Abstract
Microbial dynamics during processing and ripening of traditional cheeses such as registered designation of origin Salers cheese, an artisanal cheese produced in France, play an important role in the elaboration of sensory qualities. The aim of the present study was to obtain a picture of the dynamics of the microbial ecosystem of RDO Salers cheese by using culture-independent methods. This included DNA extraction, PCR, and single-strand conformation polymorphism (SSCP) analysis. Bacterial and high-GC% gram-positive bacterial primers were used to amplify V2 or V3 regions of the 16S rRNA gene. SSCP patterns revealed changes during the manufacturing of the cheese. Patterns of the ecosystems of cheeses that were provided by three farmers were also quite different. Cloning and sequencing of the 16S rRNA gene revealed sequences related to lactic acid bacteria (Lactococcus lactis, Streptococcus thermophilus, Enterococcus faecium, Leuconostoc mesenteroides, Leuconostoc pseudomesenteroides, Lactobacillus plantarum, and Lactobacillus pentosus), which were predominant during manufacturing and ripening. Bacteria belonging to the high-GC% gram-positive group (essentially corynebacteria) were found by using specific primers. The present molecular approach can effectively describe the ecosystem of artisanal dairy products.
The typical sensorial qualities of traditional cheese ultimately depend on several factors, including traditional cheese-making practices, feeding of dairy cows, and the dynamics of microbial communities. The qualitative and quantitative composition of the microbial flora, its evolution, and its activity during ripening play an important role in the development of hygienic and sensorial qualities. In order to better understand the functions of the microbial community, a full description of the microbial ecosystem is required. Classically, this has been addressed by enumerating members of certain microbial groups by using various culture media, followed by identification of a number of dominant isolates by phenotypic tests (16, 18, 33) or molecular techniques such as ribotyping (21), random amplified polymorphic DNA analyses, and sequencing (4, 27). However, cultivation-dependent approaches do not necessarily provide reliable information on the composition of entire microbial communities. It is therefore difficult to assess the significance of cultured microorganisms in microbial ecosystems because of the disparity between culturable and in situ diversity. Indeed, microbial communities may contain viable but nonculturable bacteria and also bacteria that would be culturable if appropriate medium had been developed. Thus, in order to study interactions between microorganisms, it is important to study the ecosystem without dissociating it. It has been shown that a dual approach, e.g., inventory by using a cultivation-dependent method and direct recovery of 16S rRNA gene sequences, can give a more objective view of the composition of a complex ecosystem than by using either approach alone (13, 19). Various PCR-based molecular typing methods have been developed for the analysis of communities: denaturating gradient gel electrophoresis (DGGE) (7, 10), temperature gradient gel electrophoresis (TGGE) (11, 12, 36), temporal temperature gel electrophoresis (TTGE) (28), terminal-restriction fragment length polymorphism (26, 29), and single-strand conformation polymorphism (SSCP). These methods provide a rapid fingerprint of a complex microbial community without cultivation. Because of the use of universal primers, SSCP and the other molecular methods can be applied without any a priori information on the species and then can give a more objective view of the microbial community. SSCP has been applied to study microbial communities in water (23), in the compost of organic agricultural substrate (30), and in anaerobic digestors (22, 37). This technique has also been used in clinical microbiology to rapidly differentiate bacteria from blood culture (34).
These direct molecular techniques have been applied to the studies of food microbiology (2, 5-7). The aim of the present study was to apply SSCP analysis to describe bacterial community dynamics during the production of registered designation of origin (RDO) Salers cheese. The RDO Salers cheese is a French farmhouse cheese produced from 15 April to 15 October, when cows are grazing. Salers cheese is produced exclusively with raw milk stored in a wooden container (the “gerle”), allowing a natural microbial flora enrichment. Consequently, its microbial flora may be high and diverse.
MATERIALS AND METHODS
Cheese sample.
The cheese samples used in the present study were obtained from three farmers who produce RDO Salers cheese by traditional methods, and the cheeses were chosen because of their sensorial diversity and qualities. Milk was neither heated nor refrigerated after the milking. No starter was added. Ripening conditions were controlled (10°C, 96% humidity). Samples were taken from the raw milk and during manufacture of cheese and ripening and then subjected immediately to classical bacteriological analysis (culture on selective media) or stored at −80°C for further SSCP analysis. Analyses were conducted on the milk and cheeses of the three farmers at 1, 8, 30, 90, and 150 days.
Extraction and purification of total bacterial DNA from cheese.
Four types of DNA extractions from curd were performed. The guanidine method has been described by Godon et al. (17). The procedure with proteolytic enzyme was established by Y. Pagot (unpublished data). The procedure with lytic enzymes has been described by Ampe et al. (2), except that a mechanical lysis with zirconium beads was added. The phenol method was derived from Delbès et al. (9): a 1-g cheese sample was resuspended in 1 ml of 4 M guanidine thiocyanate-0.1 M Tris (pH 7.5) and 125 μl of 10% N-lauroylsarcosine by grinding using a reciprocating shaker (MM2000; Kurt Retsch). A 2-ml tube containing 250 μl of the suspension was filled with 200 mg of zirconium beads, 100 μl of 20% sodium dodecyl sulfate solution, 400 μl of 0.1 M phosphate buffer (pH 8), 400 μl of 50 mM sodium acetate-10 mM EDTA (pH 6), and 400 μl of phenol-chloroform-isoamyl alcohol (25:24:1; pH 8). The tube was heated at 80°C for 3 min and shaken for 2 min in a reciprocating shaker. These two steps, shaking and heating, were repeated a second time. Two washing steps were then performed: the first with phenol-chloroform-isoamyl alcohol (25:24:1) and the second with chloroform-isoamyl alcohol (24:1). Nucleic acids were then precipitated with one-tenth of the volume of 3 M sodium acetate and 2 volumes of ethanol. The tube was gently mixed and stored at −20°C overnight. After centrifugation at 14,000 × g for 10 min, the nucleic acids pellet was washed with 70% ethanol, dried, and resuspended in 100 μl of water. DNA was then treated with 4 μl of RNase (2 mg/ml).
Amplification of DNA extracted from curd for 16S rRNA gene cloning.
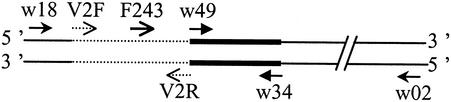
Amplification of 16S rRNA gene was carried out with either the universal primers w02 and w18 (complete gene) or with F243 and w34 for a selective amplification of high GC% gram-positive bacteria partial 16S rRNA gene (Table 1 and Fig. 1). Each reaction tube contained 10 mM Tris-HCl (pH 8.5), 25 mM KCl, 5 mM (NH4)2SO4, 2 mM MgSO4, 0.1 μM concentrations of each primer, 2 μl of purified DNA, 0.2 mM concentrations of each deoxynucleoside triphosphate, and 1 U of Pwo polymerase (Roche) and was then adjusted to a total volume of 50 μl. The reaction mixture was placed in a 9700 thermocycler (Perkin-Elmer). After an initial denaturation at 96°C for 3 min, 20 temperature cycles were performed at 96°C for 1 min, either 50°C for 1 min (primers w02 and w18) or 59.5°C for 1 min (primers F243 and w34), and 72°C for 1 min, followed by a final extension step of 72°C for 7 min. The products were electrophoresed on a 0.8% agarose gel and viewed by ethidium bromide staining. PCR products were ligated in pCR4Blunt-TOPO and transformed into Escherichia coli TOP10 OneShot as specified by the manufacturer (Invitrogen). Plasmid inserts were amplified by PCR with universal plasmid primers T3 and T7 (Table 1), as specified by the manufacturer (Invitrogen).
TABLE 1.
PCR primers used in this study
| Primer | Sequence | Targeta | Source or reference |
|---|---|---|---|
| w02 | GNTACCTTGTTACGACTT | SSU rRNA bacteria | 17 |
| w18 | GAGTTTGATCMTGGCTCAG | SSU rRNA bacteria | 17 |
| w34 | TTACCGCGGCGTGCTGGCAC | SSU rRNA bacteria | 37 |
| w49 | ACGGTCCAGACTCCTACGGG | SSU rRNA bacteria | 9 |
| F243 | GGATGAGCCCGCGGCCTA | SSU rRNA high-GC% bacteria | 20 |
| T3 | ATTAACCCTCACTAAAGGGA | Plasmid | |
| T7 | TAATACGACTCACTATAGGG | Plasmid | |
| V2F | GGCGAACGGGTGAGTAA | SSU rRNA bacteria | This study |
| V2R | ACTGCTGCCTCCCGTAG | SSU rRNA bacteria | This study |
SSU, small subunit.
FIG. 1.
Primer positions on 16S RNA gene. Dotted line, V2 region; heavy line, V3 region.
SSCP-PCR amplification.
The target DNAs amplified were either the variable region V2 or the variable region V3 of the 16S rRNA gene, which correspond to fragments of 255 and 205 bp, respectively, in the E. coli 16S rRNA gene. The PCR amplifications were made on DNA extracted from curd or on PCR products (cloned insert or product of the high-GC% gram-positive selective amplification). The primers, corresponding to conserved sequences bordering the variable regions were V2R and V2F for the V2 region and w49 and w34 for the V3 region (Table 1 and Fig. 1). Primers w34, V2R, and V2F were labeled with 5′-fluorescein phosphoramidite: (i) NED for w34 and V2R and (ii) hexachloro derivative of fluorescein (HEX) for V2F. All of the primers were synthesized by Applied Biosystems.
For PCR amplification on genomic DNA, the PCR mixture contained 50 mM biocine, 115 mM potassium acetate, 8% glycerol (wt/vol), 0.5 μM concentrations of each primer, 200 μM concentrations of each deoxynucleoside triphosphate, 1 μl of cheese or bacterial genomic DNA, and 2 U of Tth polymerase (Applied Biosystems), all adjusted to a total volume of 25 μl with water. The tubes were placed in a thermocycler (model 9700; Perkin-Elmer). An initial denaturation at 95°C for 3 min preceded 20 or 25 temperature cycles of 45 s at 95°C, 30 s at either 55°C (primers V2F and V2R) or 61°C (primers w34 and w49), and 45 s at 68°C, followed by a final extension step of 10 min at 68°C.
For PCR amplification on PCR product, the PCR mixture was the same, and 0.5 μl of PCR product was used as a template. The conditions of PCR amplification were identical except that 10 cycles were realized.
After amplification, 2.5 μl of the amplified product was run on a 0.8% agarose gel in 1× TBE (89 mM Tris base, 89 mM borate, 2 mM EDTA). DNA bands were detected by ethidium bromide staining and visualized by using UV light. PCR products were purified with StrataPrep PCR purification kit as specified by the manufacturer (Stratagene).
SSCP electrophoresis.
The SSCP-PCR products were diluted 2- to 25-fold depending on the band intensity on the 0.8% agarose gel. A 1-μl aliquot of the SSCP-PCR product was mixed with 18.5 μl of deionized formamide (Applied Biosystems) and 0.5 μl of internal DNA molecular weight standard Genescan-400HD ROX (Applied Biosystems). Samples were denatured at 95°C for 3 min and immediately cooled on ice; the DNA fragments adopt a secondary structure that depends only on the sequence of the fragment. SSCP capillary electrophoresis was performed on an ABI Prism 310 genetic analyzer (Applied Biosystems). The genetic analyzer was set up in accordance with the manufacturer's instructions. The nondenaturing polymer matrix used was 5.6% GeneScan polymer (Applied Biosystems)-10% glycerol-1× TBE. The buffer was 1× TBE-10% glycerol. The migration of the DNA fragments depends on their conformation which one depends on their sequence. A 47-cm capillary with an inner diameter of 50 μm was installed. Electrophoresis conditions were set on the instrument as follows: an injection time of 5 s, an injection voltage of 15 kV, an electrophoresis voltage of 12 kV, a syringe pump time of 300 s, a constant temperature of 32°C, and 30 min of collection time. A laser detected the DNA labeled with 5′-fluorescein phosphoramidite. The signal was automatically analyzed by using GeneScan Analysis software (Applied Biosystems). The elution time was expressed in scans (unit of the software), and the height and area of the peaks were determined. To analyze the different profiles, the ratio (i.e., the area of one peak to the sum of the areas of all of the peaks) was calculated.
Determination of the SSCP peaks corresponding to clones.
In order to assign peaks on SSCP patterns, the V3 and V2 regions of the 16S rRNA gene clone library were analyzed by SSCP. SSCP-PCR of the clones was performed, and the resulting peaks were compared to the bacterial pattern of the cheese.
Sequencing of 16S rRNA gene.
In order to assign the peak from an SSCP pattern to a 16S rRNA gene sequence, the 450 bp of the 5′ and 3′ ends of the 16S rRNA gene of 57 representative clones were sequenced. For the high-GC% gram-positive clones, only the V3 region was sequenced (22 clones). PCR products corresponding to the clones insert were sequenced by using the dye-terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase FS (Applied Biosystems) and either w18, w02, or F243 primers (Table 1). The nucleotide sequences were determined by automated DNA sequencing achieved by using an ABI Prism 310 genetic analyzer (Applied Biosystems) according to manufacturer's instructions. The results were analyzed with sequencing analysis software (Applied Biosystems).
Analysis of the sequence data.
Each sequence obtained was compared to sequences available in databases (GenBank and the Ribosomal Database Project [RDP]) by using BLASTN and Sequence_Match programs (1, 25). The CHECK_CHIMERA command (25) of the RDP facilities was used to detect chimeric sequences.
RESULTS
Comparison of the DNA extraction methods and the PCR conditions used.
The four different methods of DNA extraction were applied to a 1-day curd sample. The resulting DNA was amplified with 20 or 25 amplification cycles by the primer pairs w34 and w49, w34 and F243, and V2F and V2R. The profiles obtained from the four different templates were compared (results not shown). Based on the microbial diversity of the different patterns obtained, the phenol extraction method and 25 amplification cycles were retained.
SSCP patterns obtained with the different sets of primers: culture-independent identification of most of the peaks.
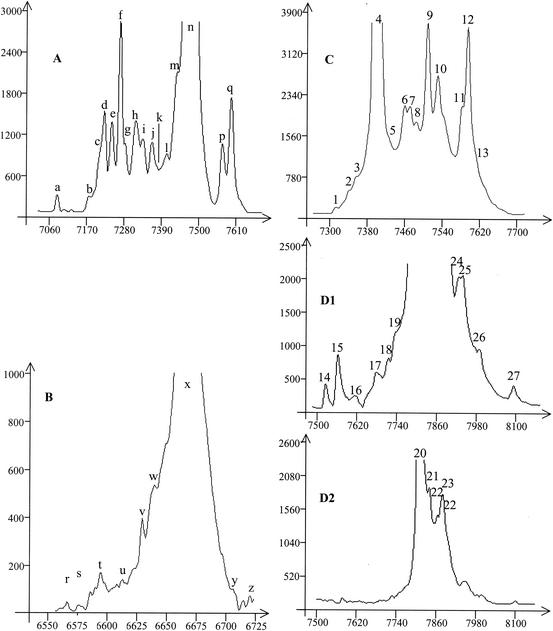
As illustrated for the curd of farmer C in Fig. 2, each DNA sample extracted from milk or cheese samples was amplified with three sets of primers, and four SSCP patterns were obtained. The pattern obtained after amplification of the V3 region with one primer labeled is shown in Fig. 2A. The patterns obtained after amplification of the V2 region with the two primers that were labeled are shown in Fig. 2C (V2F labeled with NED) and Fig. 2D1 and D2 (V2R labeled with HEX). The specific amplification of high-GC% bacterial DNA gave the pattern shown in Fig. 2B.
FIG. 2.
SSCP analysis of PCR-amplified 16S rRNA gene fragments from bacterial communities of a curd sample. (A) V3 region; (B) V2 region, forward primer labeled with NED; (C) high GC% gram-positive community; (D) V2 region, reverse primer labeled with HEX. D1 shows the SSCP-PCR product that was not diluted, and D2 shows the SSCP-PCR product that was diluted (1/5) in sterile water. y axis, fluorescence; x axis, elution in scans (unit of GeneScan software). The positions and labeling of peaks discussed in Tables 2 to 4 and in the text are indicated.
A clone library of 16S rRNA genes of the DNA extracted from the curd provided by farmer C was constructed to identify most of the peaks of the different patterns (Fig. 2 and Table 2). Most of the clones were reliably identified (homology with sequences present in the databases higher than 98%). The highest peak of the V3 pattern, peak n, originated from different sequences of lactic acid bacteria (Lactococcus lactis, Streptococcus thermophilus, Lactobacillus plantarum, and Lactobacillus pentosus) that comigrated. The closest relative corresponding to peak i was Enterococcus faecium. Peak m resulted from the coelution of two sequences, Leuconostoc pseudomesenteroides and Leuconostoc mesenteroides. The closest relative corresponding to peak h was Bacillus thuringiensis. The sequence for peak a was close to Bifidobacterium breve, with 95% homology. Other clones resulted in sequences derived from enterobacteria and corresponded to peaks b to g and peak k. The closest relative corresponding to peak c was a sequence of environmental clone (water and soil) isolated by molecular techniques. For the moment, it is not closely linked with any cultivated species. Peak d originated from a sequence of Citrobacter sp. The sequence of the clone that comigrated with peak k was close to Kingella denitrificans, with 93% homology. Sequences that were different but that comigrated in the same peak were placed in the same group. In this way, the Enterobacteriaceae groups I to IV were established as detailed in Table 3. No clone comigrated with peaks j, l, o, p, and q.
TABLE 2.
Identities of peaks obtained from SSCP analysis of the V3 region of the bacterial community
| Peaka | Relative sequences | % Identityb | GenBank accession no. |
|---|---|---|---|
| a | Bifidobacterium breve | 92 | AB006658 |
| b | Enterobacteriaceae I | See Table 3 | |
| c | Environmental clone | 98 | X85208 |
| d | Citrobacter sp. | 100 | AF025369 |
| e | Enterobacteriaceae II | See Table 3 | |
| f | Enterobacteriaceae III | See Table 3 | |
| g | Enterobacteriaceae IV | See Table 3 | |
| h | Bacillus thuringiensis | 100 | AF172711 |
| i | Enterococcus faecium | 99 | AF070223 |
| k | Kingella denitrificans | 96 | L06166 |
| m | Leuconostoc mesenteroides | 99 | AB023243 |
| m | Leuconostoc pseudomesenteroides | 99 | AB023237 |
| n | Lactococcus lactis | 100 | X64887 |
| n | Lactobacillus plantarum | 100 | M58827 |
| n | Lactobacillus pentosus | 99 | D79211 |
| n | Streptococcus thermophilus | 99 | X68418 |
TABLE 3.
Sequences forming the Enterobacteriaceae groups I to IV
| Peaka | Enterobacteriaceae group | Relative sequences | % Identityb | GenBank accession no. |
|---|---|---|---|---|
| b | I | Enterobacter aerogenes | 100 | AF395913 |
| Enterobacter absuriae | 100 | AB004744 | ||
| Citrobacter sp. | 100 | AF025368 | ||
| Klebsiella oxytoca | 100 | Y17660 | ||
| e | II | Klebsiella planticola | 99 | Y17663 |
| Klebsiella terrigena | 100 | Y17670 | ||
| Klebsiella ornithynolytica | 99 | U78182 | ||
| Enterobacter intermedius | 99 | AB004747 | ||
| Kluyvera ascorbata | 100 | AF176566 | ||
| f | III | Klebsiella trevisanii | 100 | AF390952 |
| Citrobacter werkmanii | 99 | AF025373 | ||
| Enterobacter aerogenes | 100 | AJ251468 | ||
| g | IV | Pantoea sp. | 100 | AF227860 |
| Kluyvera ascorbata | 100 | AF176567 | ||
| Enterobacter intermedius | 100 | AF310217 | ||
| Buttiauxella noackiae | 100 | AJ293689 |
The fingerprint obtained after amplification of the V2 region allowed us to distinguish some of the main lactic acid bacteria that coeluted in peak n of the V3 fingerprint. The SSCP pattern obtained with V2F labeled with NED with the attribution of the peaks is shown in Fig. 2C. Peak 4 was related to the sequences of Lactococcus lactis; peak 12 was related to coelution of two sequences of Streptococcus thermophilus and Leuconostoc pseudomesenteroides. The migration of the Lactobacillus pentosus sequence would appear to correspond to position 13. The sequences of the different enterobacteria and of Bacillus thuringiensis coeluted with the other peaks.
The SSCP patterns obtained with V2R labeled with HEX are shown in Fig. 2D1 and D2. The dilution of the PCR product before SSCP analysis (Fig. 2D2) revealed the heterogeneity of the highest peak. This analysis allowed us to distinguish the migration of the sequences corresponding to Leuconostoc pseudomesenteroides (peak 27), Streptococcus thermophilus (peak 24), and Lactobacillus pentosus (peak 26). Peak 25 was related to two sequences of Leuconostoc mesenteroides and Lactobacillus plantarum that coeluted.
We pointed out the high-GC% gram-positive bacteria by a first amplification by using F243, a primer specific for high-GC% gram-positive bacteria (downstream w49), and w34, followed by a second amplification with w49 and w34 to reveal the V3 region of high-GC% gram-positive flora (Fig. 2B and Table 4). Peak r indicates the migration of a sequence assigned to Brevibacterium avium. The closest relative corresponding to peak s was Propionibacterium freudenreichii. Peak t originated from a sequence of Corynebacterium bovis, and peak u originated from a sequence of Dermacoccus nishinomyaensis. The closest relative corresponding to peak v was Brachybacterium nesterenkovii. Peaks w and x originated from sequences of Corynebacterium variabilis, with different homology percentages, and peak y originated from a sequence corresponding to Corynebacterium afermentans. Peak z corresponded to the migration of a sequence of Corynebacterium flavescens. Some peaks remained unidentified. 16S rRNA gene of the genomic DNA of Bifidobacterium breve and Bifidobacterium longum reference strains were not amplified with primers w34 and F243.
TABLE 4.
Identities of peaks obtained from SSCP analysis of the high-GC% gram-positive bacterial community
| Peaka | Relative sequences | % Identityb | GenBank accession no. |
|---|---|---|---|
| r | Brevibacterium avium/linens | 99 | Y17962 |
| s | Propionibacterium freudenreichii | 100 | Y10819 |
| t | Corynebacterium bovis | 98 | AF311436 |
| u | Dermacoccus nishinomyaensis | 99 | X87757 |
| v | Brachybacterium nesterenkovii | 99 | X91033 |
| w | Corynebacterium variabilis | 99 | AJ222816 |
| x | Corynebacterium variabilis | 100 | AJ222816 |
| y | Corynebacterium afermentans | 99 | X82055 |
| z | Corynebacterium flavescens | 98 | X84441 |
See Fig. 2.
That is, the percentage of identical nucleotides in the sequence retrieved from the clone and the sequence of the closest relative found in GenBank and the RDP database.
Application of a SSCP-PCR method to monitor bacterial dynamics in cheese.
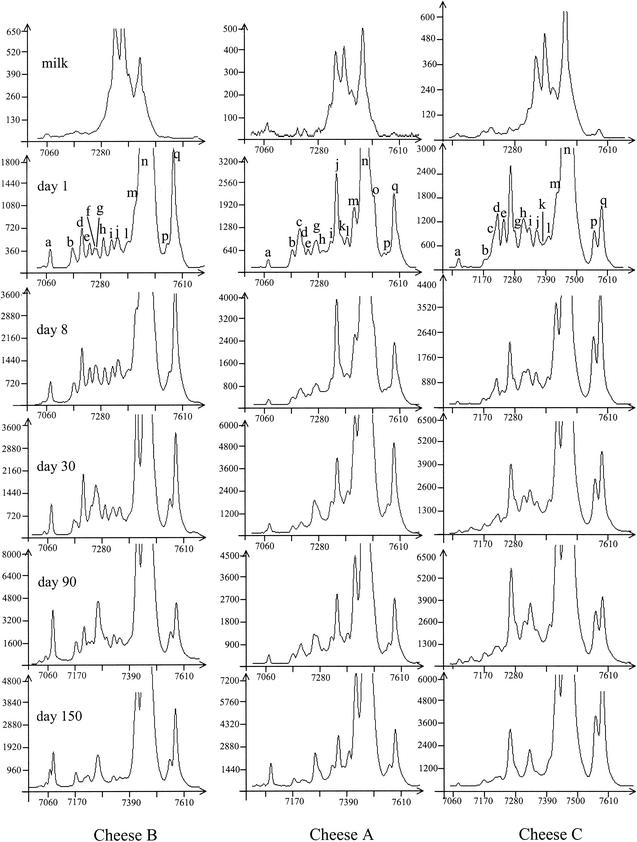
The fingerprints of the bacterial communities obtained after amplification of the V3 region of DNA isolated from milk and cheese from three producers at different times of ripening are shown as an example in Fig. 3. SSCP patterns of the DNA extracted from milk showed a limited number of bacterial populations that were relatively balanced. At 1 day, peak n (corresponding to the sequences of four lactic acid bacteria) became very high and remained predominant till the end of the ripening. During ripening, the peaks corresponding to sequences of enterobacteria became less numerous and less significant.
FIG. 3.
V3 SSCP patterns of PCR-amplified 16S rRNA gene fragments from bacterial communities of three milks and cheeses at different times of ripening. y axis, fluorescence; x axis, elution in scans (unit of GeneScan software). The positions and labeling of peaks discussed in Tables 2 and 3 and in the text are indicated.
The same kind of dynamic fingerprint as those presented in Fig. 3 were obtained for the three producers after amplification with different sets of primers V3, V2, or high GC% gram-positive. To analyze all of the different profiles, the ratio (the area of one peak to the sum of the areas of all of the peaks) was calculated for the same amplification. Indeed, such a ratio gives a semiquantitative picture of the relative abundance of the sequences in the population. The reproducibility of the method was tested on three samples. For each sample, two extractions, PCR amplification, and SSCP analysis were independently done. The results are shown in Table 5. The standard deviation was quite low. Consequently, this analysis was only performed on one sample.
TABLE 5.
Repeatability of the SSCP analysis
| Peak | Mean % repeatability of SSCP analysisa for:
|
|||||
|---|---|---|---|---|---|---|
| Sample 1
|
Sample 2
|
Sample 3
|
||||
| Mean | SD | Mean | SD | Mean | SD | |
| 1 | 0.12 | 0.02 | 0.25 | 0.09 | ||
| 2 | 0.07 | 0.02 | 0.37 | 0.23 | 2.39 | 0.32 |
| 3 | 1.24 | 0.20 | 1 | 0.19 | 1.19 | 0.10 |
| 4 | 0.71 | 0.16 | 1.46 | 0.36 | 0.81 | 0.08 |
| 5 | 1.26 | 0.23 | 1.12 | 0.36 | 2.16 | 0.12 |
| 6 | 0.52 | 0.18 | 0.68 | 0.02 | ||
| 7 | 0.61 | 0.09 | 0.57 | 0.06 | ||
| 8 | 0.38 | 0.12 | ||||
| 9 | 0.71 | 0.05 | ||||
| 10 | 91.7 | 1.29 | 90.57 | 1.51 | 86.13 | 0.87 |
| 11 | 4.9 | 0.65 | 3.28 | 0.08 | 5.81 | 0.48 |
That is, extraction of nucleic acid, PCR amplification, electrophoresis, and SSCP pattern treatment.
The changes in the ratio of some peaks of the SSCP patterns of cheeses, selected for their interest in technology and safety of cheeses, are shown in Table 6. The ratios obtained gave us an indication of the evolution of the microflora, and the values should be considered carefully. After amplification of the V3 region, whatever the producer, the proportion of sequences corresponding to Lactococcus lactis, Streptococcus thermophilus, Lactobacillus pentosus, and Lactobacillus plantarum (peak n) was high in the milk samples (33 to 40%) and increased mainly during the first day. The dynamics of peak m assigned to the sequences of Leuconostoc mesenteroides and Leuconostoc pseudomesenteroides was similar for the three cheeses. The evolution of peak e, which was related to sequences of a group of Enterobacteriaceae, was dominant in milk and then decreased rapidly at 1 day and tended to disappear during the ripening. During ripening the proportion of peak i, corresponding to the Enterococcus faecium sequence, decreased in cheeses A and C, whereas in cheese B the proportion was maximum at 30 days.
TABLE 6.
Evolution of the ratio of some peaks of the SSCP patterns of the three cheeses at different times of ripening
| Time | Ratio (%)a of peak:
|
|||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n (V3) of cheese:
|
i (V3) of cheese:
|
m (V3) of cheese:
|
e (V3) of cheese:
|
h (V3) of cheese:
|
24 (V2) of cheese:
|
27 (V2) of cheese:
|
26 (V2) of cheese:
|
x (hgc) of cheese:
|
v (hgc) of cheese:
|
|||||||||||||||||||||
| A | B | C | A | B | C | A | B | C | A | B | C | A | B | C | A | B | C | A | B | C | A | B | C | A | B | C | A | B | C | |
| Milk | 33.8 | 39.9 | 39.9 | 5.8 | 0.0 | 4.4 | 9.9 | 9.9 | 9.4 | 23.3 | 35.7 | 17.6 | 1.1 | 0.0 | 2.3 | 6.1 | 5.6 | 5.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 14.4 | 65.7 | 9.2 | 11.6 | 0.0 | 0.0 |
| Day 1 | 57.2 | 78.5 | 77.2 | 1.8 | 0.9 | 1.5 | 5.0 | 1.4 | 2.8 | 8.2 | 1.4 | 1.9 | 1.5 | 0.9 | 2.4 | 16.7 | 0.4 | 14.8 | 1.7 | 0.6 | 0.4 | 1.0 | 0.0 | 0.0 | 12.8 | 72.7 | 46.1 | 15.9 | 0.0 | 7.7 |
| Day 8 | 74.5 | 70.4 | 69.9 | 0.0 | 1.7 | 2.0 | 4.1 | 4.2 | 5.9 | 8.3 | 2.6 | 2.2 | 0.4 | 1.9 | 1.5 | 16.2 | 0.0 | 14.0 | 1.9 | 1.7 | 1.7 | 1.1 | 0.0 | 0.0 | 25.9 | 45.2 | 38.8 | 10.0 | 7.6 | 4.9 |
| Day 30 | 74.1 | 66.5 | 66.1 | 1.7 | 2.0 | 2.6 | 5.6 | 9.2 | 7.6 | 4.3 | 1.9 | 2.2 | 0.0 | 1.5 | 2.5 | 13.0 | 0.0 | 11.4 | 3.4 | 1.3 | 2.4 | 1.3 | 0.0 | 0.0 | 22.6 | 57.1 | 34.5 | 11.4 | 10.5 | 10.5 |
| Day 90 | 73.0 | 62.7 | 70.6 | 1.6 | 1.9 | 2.3 | 6.5 | 10.0 | 11.1 | 4.5 | 2.2 | 1.4 | 0.7 | 1.6 | 1.7 | 12.0 | 0.0 | 18.2 | 6.1 | 2.7 | 4.4 | 0.8 | 0.0 | 0.0 | 37.5 | 29.7 | 41.0 | 8.9 | 6.4 | 4.0 |
| Day 150 | 71.6 | 75.9 | 74.0 | 1.8 | 0.9 | 2.2 | 8.1 | 8.3 | 8.7 | 3.0 | 0.7 | 0.7 | 0.0 | 0.0 | 0.0 | 14.7 | 2.3 | 18.1 | 7.0 | 4.9 | 3.3 | 1.0 | 0.0 | 0.0 | 30.0 | 47.2 | 41.6 | 6.5 | 12.3 | 4.3 |
The percent ratio was calculated as the area of one peak to the sum of the areas of all the peaks of the pattern × 100. The organisms associated with each peak were as follows: peak n, lactic acid bacteria; peak i, Enterococcus faecium; peak m, Leuconostoc spp.; peak e, Enterobacteriaceae; peak h, Bacillus thuringiensis; peak 24, Streptococcus thermophilus; peak 27, Leuconostoc pseudomesenteroides; peak 26, Lactobacillus pentosus; peak x, Corynebacterium variabilis; and peak v, Brachybacterium nesterenkovii. SSCP amplifications are indicated in parentheses.
After amplification of the V2 region, the peak 26 (V2 pattern) corresponding to Lactobacillus pentosus sequence was only detected in cheese A. The proportion of peak 24 (V2 pattern), corresponding to the Streptococcus thermophilus sequence, was higher in cheeses A and C than in cheese B.
After amplification of high-GC% gram-positive bacterial population, the peak x (hgc pattern) assigned to the Corynebacterium variabilis sequence predominated in cheese B and tended to decrease during ripening. In cheeses A and C, the proportion of Corynebacterium variabilis increased at the end of ripening. The peak of the Brachybacterium nesterenkovii sequence (peak v on hgc pattern) was only found in milk A; it appeared during ripening in cheese B and C.
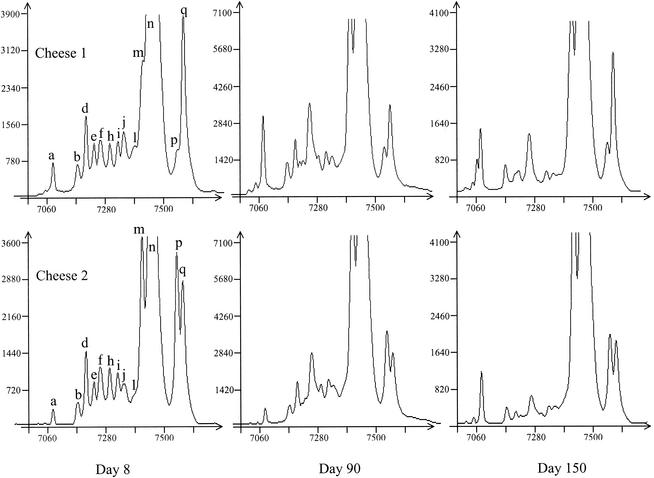
Comparison of two cheeses from the same farmer.
The stability of microbial population in time was evaluated by the analysis of cheeses from producer B manufactured at 1-month intervals. SSCP patterns obtained after V3 amplification of the DNA extracted from 8-, 90-, and 150-day-old cheeses are shown in Fig. 4. Similar profiles were obtained in terms of present peaks, but there were differences concerning the relative intensity of the peaks. For example, the proportion between peaks p and q was the opposite in cheeses 1 and 2.
FIG. 4.
V3 SSCP patterns of PCR-amplified 16S rRNA gene fragments from bacterial communities of cheeses provided by the same farmer at different times of ripening. Cheese 2 was made 1 month later than cheese 1. y axis, fluorescence; x axis, elution in scans (unit of GeneScan software).
DISCUSSION
This is the first time that SSCP analysis combined with clone library sequencing has been applied to the description and dynamics of the microbial community of cheese. The potential of this molecular approach was revealed here. SSCP analysis on gel has been applied to monitor the dynamics of bacterial population in anaerobic bioreactor (37) or in hot composting (30) or to study the fungal diversity in soils (24). SSCP analysis by capillary electrophoresis was realized to establish the dynamics of Archaea during start up of an anaerobic digestor (22). The amplification step with the use of different sets of primers as adopted in the present study constitutes a leverage to overcome the problems of coelution and to point out subdominant populations. Such a strategy was developed to separate gram-negative bacteria by the SSCP method (15). The rapidity and automatization of the SSCP method by capillary electrophoresis compared to other molecular approaches such as TGGE, DGGE, or TTGE mean that the microbial dynamics of numerous samples can be analyzed. SSCP did not require several gel conditions to separate sequences, whereas different gradients were necessary to separate sequences of low or high GC% by TGGE analysis (28). The SSCP electrophoresis conditions had been previously optimized (J. J. Godon, unpublished data). The temperature cannot be lower and the polymer concentration cannot be higher since the polymer would not be injectable. The interpretation of SSCP and TGGE profiles may be complicated by the fact that one species can be represented by different peaks because of intraspecies operon heterogeneity or because of more than one conformation of the same sequence (32). Some discrepancies appeared during the assignation of the peaks for the patterns with regard to the migration of clones. Some peaks remained unidentified, and some clones did not correspond to any peak. This bias could be caused by the use of different primers during the PCR steps. Furthermore, the SSCP analysis, due to the detection system, reveals the most frequent sequences: the fluorescent signal of a subdominant sequence would be considered as background noise. By cloning, subdominant sequences may be detected randomly. In DGGE or TTGE, the unknown bands on the gel can be sequenced directly. This is not possible with SSCP analysis by capillary electrophoresis: the volume used during the analysis is too small, and the DNA fragments cannot be recuperated. It is possible to perform an SSCP analysis on gel, but the detection threshold would be much higher.
Only three recent studies have described microbial dynamics in cheese by DGGE or TTGE analysis, and none of them included the description of high-GC% gram-positive bacteria. The microbial diversity of lactic acid bacteria in mozzarella cheese was evaluated by DGGE analysis of the V3 region (7). By calibrating the technique with reference strains, Streptococcus thermophilus, Lactococcus lactis, Enterococcus faecalis, Lactobacillus delbrueckii subsp. lactis, Lactobacillus delbrueckii subsp. bulgaricus, Enterococcus faecium, Enterococcus durans, and Leuconostoc mesenteroides were differentiated. The technique was applied to the analysis and comparison of lactic acid bacteria diversity in traditional and industrial cheeses. In another study, several bacteria present in dairy products such as Lactobacillus, Lactococcus, Leuconostoc, Enterococcus, Pediococcus, and Streptococcus spp. were detected by the TTGE technique. Gram-negative bacteria such as Buttiauxella agrestis (in raw milk Camembert cheese), Hafnia alvei (in a fresh artisan-made cheese), or Escherichia coli (in pasteurized milk Camembert cheese) were also found (28). By applying DGGE analysis on Sicilian cheeses, Randazzo et al. showed that mesophilic lactic acid bacteria (Leuconostoc spp., Lactococcus lactis) and Macrococcus caseolyticus were predominant in raw milk, whereas Streptococcus thermophilus was predominant during fermentation. Other thermophilic lactic acid bacteria (Lactobacillus delbrueckii and Lactobacillus fermentum) flourished during ripening (31). Most of these species, except for thermophilic Lactobacillus spp., were found in Salers cheese. Moreover, the SSCP analysis allowed us to establish the dynamics of the global microbial population from raw milk to ripened cheese. To our knowledge, the present work was the first report that relates the presence of coryneform bacteria in cheese core and establishes their dynamics from milk to ripened cheese. Species belonging to the coryneform bacteria group were only described as an important part of smear surface cheese bacteria (3, 21, 35).
SSCP-PCR analysis is a powerful tool for monitoring the microbial dynamics in fermented dairy products. The SSCP patterns of milk microbial flora were very different from that of curds (1 day) or ripened cheeses, qualitatively (number of peaks) as well as quantitatively (ratio between peaks). The increase was particularly important for lactic acid bacteria during the first day. All of the peaks present on milk SSCP patterns were also present on cheese SSCP patterns. Some new peaks, such as Leuconostoc pseudomesenteroides (V2 pattern), appeared in curd (1-day cheese) SSCP patterns. This does not mean that the corresponding sequences were not present in the milk but that these sequences were subdominant and therefore not detected by SSCP analysis.
The present study illustrated that each milk and cheese has it own specific dynamic microbial ecosystem. For example, cheese B was characterized by the lowest proportion of Streptococcus thermophilus and the highest proportion of Enterobacteriaceae and Corynebacterium variabilis in milk. Enterobacteriaceae and Corynebacterium variabilis presented the most important decrease in the cheese B. An increase of the ratio may occur not only because the population studied grows up but also because other populations are decreasing.
Most of the data on the microbial community of cheese relies on conventional cultivation methods and identification. Lactic acid bacteria have been the most studied group (14, 16, 33). Some of the samples analyzed in this work by SSCP analysis were also analyzed by identification of isolates picked up on different culture media for lactic acid bacteria (C. Callon, L. Millet, and M. C. Montel, unpublished data). Isolates were identified as Lactococcus lactis, Lactobacillus paracasei, Lactobacillus plantarum, Enterococcus faecalis, Enterococcus faecium, Leuconostoc mesenteroides, and Pediococcus pentosaceus. Lactobacillus sequences did not represent a high proportion of the community by SSCP analysis, and Pediococcus sequence was not detected.
In spite of the interest of SSCP-PCR, one technique cannot catch all of the diversity of a microbial community. Concerning the SSCP technique, the choice of the gene, the sampling and DNA extraction, the PCR amplification, and the pattern analysis are the key points that have to be carefully considered before drawing any conclusions (8). The conventional microbial picture depended on the more or less selective media.
It would be interesting to complete this work with a SSCP analysis of RNA to get a picture of active populations. Such data could be then linked with data available on the sensorial and aromatic profiles of the corresponding ripened cheeses.
Acknowledgments
This work was financially supported by the Direction Générale de l'Alimentation through the Aliment Qualité Santé program of the Ministère de l'Agriculture et de la Pêche. F. Duthoit was supported by an INRA-Région Auvergne thesis grant.
We thank the firms 3A and Lactalis and the Comité Interprofessionnel des Fromages for their interest. We also extend our grateful thanks to Céline Delbes, Cécile Callon, and Liliane Millet for help with this study.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ampe, F., N. ben Omar, C. Moizan, C. Wacher, and J. P. Guyot. 1999. Polyphasic study of the spatial distribution of microorganisms in Mexican pozol, a fermented maize dough, demonstrates the need for cultivation-independent methods to investigate traditional fermentations. Appl. Environ. Microbiol. 65:5464-5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennan, N. M., A. C. Ward, T. P. Beresford, P. F. Fox, M. Goodfellow, and T. M. Cogan. 2002. Biodiversity of the bacterial flora on the surface of a smear cheese. Appl. Environ. Microbiol. 68:820-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cibik, R., E. Lepage, and P. Talliez. 2000. Molecular diversity of Leuconostoc mesenteroides and Leuconostoc citreum isolated from traditional french cheeses as revealed by RAPD fingerprinting, 16S rDNA sequencing, and 16S rDNA fragment amplification. Syst. Appl. Microbiol. 23:267-278. [DOI] [PubMed] [Google Scholar]
- 5.Cocolin, L., L. F. Bisson, and D. A. Mills. 2000. Direct profiling of the yeast dynamics in wine fermentations. FEMS Microbiol. Lett. 189:81-87. [DOI] [PubMed] [Google Scholar]
- 6.Cocolin, L., M. Manzano, C. Cantoni, and G. Comi. 2001. Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl. Environ. Microbiol. 67:5113-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coppola, S., G. Blaiotta, D. Ercolini, and G. Moschetti. 2001. Molecular evaluation of microbial diversity occurring in different types of mozzarella cheese. J. Appl. Microbiol. 90:414-420. [DOI] [PubMed] [Google Scholar]
- 8.Dahllof, I. 2002. Molecular community analysis of microbial diversity. Curr. Opin. Biotechnol. 13:213-217. [DOI] [PubMed] [Google Scholar]
- 9.Delbès, C., J. J. Godon, and R. Moletta. 1998. 16S rDNA sequence diversity of a culture-accessible part of an anaerobic digestor bacterial community. Environ. Microbiol. 4:267-275. [DOI] [PubMed] [Google Scholar]
- 10.Diez, B., C. Pedros-Alio, T. L. Marsh, and R. Massana. 2001. Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblages and comparison of DGGE with other molecular techniques. Appl. Environ. Microbiol. 67:2942-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felske, A. 1999. Reviewing the DA001-files: a 16S rRNA chase on suspect #X99967, a bacillus and Dutch underground activist. J. Microbiol. Methods 36:77-93. [DOI] [PubMed] [Google Scholar]
- 12.Felske, A., B. Engelen, U. Nubel, and H. Backhaus. 1996. Direct ribosome isolation from soil to extract bacterial rRNA for community analysis. Appl. Environ. Microbiol. 62:4162-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felske, A., A. Wolterink, R. van Lis, W. M. de Vos, and A. D. L. Akkermans. 1999. Searching for predominant soil bacteria: 16S rDNA cloning versus strain cultivation. FEMS Microbiol. Ecol. 30:137-145. [DOI] [PubMed] [Google Scholar]
- 14.Fitzsimons, N. A., T. M. Cogan, S. Condon, and T. Beresford. 1999. Phenotypic and genotypic characterization of non-starter lactic acid bacteria in mature cheddar cheese. Appl. Environ. Microbiol. 65:3418-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghozzi, R., P. Morand, A. Ferroni, J. L. Beretti, E. Bingen, C. Segonds, M. O. Husson, D. Izard, P. Berche, and J. L. Gaillard. 1999. Capillary electrophoresis-single-strand conformation polymorphism analysis for rapid identification of Pseudomonas aeruginosa and other gram-negative nonfermenting bacilli recovered from patients with cystic fibrosis. J. Clin. Microbiol. 37:3374-3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gobbetti, M., B. Folkertsma, P. F. Fox, A. Corsetti, E. Smacchi, M. De Angelis, J. Rossi, K. Kilcawley, and M. Cortini. 1999. Microbiology and biochemistry of Fossa (pit) cheese. Int. Dairy J. 9:763-773. [Google Scholar]
- 17.Godon, J. J., E. Zumstein, P. Dabert, F. Habouzit, and R. Moletta. 1997. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 63:2802-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatzikamari, M., E. Litopoulou-Tzanetaki, and N. Tzanetakis. 1999. Microbiological characteristics of anevato: a traditional Greek cheese. J. Appl. Microbiol. 87:595-601. [DOI] [PubMed] [Google Scholar]
- 19.Hengstmann, U., K. J. Chin, P. H. Janssen, and W Liesack. 1999. Comparative phylogenetic assignment of environmental sequences of genes encoding 16S rRNA and numerically abundant culturable bacteria from an anoxic rice paddy soil. Appl. Environ. Microbiol. 65:5050-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heuer, H., M. Kresk, P. Baker, K. Smalla, and E. M. H. Wellington. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irlinger, F. 2000. Caractérisation phénotypique et moléculaire de la diversité des bactéries d'intérêt technologique, de la surface des fromages. Ph.D. thesis. Institut National Agronomique Paris-Grignon, Paris, France.
- 22.Leclerc, M., C. Delbes, R. Moletta, and J. J. Godon. 2001. Single-strand conformation polymorphism of 16S rDNA Archae during start-up of an anaerobic digester. FEMS Microbiol. Ecol. 34:213-220. [DOI] [PubMed] [Google Scholar]
- 23.Lee, D. H., Y. G. Zo, and S. J. Kim. 1996. Nonradioactive method to study genetic profiles of natural bacterial communities by PCR-single-strand-conformation polymorphism. Appl. Environ. Microbiol. 62:3112-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowell, J. L., and D. A. Klein. 2001. Comparative single-strand conformation polymorphism (SSCP) and microscopy-based analysis of nitrogen cultivation interactive effects on the fungal community of a semiarid steppe soil. FEMS Microbiol. Ecol. 36:85-92. [DOI] [PubMed] [Google Scholar]
- 25.Maidak, B. L., J. R. Cole, C. T. Parker, G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moeseneder, M. M., J. M. Arrieta, G. Muyzer, C. Winter, and G. J. Herndl. 1999. Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:3518-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morea, M., F. Baruzzi, and P. S. Cocconcelli. 1999. Molecular and physiological characterization of dominant bacterial populations in traditional mozzarella cheese processing. J. Appl. Microbiol. 87:574-582. [DOI] [PubMed] [Google Scholar]
- 28.Ogier, J. C., O. Son, A. Gruss, P. Tailliez, and A. Delacroix-Buchet. 2002. Identification of the bacterial microflora in dairy products by temporal temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 68:3691-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osborn, A. M., E. R. B. Moore, and K. N. Timmis. 2000. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39-50. [DOI] [PubMed] [Google Scholar]
- 30.Peters, S., S. Koschinsky, F. Schwieger, and C. C. Tebbe. 2000. Succession of microbial communities during hot composting as detected by PCR-single-strand-conformation-polymorphism-based genetic profiles of small-subunit rRNA genes. Appl. Environ. Microbiol. 66:930-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Randazzo, C. L., S. Torriani, A. D. L. Akkermans, W. M. de Vos, and E. E. Vaughan. 2002. Diversity, dynamics, and activity of bacterial communities during production of an artisanal sicilian cheese as evaluated by 16S rRNA analysis. Appl. Environ. Microbiol. 68:1882-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmalenberger, A., F. Schwieger, and C. C. Tebbe. 2001. Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl. Environ. Microbiol. 67:3557-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tavaria, F. K., and F. X. Malcata. 1998. Microbiological characterization of Serra da Estrela cheese throughout its appellation d'origine protegee region. J. Food Protein 61:601-607. [DOI] [PubMed] [Google Scholar]
- 34.Turenne, C. Y., E. Witwicki, D. J. Hoban, J. A. Karlowsky, and A. M. Kabani. 2000. Rapid identification of bacteria from positive blood cultures by fluorescence-based PCR-single-strand conformation polymorphism analysis of the 16S rRNA gene. J. Clin. Microbiol. 38:513-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valdes-Stauber, N., S. Scherer, and H. Seiler. 1997. Identification of yeasts and coryneform bacteria from the surface microflora of brick cheeses. Int. J. Food Microbiol. 34:115-129. [DOI] [PubMed] [Google Scholar]
- 36.Zoetendal, E. G., A. D. Akkermans, and W. M. De Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zumstein, E., R. Moletta, and J. J. Godon. 2000. Examination of two years of community dynamics in an anaerobic bioreactor using fluorescence polymerase chain reaction (PCR) single-strand conformation polymorphism analysis. Environ. Microbiol. 2:69-78. [DOI] [PubMed] [Google Scholar]