using oligonucleotide hybridization probes. Appl. Environ. Microbiol. 64:2503-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De los Reyes, M. F., D. B. Oerther, F. de los Reyes, L. Hernandez, and L. Rakin. 1998. Characterization of filamentous foaming in activated sludge systems using oligonucleotide hybridization and antibody probes. Water Sci. Technol. 37:485-493. [Google Scholar]
- 9.Dukan, S., and D. Touati. 1996. Hypochlorous acid stress in Escherichia coli: resistance, DNA damage, and comparison with hydrogen peroxide stress. J. Bacteriol. 178:6145-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaten, A. D., L. S. Clesceri, and A. E. Greenberg (ed.). 1995. Standard methods for the examination of water and wastewater, 19th ed. American Public Health Association, Washington, D.C.
- 11.Erhart, R., D. Bradford, R. Seviour, R. Amann, and L. Blackall. 1997. Development and use of fluorescent in situ hybridization probes for the detection and identification of Microthrix parvicella in activated sludge. Syst. Appl. Microbiol. 20:310-318. [Google Scholar]
- 12.Giovannoni, S. J., E. F. DeLong, G. J. Olsen, and N. R. Pace. 1988. Phylogenetic group specific oligodeoxynucleotide probes for identification of single microbial cells. J. Bacteriol. 170:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howgrave-Grahan, A. R., and P. L. Steyn. 1988. Application of the fluorescent-antibody technique for the detection of Sphaerotilus nalans in activated sludge. Appl. Environ. Microbiol. 54:799-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, R. C., C. A. Ball, D. Pfeffer, and M. I. Simon. 1988. Isolation of the gene encoding the Hin recombinational enhancer binding protein. Proc. Natl. Acad. Sci. USA 85:3484-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kappeler, J., and W. Gujer. 1992. Estimation of kinetic parameters of heterotrophic biomass under aerobic conditions and characterization of wastewater for activated sludge modeling. Water Sci. Technol. 25:125-139. [Google Scholar]
- 16.Koch, C., J. Vandekerckhove, and R. Kahmann. 1988. Escherichia coli host factor for site-specific DNA inversion: cloning and characterization of the fis gene. Proc. Natl. Acad. Sci. USA 85:4237-4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krska, J., T. Elthon, and P. Blum. 1993. Monocolonal antibody recognition and functions of a DnaK (HSP70) epitope found in gram-negative bacteria. J. Bacteriol. 175:6433-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeChevallier, M. W., C. D. Cawthon, and R. G. Lee. 1988. Factors promoting survival of bacteria in chlorinated water supplies. Appl. Environ. Microbiol. 54:649-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lisle, J. H., S. C. Broadaway, A. M. Prescott, B. H. Pyle, C. Fricker and G. A. McFeters. 1998. Effects of starvation on physiological activity and chlorine disinfection resistance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 64:4658-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomoskaya, O. L., J. P. Kidwell, and A. Martin. 1994. Characterization of the σ38-dependent expression of a core Escherichia coli starvation gene, pexB. J. Bacteriol. 176:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K. H. Schleifer. 1992. Phylogenetic oligooxynuleotide probes for the major subclass of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 22.Manz, W., U. Szewzyk, P. Ericsson, R. Amaan, K. H. Schleifer, and T. A. Stenstrom. 1993. In situ identification of bacteria in drinking water and adjoining biofilms by hybridization with 16S and 23S rRNA-directed fluorescent oligonucleotide probes. Appl. Environ. Microbiol. 59:2293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mason, C., and T. Egli. 1993. Dynamics of microbial growth in the decelerating and stationary phase of batch culture, p. 81-102. In S. Kjelleber (ed.), Starvation in bacteria. Plenum Press, New York, N.Y.
- 24.Matin, A. 1991. The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Mol. Microbiol. 5:3-10. [DOI] [PubMed] [Google Scholar]
- 25.Matin, A., and S. Harakeh. 1990. Effect of starvation on bacterial resistance to disinfectants, p. 88-103. In G. A. McFeters (ed.), Drinking water microbiology. Springer-Verlag, New York, N.Y.
- 26.Mittleman, M. W., M. Habash, J. M. Lacroix, A. E. Khoury, and M. Krajden. 1997. Rapid detection of Enterobacteriaceae in urine by fluorescent 16S rRNA in situ hybridization on membrane filters. J. Microbiol. Methods 30:153-160. [Google Scholar]
- 27.Nilsson, L., H. Verbeek, E. Vijgenboom, C. van Drunen, A. Vanet, and L. Bosch. 1992. FIS-dependent trans activation of stable RNA operons of Escherichia coli under various growth conditions. J. Bacteriol. 174:921-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Notley, L., and T. Ferenci. 1996. Induction of RpoS-dependent functions in glucose-limited continuous culture: what level of nutrient limitation induces the stationary phase of Escherichia coli? J. Bacteriol. 178:1465-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oerther, D. B., F. L. de los Reyes, and L. R Askin. 1999. Interfacing phylogenetic oligonucleotide probe hybridization with representations of microbial populations and specific growth rates in mathematical models of activated sludge process. Water. Sci. Technol. 39:11-20. [Google Scholar]
- 30.Ogoshi, M., Y. Suzuki, and T. Asano. 2001. Water reuse in Japan. Water Sci. Technol. 43:17-23. [PubMed] [Google Scholar]
- 31.Olsen, B., and M. Stewart. 1987. Factors that change bacterial resistance to disinfection, p. 885-904. In R. L. Jolley, L. W. Condie, J. D Johnson, S. Katz, R. A. Minear, J. S. Mattice, and V. A Jacobs (ed.), Water chlorination: chemistry, environmental impact and health effects, vol. 6. Lewis Publishers, Inc., Chelsea, Mich.
- 32.Olsen, G. J., D. J. Lane, S. J. Giovannoni, N. R. Pace, and D. A. Stahl. 1986. Microbial ecology and evolution; a ribosomal RNA approach. Annu. Rev. Microbiol. 40:337-365. [DOI] [PubMed] [Google Scholar]
- 33.Onuki, M., H. Satoh, and T. Mino. 2002. Analysis of microbial community that performs enhanced biological phosphorus removal in activated sludge fed with acetate. Water Sci. Technol. 46:145-154. [PubMed] [Google Scholar]
- 34.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 35.Poulsen, L. K., G. Ballard, and D. A. Stahl. 1993. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 59:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preez, M., R. Kfir, and P. Coubrough. 1995. Investigation of injury of enterobacteria after chlorination. Water Sci. Technol. 31:115-118. [Google Scholar]
- 37.Rittman, B. E., and P. L. McCarty (ed.). 2001. Environmental biotechnology: principles and applications. McGraw-Hill Higher Education. New York, N.Y.
- 38.Rockabrand, D., T. Arthur, G. Korinek, K. Livers, and P. Blum. 1995. An essential role of the Escherichia coli DnaK protein in starvation-induced thermotolerence, H2O2 resistance, and reductive division. J. Bacteriol. 177:3695-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rockabrand, D., K. Livers, T. Austin, R. Kaiser, D. Jensen, R. Burgess, and P. Blum. 1998. Roles of DnaK and RpoS in starvation-induced thermotolerance of Escherichia coli. J. Bacteriol. 180:846-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rockabrand, D., T. Austin, R. Kaiser, and P. Blum. 1999. Bacterial growth state distinguished by single-cell protein profiling: does chlorination kill coliforms in municipal effluent? Appl. Environ. Microbiol. 65:4181-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuppler, M., M. Wagner., G. Schon, and U. B. Gobel. 1998. In situ hybridization of nocardioforms in activated sludge using fluorescent rRNA targeted oligonucleotide probes. Microbiology 144:249-259. [DOI] [PubMed] [Google Scholar]
- 42.Standridge, J. H., and J. J. Delfino. 1981. A-1 medium: alternative technique for fecal coliform organism enumeration in chlorinated wastewaters. Appl. Environ. Microbiol. 42:918-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart, M., and B. Olson. 1992. Physiological studies of chloramine resistance developed by Klebsiella pneumoniae to chloramines. Appl. Environ. Microbiol. 58:2649-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szewzyk, U., W. Manz, R. Amaan, K. H. Schleifer, and T. A. Stenstrom. 1994. Growth and in situ detection of a pathogenic Escherichia coli in biofilms of a heterotrophic water-bacterium by use of 16S and 23S rRNA-directed fluorescent oligonucleotide probes. FEMS Microbiol. Ecol. 13:169-176. [Google Scholar]
- 45.Thompson, J. F., L. Moitoso de Vargas, D. Kock, R. Kahmann, and A. Landy. 1987. Cellular factors couple recombinations with growth phase: characterization of a new component in the λ site-specific recombination pathway. Cell 50:901-908. [DOI] [PubMed] [Google Scholar]
- 46.U.S. Environmental Protection Agency. 1995. National pollutant discharge elimination system permit application requirements for publicly owned treatment works and other treatment works treating domestic sewage. Fed. Regist. 60:62562-62569.
- 47.Villanueva, C., M. Kogevinas, and J. Grimalt. 2001. Chlorination of drinking water in Spain and bladder cancer. Gac. Sanit. 15:48-53. [DOI] [PubMed] [Google Scholar]
- 48.Volsch, A., W. F. Nader, H. K. Geiss, G. Nebe, and C. Birr. 1990. Detection and analysis of two serotypes of ammonia-oxidizing bacteria in sewage plants by flow cytometry. Appl. Environ. Microbiol. 56:2430-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner, M., R. Amann, H. Lemmer, and K. H. Schleifer. 1993. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl. Environ. Microbiol. 59:1520-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner, M., R. Amaan, H. Lemmer, W. Manz, and K. H. Schleifer. 1994. Probing activated sludge with fluorescently labeled rRNA targeted oligonucleotides. Water Sci. Technol. 29:15-23. [Google Scholar]
- 51.Wagner, M., R. Erhart, W. Manz, R. Amann, H. Lemmer., D. Wedi, and K. H. Schleifer. 1994. Development of an rRNA targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl. Environ. Microbiol. 60:792-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wintzingercode, F. V., U. B. Gobel, and E. Stackebrabdt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 53.Xu, P., F. Brissaud, J. C. Maihol, F. Valette, and V. Lazarova. 2002. Design of climate-dependent water reuse project. Water Sci. Technol. 46:289-296. [PubMed] [Google Scholar]
- 54.Zarda, B., R. Amann, G. Wallner, and K. H. Schliefer. 1991. Identification of single bacterial cells using digoxigenin-labelled rRNA-targeted oligonucleotides. J. Gen. Microbiol. 137:2823-2830. [DOI] [PubMed] [Google Scholar]