Abstract
We describe here the identification and characterization of a functional downstream element in the human adult β-globin promoter. The existence of this element was indicated by two mutations at +22 and +33 downstream of the β-globin transcriptional start site in humans with β-thalassemia. In vitro transcriptional analysis of these mutants, plus a third at +13, indicates that all three decrease transcription from the β-globin promoter. Scanning mutagenesis from +10 to +45 indicates that this region contains a functional cis element(s) in vitro, and we designated this element the DCE (downstream core element). The DCE functions in concert with the β-globin CATA box and initiator element, as well as in a heterologous, TATA-less context. A second set of mutants indicates that a particular geometry of the DCE and core promoter is necessary for promoter function. Lastly, DCE mutants show reduced affinity for transcription factor IID (TFIID). These data indicate that TFIID makes sequence-specific contacts to the DCE and that TFIID binding is necessary for DCE function.
The focal point of eukaryotic transcriptional regulation is the core promoter. Its integrity is required for the assembly of the general machinery in response to a transcriptional activator. The core promoter consists of several cis elements. The TATA box, located approximately 30 base pairs upstream of the initiation site, is thought to dictate the position of initiation in higher eukaryotes (1). A second element, the initiator (Inr), is centered around the transcription start site. The Inr, as exemplified by the TdT Inr, was originally described in a TATA-less promoter, where it establishes correct initiation in the absence of a TATA box (2). Since the discovery of the TdT Inr, a variety of promoters have been found to contain Inr elements, including TATA-dependent promoters. Although the role of the Inr in the presence of a TATA box remains to be fully defined, it appears to contribute to the magnitude of transcription (1, 3).
Several examples of a third element, found downstream of the transcriptional start site, have been reported. Regions downstream of transcriptional start sites that affect transcription were first described in the adenovirus major late promoter, from +7 to +33 (4). A second report described a downstream region in the human glial fibrillary acidic protein promoter extending from +10 to +50 (5). In the TATA-less TdT promoter, a downstream region lies from +33 to +59, and its deletion resulted in a reduction in transcription (2, 6). Burke and Kadonaga describe detailed analysis of a downstream element identified in several Drosophila and human TATA-less promoters. This element, which is centered at +30, stimulates transcription and its function is redundant to a TATA box in a heterologous context (7, 8). The existence of downstream elements has also been inferred from transcription factor IID (TFIID) footprints on various promoters, extending 30–40 base pairs downstream of the transcriptional start site (9–17), but it is unclear whether downstream protections reflect sequence-specific contacts (10).
Members of the TFIID complex have been shown to be necessary for function of core promoter elements. Well documented is the interaction of the TATA box with TBP (18). Two members of the TFIID complex, hTAFII250 and hTAFII150, have been shown to interact with initiator and downstream elements (16, 17, 19). Drosophila TFIID was found to footprint the downstream promoter element (DPE), and mutations in the DPE correlate with reduced TFIID binding. Furthermore, the DPE can be crosslinked to two components of Drosophila TFIID complex, dTAFII40 and dTAFII60 (7, 8).
Hemoglobin consists of a tetramer of two α and two β polypeptides. The ratio of α and β chains must be precisely balanced in red blood cells; imbalances result in human thalassemia disease. A variety of mutations in the β-globin gene decrease expression of the β-globin protein, resulting in human β-thalassemia. The free, excess α-globin protein precipitates in erythroblasts and inhibits their maturation. This leads to anemias of varying severity depending on the β-globin expression defect. Some β-thalassemia disease mutations lie in the major regulatory elements of the β-globin promoter: the CACC box, the TATA box, and the initiator element. Point mutations in the CACC box and TATA box typically show a reduction in β-globin expression of 70–80% of wild-type levels (20).
Two uncharacterized β-thalassemia mutations lie in the βglobin 5′ untranslated region, at positions +22 (21, 22) and +33 (23) downstream of the β-globin promoter transcriptional start site. One patient, heterozygous for a G-A transition at position +22, was described as exhibiting a typical β-thalassemia trait (21). Ho et al. (23) reported four individuals heterozygous for a C-G transversion at position +33. This mutation creates a site for the NlaIV restriction enzyme and permits a semiquantitative analysis of expression from the mutant and normal alleles. All four individuals showed approximately 30% expression from the mutant as compared with the normal allele.
The existence of these β-thalassemia mutations at +22 and +33 prompted us to ask whether they impair function of a downstream element. Here we describe our characterization of these mutations and the identification of a downstream core promoter element (DCE). The DCE extends from +10 to +40, functions in the context of the β-globin TATA box and initiator element, and also functions in a heterologous, TATA-less context. Spacing mutants indicate that a unique geometry of the DCE is important for promoter activity. Lastly, gel shift analysis of DCE mutants indicates reduced affinity for the human TFIID complex. This correlation indicates that TFIID binds to the DCE in a sequence-specific manner and that TFIID, at least in part, is necessary for promoter function via the DCE.
Materials and Methods
In Vitro Transcriptions.
In vitro transcriptions and MEL nuclear extracts were as described (24). Degradation assays were performed as indicated below, and essentially as described (8). Transcription reactions were incubated for 30 min at 30°C. At this time, α-amanitin was added to 2 μg/ml final concentration. Transcription reactions were then stopped at the indicated time points with the addition of stop buffer (24). All quantitations of primer extension products were performed by using a Molecular Dynamics PhosphorImager. All transcriptions were done at least twice with separate nuclear extracts to ensure reproducibility.
Agarose/TFIID Gel Shifts.
Agarose gel shifts of TFIID were done as described (12, 25). Purified HA-tagged TFIID and rTFIIA (26) were incubated with either wild-type or mutant β-globin promoter 32P-labeled probes. These 210-bp probes were generated by PCR using a 32P end-labeled primer. Thus, all probes had the same specific-activity. Binding conditions and electrophoresis were as described (12). Gel shifts were done at least three times to ensure reproducibility. Regression analysis of the transcriptional data and TFIID binding data were done by using Microsoft excel. Transcriptional data for the plot was derived from Figs. 2 and 5. TFIID binding data are derived from Fig. 6 A and B, with the exception of the +37/39 mutant (data not shown).
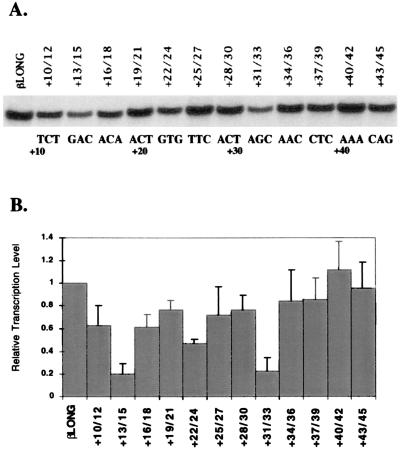
Figure 2.
Scanning mutagenesis of the region downstream of the β-globin initiator element indicates the presence of a downstream core promoter element. (A) Representative in vitro transcriptional analysis of triplet mutations from +10 to +45 of the β-globin promoter. The sequence of the wild-type β-globin downstream region is indicated underneath the autoradiograph. (B) Graphical presentation of the scanning mutagenesis. Bars indicate the mean of three experiments with standard deviations indicated by error bars.
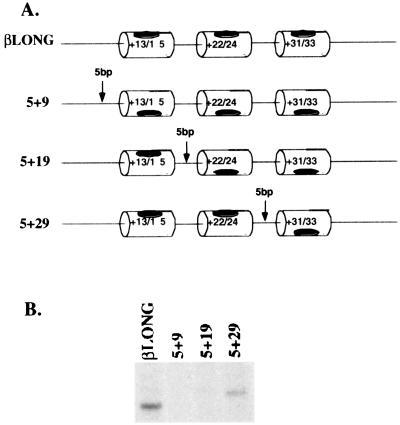
Figure 5.
Five base pair insertions between DCE subelements severely disrupt β-globin transcription in vitro. (A) Schematic diagram showing the design of the spacing mutants. βLONG is the wild-type β-globin promoter. The three subelements are as indicated using a black patch to indicate their orientation relative to each other. The arrows indicate the position of the 5-bp insertion. This position is also indicated in the name of each template (i.e., the 5 + 9 mutant has a 5-bp insertion starting at position + 9 of the wild type promoter). (B) In vitro transcriptional analysis of the spacing mutants. Wild-type and mutant promoter templates were incubated with a MEL nuclear extract. The resulting RNA product was detected by primer extension analysis, was run on an 8% sequencing gel, and was autoradiographed.
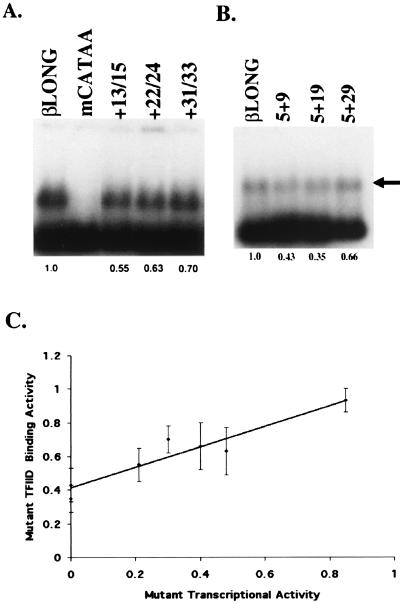
Figure 6.
The TFIID complex shows a reduced affinity for DCE mutants; a linear correlation exists between DCE mutant transcriptional activity and TFIID binding activity. Binding of DCE mutants (A) or the spacing mutants (B) to TFIID was assayed by agarose gel electrophoresis using rTFIIA and immunopurified HA-tagged HeLa TFIID. The arrow indicates the position of the rTFIIA/TFIID complex. Free probe runs at the bottom of the gel. Probes extend from −110 to +100 of the β-globin promoter and were PCR amplified by using a 32P-labeled −110 primer to ensure that all probes had the same specific activity. Quantitation under each lane is relative to the wild-type βLONG TFIIA/TFIID complex and represents the mean of three to five experiments. (C) Regression analysis of the relationship between the transcriptional activity of the DCE mutants (+13/15, +22/24, +31/33, and +37/39 in Fig. 2), and the spacing mutants (Fig. 5), and their respective abilities to bind TFIID (Fig. 6 A and B).
Template Construction.
β-globin promoter templates and mutants were prepared as described (24). The scanning mutagenesis templates in Fig. 2 were constructed by using a similar strategy as in (24). The wild-type to mutant conversions are as follows: +10/12 TCT to AAA, +13/15 GAC to TTT, +16/18 ACA to TTT, +19/21 ACT to GGG, +22/24 GTG to CCC, +25/27 TTC to AAA, +28/30 ACT to GGG, +31/33 AGC to TTT, +34/36 AAC to TTT, +37/39 CTC to GGG, +40/42 AAA to GGG, +43/45 CAG to TTT. Spacing mutants were assembled in a similar manner but with the insertion of a five base pair sequence (TTTAA), starting at one of three wild-type promoter sequence positions: +9, +19, or +29. The Sp1 templates were constructed by inserting a double-stranded oligomer into the SmaI site of pSp1 (2, 24). Sp1/β-globin Inr (βInr) has been described in ref. 24. The Sp1/βInrDCE consists of β-globin promoter sequence from −8 to +40. The Sp1/βInr DCE+22 template contains the +22 β-thalassemia point mutation (21, 22). The Sp1/2,3DCE template contains the CA to GG double point mutation at positions +2 and +3 (24).
Results
β-Thalassemia Mutations at +22 and +33 Are β-Globin Promoter Defects.
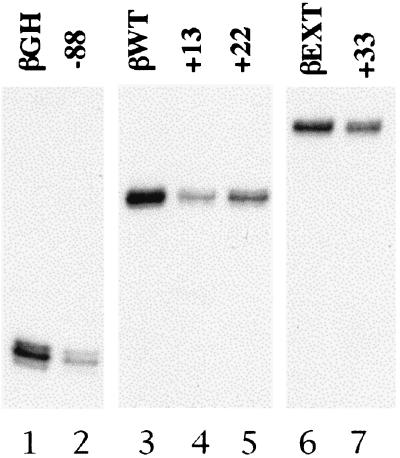
An advantage of studying mutations found in human β-thalassemia is that their physiological significance is established by the in vivo phenotype. The first experiments therefore assayed the effects of these mutations in vitro. We incorporated the +22 and +33 β-thalassemia mutations (21–23) into two β-globin promoter templates for in vitro transcription assays. We also incorporated a point mutation at +13 that showed a 40% reduction in a transient transfection reporter assay in cells (27). We compared the level of expression from these mutant promoters to a fourth mutation serving as a positive control. This template contains a β-thalassemia point mutation at −88 in the proximal CACC box (28). The CACC box binds the erythroid-specific transcription factor EKLF, which is required for βglobin promoter activity (29–31). Expression from the −88 mutant promoter in vitro is 40% of wild-type activity (Fig. 1, lanes 1 and 2; Table 1). Expression levels of the +13, +22, and +33 mutants are at 42, 57, and 50% of wild-type promoter activity, respectively (Fig. 1, lanes 3–5 and 6 and 7; Table 1). The approximately two-fold reduction is consistent with mild β-thalassemia (21–23). Thus, the two β-thalassemia mutations at +22 and +33, and a third mutation at +13, affect transcription of the β-globin promoter in vitro. Furthermore, these mutations suggest the existence of an additional core promoter element(s) downstream of the β-globin initiator element.
Figure 1.
β-Thalassemia point mutations downstream of the transcriptional start site decrease transcription from the human β-globin promoter in vitro. Lanes 1–7 are primer extension assays of in vitro transcriptions using various β-globin promoter templates. The mutations are indicated above each lane. The templates in lanes 1 and 2, 3–5, and 6 and 7 are based on three wild-type promoter templates, βGH, βWT, and βEXT, extending to +18, +35, and +45, respectively.
Table 1.
Relative transcription levels of templates in Fig. 1
| Templates | Relative transcription levels |
|---|---|
| βGH | 1.0 |
| −88 | 0.42 (2) |
| βWT | 1.0 |
| +13 | 0.42 ± 0.07 |
| +22 | 0.58 ± 0.22 |
| βEXT | 1.0 |
| +33 | 0.46 ± 0.12 |
The Human β-Globin Promoter Contains a Core Promoter Element Downstream of the Start Site of Transcription.
To more accurately define the region downstream of the start site, we constructed β-globin promoter templates containing three base pair substitutions (see Materials and Methods). For example, the +10/12 template has mutations at positions +10, +11, and +12. Fig. 2A shows a representative assay, and Fig. 2B provides the quantitation from three experiments. Transcription is reduced by most of the mutations in the region from +10 to +45, as compared with the wild-type βLONG template, with the exception of +40/42. The distribution of the effects of the mutations is striking in that the maximal effects occur at 10 base pair intervals, equivalent to one helical turn of the DNA. The most drastic effects are at +13/15, +22/24, and +31/33, coinciding with the positions of the two β-thalassemia mutations at +22 and +33. The scanning mutagenesis of the region from +10 to +45 revealed the existence of a functional downstream region that we refer to as the DCE (downstream core element) and refer to the three affected regions at +13/15, +22/24, and +31/33 as DCE subelements.
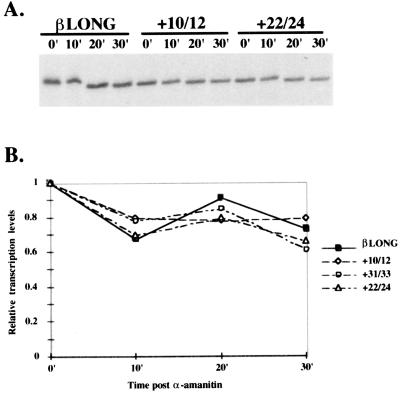
It was possible that the effects observed were not caused by changes in transcription. Instead, mutations in the DCE might affect RNA stability in vitro because they are present in the RNA transcripts. Therefore, to ask whether these mutations were increasing the degradation of the transcribed RNA, we modified the in vitro transcription assay. Transcription was permitted for 30 min, and then α-amanitin was added to prevent any further transcription. Transcription products were assayed at various time points (8). As shown in Fig. 3, no accelerated degradation of +10/12, +22/24, or +31/33 transcripts was observed. In addition, the +13/15 subelement shows similar kinetics (data not shown). Thus, the mutations in the DCE affect the transcriptional activity of the β-globin promoter rather than transcript stability.
Figure 3.
Degradation control assays indicate that DCE mutants do not significantly affect RNA stability. (A) Results of an in vitro transcription assay to analyze RNA stability of various DCE mutants at times after the addition of α-amanitin. (B) Graph of primer extension signal intensity relative to the 0 min data point. Each mutant template was normalized to a relative transcription level of 1. The data points in the graph are the mean of three experiments.
The DCE Functions in a Heterologous Context and in the Absence of a TATA Box.
To further characterize the DCE and the promoter context in which it might function, we placed the DCE in a heterologous context. We based our analysis on the Sp1/βInr template, which contains the β-globin Inr element (from −8 to +13 of the β-globin promoter) downstream of six Sp1 sites. We previously showed that this template is accurately transcribed in vitro (24) (Fig. 4, lane 1). Using the same Sp1 background plasmid (2), we extended the region downstream of the initiator element to +40 (Fig. 4, lane 2). The Sp1/βInr DCE template showed a higher transcription level than the shorter Sp1/βInr template (Fig. 4). Thus, the DCE appears to contribute to the strength of a βInr-dependent template. To be certain that the DCE functions in this context, we incorporated the +22 β-thalassemia mutation into the Sp1/βInrDCE template. In this context, this mutation is more deleterious than within the β-globin promoter, as transcription was reduced, in two experiments, 3- and 8-fold (Fig. 4, lane 3). We also incorporated a double point mutation at positions +2 and +3 in the βInr to ask whether the DCE functions in the absence of the βInr. This mutation completely destroyed βInr activity when assayed in the context of the shorter Sp1/βInr template (24). As can be seen in Fig. 4, lane 4, the DCE is at best capable of weakly initiating transcription. Curiously, transcription is correctly initiated, suggesting that, at some low level, the DCE can dictate an initiation site. Clearly, however, the DCE can cooperate with the βInr element to increase transcription in a TATA-less context.
Figure 4.
The DCE functions in a heterologous context to increase transcription from a βInr-dependent template. Lane 1 is primer extension analysis of an in vitro transcription assay using the Sp1/βInr described previously (24). The correctly initiated transcript is indicated by the arrow to the left of lane 1 (24). Lane 2 is an in vitro transcription assay using the longer Sp1/βInrDCE template extending to +40. In two experiments, it was expressed 3× and 8× higher than the Sp1/βInr template in lane 1. The template in lane 3 contains the β-thalassemia mutation at +22, and lane 4 contains a double point mutation in the βInr element (20). These templates are otherwise identical to Sp1/βInrDCE in lane 2. The arrow to the right of lane 4 indicates the position of the correctly initiated transcript.
Spacing/Phasing Mutants Severely Affect Transcription from the β-Globin Promoter in Vitro.
One interpretation of the scanning mutagenesis is that the major activity of the DCE is derived from three subelements, at positions +13/15, +22/24, and +31/33. These subelements occur at roughly 10-base pair intervals, which is equivalent to one helical turn of the DNA. To test whether the spacing/phasing of the three subelements is important for β-globin transcription, we made five base pair insertions at three positions in the DCE. The three insertions, at +9, +19, and +29, lie outside of the DCE subelements and serve to rotate one or more subelements 180° relative to elements upstream of the insertion (Fig. 5A). For example, the insertion at +9 (construct 5 + 9) flips the entire DCE relative to the other core promoter elements upstream.
In vitro transcriptions using these three templates (Fig. 5B) show that these mutations have drastic effects on β-globin promoter activity. The 5 + 9 and 5 + 19 insertions completely abrogate transcription (Fig. 5B) whereas the 5 + 29 insertion shows 40% of wild-type activity. Although the transcriptional start site appears to have shifted a short distance upstream, this is attributable to the five-base pair insertion. This experiment suggests that the spacing mutations affect transcriptional activity by disrupting the spacing or geometry between the subelements themselves and/or the upstream core promoter elements.
DCE Mutants Show a Reduced Affinity for the TFIID Complex.
Prior work demonstrated that TFIID footprints regions downstream of the transcriptional start sites of several promoters (9–15). In addition, the well characterized downstream region, the DPE, has been shown to bind the Drosophila histone-like TAFs p42 and p62 (8). These data implicate a role of TFIID in downstream element function and thus led us to assay whether DCE mutations showed a reduced affinity for TFIID.
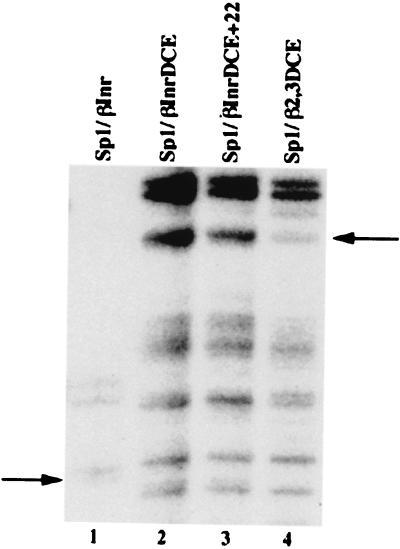
We performed agarose gel shift assays as described (25, 32) by using recombinant TFIIA and HA-tagged HeLa TFIID. βglobin promoter fragments from −110 to +100 were labeled and amplified by PCR. As a positive control, we used a template containing four mutations in the β-globin TATA box (mCATAA). This mutation completely destroys transcription from the β-globin promoter (ref. 24; data not shown) and, as expected, does not bind the rTFIIA/TFIID complex. We assayed the three mutants most severely affected transcriptionally (+13/15, +22/24, and +31/33; see Fig. 2) and another that was mildly affected (+37/39). As seen in Fig. 6A, rTFIIA/TFIID complex binding to the three mutant subelements is reduced (0.55 ± 0.10, 0.63 ± 0.14, and 0.70 ± 0.08, respectively) relative to the wild-type βLONG probe whereas the +37/39 mutant DNA shows only a slight decrease in binding the TFIIA/TFIID complex (0.93 ± 0.07).
We next assayed the spacing mutants in Fig. 5 for TFIID binding by agarose gel shift assays. Fig. 6B shows that TFIID binding to all three spacing mutants is reduced. The 5 + 9 and 5 + 19 spacing mutants show no transcriptional activity and binding activities of 0.43 ± 0.10 and 0.35 ± 0.08, respectively (Fig. 6B). This remaining binding activity can be attributed to the presence of the β-globin TATA box (CATA) and initiator element. The 5 + 29 mutant's transcriptional level is 40% of wild-type βLONG template activity (Fig. 5) and its binding activity 0.66 ± 0.14 (Fig. 6B). These spacing mutants provide a second set of DCE mutants that show both reduced promoter activity and a reduced affinity for TFIID.
To establish whether any correlation existed between the transcriptional activity of the various DCE mutants and TFIID binding, transcriptional activity (Figs. 2 and 5) was plotted versus TFIID binding activity (Fig. 6 A and B). The regression analysis shows a precise linear correlation between transcriptional activity and TFIID binding (Fig. 6C). The P value for the analysis is 0.001 and shows that the correlation is highly significant. These data indicate that TFIID interacts with the DCE sequence-specifically and that there is a linear correlation between a mutant's transcriptional activity and its affinity for TFIID.
Discussion
We present here the detailed characterization of a functional downstream region within a TATA- and Inr-dependent promoter, which we refer to as the DCE, for downstream core element. The existence of the DCE was suggested by two mutations at +22 and +33 found in β-thalassemia patients, and a third point mutation at +13 (21–23, 27). Incorporation of the β-thalassemia mutations into promoter constructs led to decreases in transcription in vitro (Fig. 1). This interpretation was supported by scanning mutagenesis of the DNA downstream of the transcriptional start site, which revealed that positions +13, +22, and +33 were part of a larger functional element with maximal decreases in transcription at positions +13–15, +22–24, and +31–33 (Fig. 2).
The DCE is functionally unique as compared with the only other characterized downstream element, the DPE (7, 8). The DPE is centered around +30 and requires a minimum of four base pairs to function (7, 8). The DCE, however, extends over 30 base pairs and consists of three regions, or subelements, as defined by scanning mutagenesis (Fig. 2). In addition, the core promoter context within which the DCE and DPE function differs. The DPE is redundant in the context of a TATA-containing promoter (7). DPE function is only observed when coupled to an initiator region. In contrast, the DCE functions in its natural context in the presence of both a TATA box (albeit a nonconsensus TATA box, CATA) and the βInr element (24) (Fig. 2) and in a heterologous, TATA-less context (Fig. 4). Hence, the DCE appears to be much more flexible with regard to the context in which it can function.
Prior evidence indicates that the TFIID complex footprints and can be crosslinked to DNA downstream of the transcriptional start site of several promoters (7–15, 17, 33). These data prompted us to test for a disruption of TFIID binding to various DCE mutations (Fig. 6). We began with the hypothesis that the β-globin core promoter represents five TFIID binding sites: the CATA box, the βInr element, and the three DCE subelements. Second, considering the mild effects of the DCE scanning mutagenesis (Fig. 2), it was presumed that any loss of TFIID binding would be of a similar magnitude. TFIID gel shift analysis of the three DCE subelements (+13/15, +22/24, and +31/33; Fig. 2) indicated that this was indeed the case (Fig. 6A). Because the two spacing mutants, 5 + 9 and 5 + 19, showed the most severe transcriptional defect, we expected those mutants to show a further reduction in TFIID binding. From Fig. 6B, it is apparent that the 5 + 9 and 5 + 19 spacing mutants bind TFIID at approximately 40% of the wild-type promoter. Residual binding activity is attributable to TFIID binding to the CATA box and βInr element. Regression analysis of the relationship between the transcriptional activity of the DCE mutants and their TFIID binding activity revealed a statistically significant, linear correlation between the two (Fig. 6C). The analysis also suggests that, for any DCE mutant having no transcriptional activity, a TFIID binding activity of 40% of wild-type would be anticipated. This is consistent with our interpretation that residual binding is attributable to the CATA box and βInr element. These data are consistent with work by others showing that TFIID binding is required for the function of the downstream elements in the hsp70 promoter, the adenovirus MLP, and the DPE (7, 8, 14, 15, 19). The residual 40% binding of TFIID to the transcriptionally inactive 5 + 9 and 5 + 19 mutants is similar to the 35% binding of TFIID to a TATA-Inr template that was not transcribed because of a mutated Inr element (12). The existence of multiple subelements is also consistent with data showing a progressive decrease of TFIID binding to a series of deletions 3′ to the Drosophila hsp70 start site (34).
We suggest that the DCE, along with the CATA box and βInr, serves as a TFIID binding site. This combination of elements serves to maximize TFIID stability on the promoter, thereby contributing to promoter strength. Scanning mutagenesis suggests that these are sequence-specific contacts. Maximal decreases in transcription occur at approximately 10-base pair intervals, equivalent to one helical turn of the DNA. Thus, the three subelements at +13/15, +22/24, and +31/33 may lie on the same “side” of the helix. This spacing is reminiscent of the nucleosome structure in which the DNA makes contacts every 10 base pairs as it wraps around a histone H2A/H2B and/or H3/H4 heterodimer (35). As such, these DCE contacts may be mediated by a histone-fold containing TAF (36–38). In addition, the spacing mutants (Fig. 6) imply that the geometry between the DCE and the CATA box/βInr is essential for promoter function. We suggest that there is a distinct core promoter architecture consisting of the DCE, βInr element, and β-globin CATA box and TFIID. This architecture is partially dictated by sequence-specific contacts in the DCE. Although these DNA contacts are sequence-specific, it is possible that they may not reflect a protein/base interaction. Instead, a particular sequence may induce a DNA structure (for example, a bend) that is recognized by TFIID. Possible proteins mediating these sequence-specific contacts include the histone-fold TAFs, TAFII250, and TAFII150, all of which have been shown to interact with downstream element DNA (7, 8, 16, 17, 19)
The existence of a downstream element in a promoter containing a TATA box and initiator is perhaps counter to a conventional view of core promoters. Initiators and downstream elements help to account for the activity of TATA-less promoters, yet the β-globin promoter already has a TATA box and Inr. One explanation is that the DCE simply contributes to the strength of the β-globin promoter. A requirement for the DCE may reflect the relative weakness of the β-globin nonconsensus TATA (CATA) box and weaker Inr element (24). In support of this, preliminary data indicates that DCE mutants are defective in the formation of preinitiation complexes and are slower to reinitiate (B.A.L., unpublished data). TATA box mutants exhibit identical properties of preinitiation complex formation and reinitiation (39), thereby suggesting a similar role of TFIID in DCE function. It will be interesting to ascertain whether other downstream regions define other types of downstream elements or whether they fall into the DCE or DPE class of downstream elements. A second explanation is one of specificity. The DCE and DPE represent two classes of downstream elements both in terms of sequence and function. This is in contrast to the apparently ubiquitous TATA box and initiator elements and raises the question of whether downstream elements confer a specificity to the promoter that is not supplied by other core promoter elements. Consistent with this is data showing that hTAFII250 (and the yeast homologue TAFII145) and hTAFII150 play a role in core promoter selectivity and downstream element function (40, 41).
Several conclusions emerge from this study. First, seemingly small changes in the transcription levels of a promoter may have significant in vivo effects. Second, downstream elements differ with respect to sequence and functional properties. These differences may have important consequences in promoter regulation and specificity. Third, TFIID binding to the DCE may account for DCE function. It will be important to show how TFIID can mediate both DPE and DCE function. Finally, this work elucidates the emerging complexity and significance of core promoters and their role in vivo.
Acknowledgments
We thank David Gilmour, Jeff Parvin, Anne Deconinck, Richard Wells, and other members of the Orkin lab for critical reading of the manuscript and many helpful discussions, Sam Katz for regression analysis, and Paul Liebermann for advice on TFIID gel shift assays. B.A.L. especially thanks Danny Reinberg for his interest and generous supply of resources. B.A.L. was, in part, supported by a fellowship from the Cooley's Anemia Foundation. S.H.O. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- TFIID
transcription factor IID
- DCE
downstream core element
- DPE
downstream promoter element
- Inr
initiator element
- βInr
β-globin initiator element
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.120181197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.120181197
References
- 1.Smale S T. In: Transcription: Mechanisms and Regulation. Conaway R C, Conaway J W, editors. Vol. 3. New York: Raven; 1994. [Google Scholar]
- 2.Smale S T, Baltimore D. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 3.Weis L, Reinberg D. FASEB J. 1992;6:3300–3309. doi: 10.1096/fasebj.6.14.1426767. [DOI] [PubMed] [Google Scholar]
- 4.Lewis E D, Manley J L. Mol Cell Biol. 1985;5:2433–2442. doi: 10.1128/mcb.5.9.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakatani Y, Horikoshi M, Brenner M, Yamamoto T, Besnard F, Roeder R G, Freese E. Nature (London) 1990;348:86–88. doi: 10.1038/348086a0. [DOI] [PubMed] [Google Scholar]
- 6.Martinez E, Chiang C M, Ge H, Roeder R G. EMBO J. 1994;13:3115–3126. doi: 10.1002/j.1460-2075.1994.tb06610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke T W, Kadonaga J T. Genes Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- 8.Burke T W, Kadonaga J T. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sawadogo M, Roeder R G. Cell. 1985;43:165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- 10.Nakajima N, Horikoshi M, Roeder R G. Mol Cell Biol. 1988;8:4028–4040. doi: 10.1128/mcb.8.10.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Q, Lieberman P M, Boyer T G, Berk A J. Genes Dev. 1992;6:1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]
- 12.Kaufmann J, Smale S T. Genes Dev. 1994;8:821–829. doi: 10.1101/gad.8.7.821. [DOI] [PubMed] [Google Scholar]
- 13.Oelgeschlager T, Chiang C-M, Roeder R G. Nature (London) 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 14.Purnell B A, Gilmour D S. Mol Cell Biol. 1993;13:2593–2603. doi: 10.1128/mcb.13.4.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purnell B A, Emanuel P A, Gilmour D S. Genes Dev. 1994;8:830–842. doi: 10.1101/gad.8.7.830. [DOI] [PubMed] [Google Scholar]
- 16.Verrijzer C P, Yokomori K, Chen J L, Tjian R. Science. 1994;264:933–941. doi: 10.1126/science.8178153. [DOI] [PubMed] [Google Scholar]
- 17.Sypes M A, Gilmour D S. Nucleic Acids Res. 1994;22:807–814. doi: 10.1093/nar/22.5.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moncollin V, Roy R D W, Egly J-M. In: Transcription: Mechanisms and Regulation. Conaway R C, Conaway J W, editors. Vol. 3. New York: Raven; 1994. [Google Scholar]
- 19.Verrijzer C P, Chen J, Yokomori K, Tjian R. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 20.McDonagh K T, Nienhuis A W. In: Hematology of Infancy and Childhood. Nathan D G, Oski F A, editors. Philadelphia: Saunders; 1993. [Google Scholar]
- 21.Cai S P, Eng B, Francombe W H, Olivieri N F, Kendall A G, Waye J S, Chui D H. Blood. 1992;79:1342–1346. [PubMed] [Google Scholar]
- 22.Oner R, Agarwal S, Dimovski A J, Efremov G D, Petkov G H, Altay C, Gurgey A, Huisman T H J. Hemoglobin. 1991;15:67–76. doi: 10.3109/03630269109072485. [DOI] [PubMed] [Google Scholar]
- 23.Ho P J, Rochette J, Fisher C A, Wonke B, Jarvis M K, Yardumian A, Thein S L. Blood. 1996;87:1170–1178. [PubMed] [Google Scholar]
- 24.Lewis B, Orkin S H. J Biol Chem. 1995;270:28139–28144. doi: 10.1074/jbc.270.47.28139. [DOI] [PubMed] [Google Scholar]
- 25.Lieberman P, Berk A J. Genes Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 26.Maldonado E, Drapkin R, Reinberg D. Methods Enzymol. 1996;274:72–100. doi: 10.1016/s0076-6879(96)74009-0. [DOI] [PubMed] [Google Scholar]
- 27.Cowie A, Myers R M. Mol Cell Biol. 1988;8:3122–3128. doi: 10.1128/mcb.8.8.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orkin S H, Antonarakis S E, Kazazian H H., Jr J Biol Chem. 1984;259:8679–8681. [PubMed] [Google Scholar]
- 29.Miller I J, Bieker J J. Mol Cell Bol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Nature (London) 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 31.Perkins A C, Sharpe A H, Orkin S H. Nature (London) 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 32.Kaufmann J, Verrijzer C P, Shao J, Smale S T. Genes Dev. 1996;10:873–886. doi: 10.1101/gad.10.7.873. [DOI] [PubMed] [Google Scholar]
- 33.Nakatani Y, Brenner M, Freese E. Proc Natl Acad Sci. 1990;87:4289–4293. doi: 10.1073/pnas.87.11.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emanuel P A, Gilmour D S. Proc Natl Acad Sci USA. 1993;90:8449–8453. doi: 10.1073/pnas.90.18.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Nature (London) 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 36.Xie X, Kokubo T, Cohen S, Mirza U A, Hoffmann A, Chait B T, Roeder R G, Nakatani Y, Burley S K. Nature (London) 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- 37.Hoffman A, Chiang C-M, Oelgeschlager T, Xie X, Burley S K, Nakatani Y, Roeder R G. Nature (London) 1996;380:356–359. doi: 10.1038/380356a0. [DOI] [PubMed] [Google Scholar]
- 38.Burley S K, Roeder R G. Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 39.Yean D, Gralla J. Mol Cell Biol. 1997;17:3809–3816. doi: 10.1128/mcb.17.7.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen W-C, Green M R. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- 41.Verrijzer C P, Chen J L, Yokomori K, Tjian R. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]