Abstract
Appropriate interpretation of a positive reverse transcription-PCR is an important issue for virus-related health hazard assessment because viral genomes and infectious viruses exhibit different behavior patterns in water. In this context, using Poliovirus 1 and Feline calicivirus f9 as examples of enteric viruses, first we demonstrated that the stability of infectious viruses is greatly affected by the temperature of mineral water (10, 20, and 35°C) and that, in contrast, temperature has little effect on the corresponding genomes. Second, we demonstrated that infectious particles are degraded more rapidly than viral genomes at all temperatures studied. At 35°C, Poliovirus 1 infectivity was reduced 4 logs after only 19 days, while an equivalent reduction would have taken 75 years (according to the model applied) for the viral genome. Contradictory conclusions can also be drawn concerning the sensitivity of viral serotypes depending on whether the infectious virus or the viral genome is considered. The Feline calicivirus f9 genome is more resistant than the Poliovirus 1 genome, whereas the opposite is true for the corresponding infectious viruses. Thus, we concluded that a positive test for a viral genome in mineral water must be interpreted with utmost caution because of the lack of a correlation between the presence of viral genomes and viral infectivity. Detection of viral genomes may be necessary to identify infectious risk for the human population, but it cannot be considered sufficient.
Norovirus (NV) members of the family Caliciviridae are the leading cause of viral gastroenteritis in adults (11). Needless to say, the detection of a fragment of an NV genome in certain mineral waters (3) described previously (2) hit the scientific community like an earthquake. However, is there scientific evidence demonstrating that the presence of viral genomes in water should be interpreted as an infectious risk? Available qualitative or semiquantitative data suggest that viral genomes and truly infectious viruses exhibit different behavior patterns (8, 10, 12, 18, 26).
Over the last 45 years, the infectivity of viruses has been determined with cell cultures, a high-performance technique widely used to detect enteric viruses. Consequently, the infectious viral unit, defined as the viral unit capable of replication in a sensitive cell, has been the only standard available for study. It has been used in a wide range of research, including analyses of minimal infectious doses, viral survival in the environment, levels of water contamination, the effects of environmental factors or water treatments, and virus concentration-extraction methodology. Although cell culture is a fundamental tool, it is a long costly procedure and is unable to detect certain viruses, such as Norovirus and Sapovirus, which are frequently the causal agent of gastroenteritis epidemics. Thus, when reverse transcription-PCR (RT-PCR) methods became available (24), environmental virology researchers rapidly switched to detecting viral genomes with this rapid, sensitive, specific, quantitative technique, which is well adapted to all viruses and has been presented as the ideal method. Enteric virus genomes have been detected in seafood (17), in residual sludge (20), and in all types of water, including raw wastewater (13), treated wastewater (9), river water (23), seawater (14), and, most recently, mineral water (3). This molecular tool does, however, have a few drawbacks, including a high risk of false-positive results due to laboratory contamination and a high risk of false-negative results due to enzyme inhibition. Moreover, RT-PCR detection of a virus genome does not provide any information about the infectious nature of the virus isolated. This fundamental point has led to much debate. According to some workers, genomic RNA degrades rapidly in water so the genome detected has to be encapsidated and thus corresponds to a complete potentially pathogenic virus (3, 16). Other workers recall that little is known about what happens to genomic RNA in water and, most importantly, that the presence of a capsid is an insufficient criterion to determine the infectious nature of a virus. It is well known that capsids can be damaged, rendering the virus incapable of binding to sensitive cells, an essential step required before cell infection (21).
We have a good deal of well-documented evidence on infectious viral particles which allows reasonable estimation of the public health hazard. For viral genomes, however, data are scarce and often fragmentary. Thus, before drawing conclusions about genomes detected in mineral waters, we have to acquire more precise information on the behavior of viral genomes and viral particles in water.
To illustrate this problem, we monitored infectious viruses and their genomes in mineral water at three temperatures (10, 20, and 35°C). Three single-stranded RNA viruses were studied, one member of the Picornaviridae (Poliovirus 1 [PV1]) and two members of the Caliciviridae (Norovirus Ober.98 [NV.Ober.98] and Feline calicivirus f9 [FCV-f9]). PV1 and FCV-f9 replicate in cell cultures (Buffalo green monkey [BGM] and feline fibroblast cells [FEA], respectively), but the NV genogroup I representative NV.Ober.98 does not. After simulating mineral water contamination by PV1, FCV-f9, and NV.Ober.98, we monitored the infectivities of PV1 and FCV-f9 by using cell cultures and the persistence of the genomes of all three viruses by fluorogenic quantitative RT-PCR.
MATERIALS AND METHODS
Virus and cells.
Stools containing NV.Ober.98 (98% homologous with Caliciviridae strain Oberschleissheim [GenBank accession number :gi13194695]), which were collected from children involved in outbreaks, were diluted to obtain a 10% suspension in phosphate-buffered saline, mixed with an equal volume of 1,1,2-trichloro-1,2,2-trifluoroethane, and clarified by centrifugation at 500 × g as previously described (1). Preparations were stored at −70°C.
PV1 (Lsc2ab strain) was propagated in BGM cells growing in minimum essential medium (MEM) (catalog no. M5650; Sigma) containing 5% fetal calf serum and 1% l-glutamine (catalog no. G7513; Sigma).
FCV-f9 was propagated in a monolayer of feline embryo-derived (FEA) cells in MEM containing 10% of fetal calf serum and 1% l-glutamine.
For each cultivable strain, viral advanced cytopathic effects were observed after 24 to 48 h of incubation at 37°C. Stocks of viral inocula were prepared by freezing and thawing them three times and were centrifuged (10,000 × g, 60 min, 4°C). Each supernatant, which was stored at −70°C, constituted a viral stock. Virus quantities were expressed in most probable numbers of cytopathogenic units (MPNCU).
Water samples.
A mineral groundwater with the autochthonous microbial flora was used for all experiments. The conductivity of this water was 645 μS/cm, the pH was 7.51, and the water contained no CO3−. The concentrations of ions and compounds were as follows: Ca2+, 97 mg/liter; Mg2+, 21 mg/liter; Na+, 8.68 mg/liter; K+, 5.24 mg/liter; Fe2+, 0.390 mg/liter; Mn2+, 0.040 mg/liter; SO42−, 128 mg/liter; HCO3−, 256 mg/liter; Cl−, 4.6 mg/liter; PO43−, <0.01 mg/liter; SiO2, 8.1 mg/liter; NO3−, <0.1 mg/liter; NO2−, <0.01 mg/liter; and NH4+, 0.01 mg/liter. The physicochemical composition of this mineral water is similar to that of other commercial bottled mineral waters. This water comes from a deep well in the Bundsandstein sandstone in northeastern France. According to the mineral water classification scheme, this water is a bicarbonate-sulfatocalcic water with Ca, HCO3−, and SO42− domination.
Viral seeding of water samples.
One hundred milliliters of water was contaminated with 10-fold serial dilutions of viral stocks to obtain a sample containing 105 MPNCU of PV1 per ml and NV.Ober.98 in the final 100-fold dilution. A second 100-ml water sample was contaminated with 105 MPNCU of FCV-f9 per ml and then with the same 100-fold dilution used for NV.Ober.98.
The two 100-ml water samples were homogenized for 15 min at 25°C and distributed into 1-ml polypropylene tubes, which where placed in the dark at 10, 20, and 35°C.
At each sampling time, tubes were removed and stored at −70°C until they were analyzed by cell culture and fluorogenic quantitative RT-PCR methods.
Quantification of infectious virus by cell culture.
Each sample was treated with an antibiotic and antimycotic solution (catalog no. A5955; Sigma) for 2 h at 37°C. Infectious PV1 and infectious FCV-f9 were quantified in 96-well microplates by using BGM cells and FEA cells, respectively. Fifty-microliter portions of three successive logarithmic dilutions of each sample were added to 200-μl preparations containing 7.5 × 104 BGM or FEA cells ml−1 in MEM containing 2% fetal calf serum. Each dilution was seeded in 40 wells. The plates were incubated for 6 days at 37°C in a 5% CO2-95% air atmosphere and then examined for cytopathogenic effects. The numbers of wells with cytopathogenic effects were counted, and the MPNCU was calculated (19).
Viral genome detection. (i) Viral RNA extraction.
RNA of viruses (PV1, FCV-f9, and NV.Ober.98) were extracted from 140 μl of seeded water with a Qiamp viral RNA kit (catalog no. 52904; Qiagen) used according to the manufacturer's instructions. Sixty microliters of extract product was obtained after this step.
(ii) Primers and probes.
The primers and probes used for each virus in TaqMan assays were designed with Primer Express software (version 1.0). Each oligonucleotide was selected for specific sequences (GenBank accession number gi 61257 for PV1 and GenBank accession number gi323877 for FCV-f9). For NV.Ober.98, the specific sequence of the virus was defined before primers and a probe were designed. The different primer sets and probes are shown in Table 1.
TABLE 1.
Primers and probes designed with Primer Express software (version 1.0) and used in the three fluorogenic RT-PCR assays
| Virus | Primer or probe | Sequence (5′-3′)a | Localization |
|---|---|---|---|
| FCV-f9 | FCV-r | 5′-GATCGGAAAAGTAACGAAGGATGT-3′ | 4547-4524 |
| FCV-f | 5′-GATAGCCCCAGCGTCGAAG-3′ | 4406-4424 | |
| FCV-p | 5′-f-TCGACCCAATCGCCTCGTGTCA-t-3′ | 4487-4508 | |
| PV1 | ENT-r | 5′-CAAGATTGGTTCCTGCTTGATCTT-3′ | 5648-5625 |
| ENT-f | 5′-AGCATTGTGATCGATGGCAA-3′ | 5573-5592 | |
| ENT-p | 5′-f-AGATCTTGGATGCCAAAGCGCTCG-t-3′ | 5601-5624 | |
| NV.Ober.98b | NV-r | 5′-CCATAGAAGGAGAAACAGGAATGAG-3′ | 284-260 |
| NV-f | 5′-CGATAGCACACTGGATCCTAACCT-3′ | 188-211 | |
| NV-p | 5′-f-ATCAGGCCTTTCACCAGATGTTGTCCA-t-3′ | 240-257 |
f, 6-carboxyfluorescein reporter dye; t, 6-carboxytetramethyl rhodamine quencher dye.
NV.Ober.98 was 98% homologous to the Oberschleissheim 102/1999 strain.
(iii) cDNA synthesis.
cDNA was synthesized for each virus from the extracted RNA by using the reverse primer at a final concentration of 0.5 μM in a 20-μl mixture containing 4 μl of 5× reverse transcription buffer (250 mM Tris-HCl [pH 8.4], 50 mM MgCl2, 350 mM KCl, 15 mM dithiothreitol, 2.5 mM spermidine), 40 U of RNase inhibitor (catalog no. N2111; Promega), each deoxynucleoside triphosphate (catalog no. N8080260; Perkin-Elmer) at a concentration of 0.25 μM, 10 U of reverse transcriptase (catalog no. M5108; Promega), 6 μl of DNase-free, RNase-free water (catalog no. W4502; Sigma), and 5 μl of extracted product heated for 3 min at 95°C. RT was performed at 42°C for 60 min. RNA-DNA hybrids were denatured, and reverse transcriptase was inactivated by heating the preparation at 95°C for 5 min. The resulting cDNA was then amplified by PCR.
(iv) PCR amplification.
For each virus a PCR assay was done separately by using 5 μl of cDNA in a 50-μl (final volume) mixture containing 25 μl of TaqMan universal master mixture (catalog no. 4304437; PE Biosystems), 14.5 μl of nuclease-free water, forward and reverse primers (each at a final concentration of 0.5 μM), and TaqMan probe at a final concentration of 0.2 μM for NV.Ober.98, 0.25 μM for FCV-f9, and 0.3 μM for PV1.
Amplification and detection were performed with an ABI Prism 7700 sequence detection system (Perkin-Elmer Inc.). The amplification procedure included two hold programs, (i) 2 min at 50°C to activate the uracil N′-glycosylase and then (ii) 10 min at 95°C to release the activity of the hot start DNA polymerase, followed by 50 cycles consisting of 15 s at 95°C and 60 s at 60°C. Real-time fluorescence measurements were obtained and analyzed directly with the ABI Prism 7700 sequence detection system software. The threshold cycle value for each sample generated was calculated by determining the point at which fluorescence exceeded a threshold limit.
RESULTS
The persistence of infectious viruses (PV1 and FCV-f9) and viral genomes (PV1, FCV-f9, and NV.Ober.98) was monitored in mineral water at 10, 20, and 35°C.
Reductions in the levels of infectious viruses in mineral water.
Survival of infectious viruses was monitored by cell culturing for PV1 and FCV-f9 until a reduction in the virus level of at least 1 log was obtained for each temperature. For PV1, inactivation of the infectious viral particles exhibited classical linear kinetics fitting the following equation: log (Nx/N0) = −aT (equation 1), where Nx/N0 is the ratio of the concentration of infectious viral particles at time x to the concentration of infectious viral particles at time zero, T is the time (in days), and a is the coefficient of inactivation (Fig. 1). This allowed calculation of the T90 (time after which the viral titer had decreased by 90%) values shown in Table 2. As expected, temperature had a potent effect. There was little variation (<1 log) at 10°C over a 262-day period, whereas at 35°C the reduction was more than 3.5 logs in only 16 days. The observed T90 values were 345, 59, and 4.7 days for 10, 20, and 35°C, respectively. For FCV-f9, the inactivation kinetics also seemed to follow equation 1, but the reduction in the virus level was so rapid that there were too few experimental points at 35 and 20°C for plotting. The T90 of FCV-f9 was 11.5 days at 10°C and was estimated to be 1.1 days at 20°C and 0.3 day at 35°C. The reduction in the virus level in mineral water was significantly greater for FCV-f9 than for PV1 (P < 0.05, as determined by the Student t test).
FIG. 1.
Persistence of infectious PV1 in mineral water at 10, 20, and 35°C over time.
TABLE 2.
Persistence of infectious viruses (PV1 and FCV-f9) in mineral water at different temperatures (10, 20, and 35°C)
| Infectious virus | Initial viral titer (MPNCU/ml) | Storage temp (°C) | No. of experi- mental points | Duration of exper- iment (days) | r2 (linear regres- sion)a | T90 (days) |
|---|---|---|---|---|---|---|
| PV1 | 105 | 10 | 15 | 262 | 0.60 | 345 |
| 20 | 15 | 140 | 0.90 | 59 | ||
| 35 | 15 | 16 | 0.94 | 4.7 | ||
| FCV-f9 | 105 | 10 | 7 | 24 | 0.74 | 11.5 |
| 20 | 3 | 4 | —b | 1.1c | ||
| 35 | 2 | 2 | —b | 0.3d |
See equation 1.
—, the number of experimental points was insufficient to calculate r2.
Estimate based on three points (equation 1).
Estimate based on two points (equation 1).
Reduction in the levels of viral genomes in mineral water.
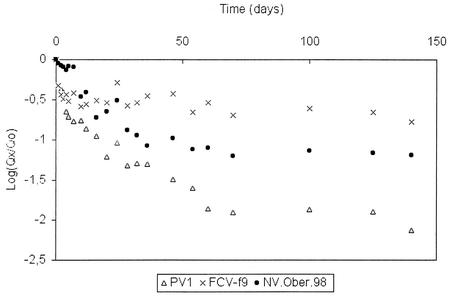
Reductions in the levels of viral genomes were monitored by quantitative RT-PCR for PV1, FCV-f9, and NV.Ober.98 at three temperatures. At 10°C no significant decrease was observed for the three viral genomes over a 262-day period. At 20°C no significant decrease was observed over a 140-day period. These results show that temperatures from 10 to 20°C have very little effect on the decreases in the quantities of viral genomes in mineral water medium. The reductions became significant only at 35°C for the three viruses (Fig. 2). The FCV-f9 genome was the most resistant, followed by the NV.Ober.98 genome and finally by the PV1 genome, which was the least resistant (P < 0.05, as determined by the Student t test).
FIG. 2.
Persistence of PV1, FCV-f9, and NV.Ober.98 genomes in mineral water at 35°C over time.
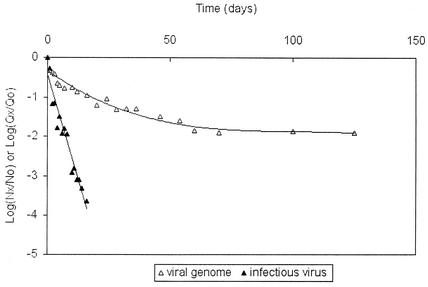
Conversely, the disappearance of genomes of viruses (FCV-f9, PV1, and NV.Ober.98) followed a log T curve-fitting equation: log (Qx/Q0) = −b · log T (or Qx/Q0 = T−b) (equation 2), where Qx/Q0 is the ratio of the concentration of viral genomes at time x to the concentration of viral genomes at time zero, T is the time (in days), and b is the coefficient of disappearance. Therefore, when the disappearance of viral genomes and the disappearance of infectious viruses at 35°C were compared (Fig. 3), PV1 infectious particles exhibited a 4-log reduction after only 19 days, a level of reduction that would have been reached only after 75 years (according to the model used) for the viral genome. The same phenomenon was observed for FCV-f9, for which the differences were greater because of a very quick decrease in the number of infectious particles at each temperature (Fig. 2 and Table 2).
FIG. 3.
Persistence of the PV1 genome (viral genome) and infectious PV1 (infectious virus) in mineral water at 35°C over time as described by equations 2 and 1, respectively.
DISCUSSION
This study was undertaken to determine differences in the behavior patterns of viral genomes and infectious viruses, which appears to be an important issue for correct interpretation of positive RT-PCR results for the environment in terms of risk assessment.
As NV genomes have recently been detected in mineral water (3), we focused on this kind of water. In addition, since NV are unable to grow in cell cultures, we investigated two other cultivable virus models: PV1, which is a well-known enteric virus, and FCV-f9, which, like NV, belongs to the family Caliciviridae but is a respiratory virus.
Our results show that there is a great difference in heat resistance between infectious viruses and viral genomes. Heat is known to have a potent effect on infectious particles. Thus, for PV1 the T90 values were 345, 59, and 4.7 days for 10, 20, and 35°C, respectively, while for FCV-f9 the T90 values were significantly shorter but decreased similarly with temperature. These expected results confirmed previous findings describing viral sensitivity to heat (4, 6, 7, 15, 30). An increased temperature can modify the viral protein capsid, resulting in inactivation (5, 22, 29).
In contrast, temperature seems to have little effect on the viral genome. Thus, in mineral water after 262 days at 10°C and after 140 days at 20°C, there was no quantitative variation in the viral genome (<0.5 log) for any of the three viruses studied (FCV-f9, PV1, and NV.Ober.98). Temperatures from 10 to 35°C were expected to have little direct effect on viral RNA (25). Nevertheless, at 35°C after 140 days the quantity of the viral genome remaining was lower, and there were significant differences among the three viruses. The difference in degradation mechanisms between infectious viruses and viral genomes was underlined by the use of two different mathematical models to describe the kinetics of disappearance. Inactivation of the infectious viral particles (FCV-f9 and PV1) exhibited classical linear kinetics over time, while the disappearance of viral genomes (FCV-f9, PV1, and NV.Ober.98) exhibited linear kinetics as a function of the logarithm of time. According to these models, infectious viral particles are degraded more rapidly than viral genomes. At 20°C the difference in behavior led to a significant reduction (>2 logs) in the level of infectious PV1 at 140 days, while no variation in the level of the PV1 genome was observed over the same period. Similar results were obtained at 35°C.
Therefore, the degradation observed at 35°C could have been due to the presence of RNases in the media studied, which were able to digest viral RNA as soon as it became available outside the viral capsid (5, 8, 27, 28, 29). Such RNases could have had different origins, such as the stool extract which constituted the NV stock, the PV1 or FCV-f9 suspension obtained from a cell culture supernatant, or the mineral water. If the hypothesis that RNase was present is accepted, the stability of the genome at 10 and 20°C can only be explained by protection of the RNA inside the capsid, protection that disappeared partially with time at 35°C. Inactivated viral particles inside which RNA is protected against RNase have been described previously (21).
Furthermore, contradictory conclusions concerning the sensitivities of two viruses, PV1 and FCV-f9, can be made depending on whether the genome or the infectious virus is considered the standard. If the infectious virus is the standard, FCV-f9 was found to be a very fragile virus (the T90 values were 11.5 and 0.3 days for 10 and 35°C, respectively), while PV1 survived significantly longer at all three temperatures tested (the T90 values were 345 and 4.7 days for 10 and 35°C, respectively). However, if the viral genome is used as the standard, FCV-f9 exhibited significantly greater resistance than PV1 for 140 days at 35°C. Under these conditions and notwithstanding the fact that degradation of the NV.Ober.98 genome followed a curve located between the curves for the two viral models used (PV1 and FCV-f9), no conclusion can be drawn concerning the infectious behavior of NV.Ober.98.
In light of evidence available today, detection of viral genomes may be necessary but not sufficient for assessment of the infectious risk for a human population.
REFERENCES
- 1.Ando, T., S. S. Monroe, J. R. Gentsch, Q. Jin, D. C. Lewis, and R. I. Glass. 1995. Detection and differentiation of antigenically distinct small round-structured viruses (Norwalk-like viruses) by reverse transcription-PCR and Southern hybridization. J. Clin. Microbiol. 33:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 2002. Mineral water contamination claim. Nature 46:IX. [Online.] http://www.nature.com/nsu.
- 3.Beuret, C., D. Kohler, A. Baumgartner, and T. M. Luthi. 2002. Norwalk-like virus sequences in mineral waters: one-year monitoring of three brands. Appl. Environ. Microbiol. 68:1925-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biziagos, E., J. Passagot, J. M. Crance, and R. Deloince. 1988. Long-term survival of hepatitis A virus and poliovirus type 1 in mineral water. Appl. Environ. Microbiol. 54:2705-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breindl, M. 1971. The structure of heated poliovirus particles. J. Gen. Virol. 11:147-156. [DOI] [PubMed] [Google Scholar]
- 6.Doultree, J. C., J. D. Druce, C. J. Birch, D. S. Bowden, and J. A. Marshall. 1999. Inactivation of feline calicivirus, a Norwalk virus surrogate. J. Hosp. Infect. 41:51-57. [DOI] [PubMed] [Google Scholar]
- 7.Enriquez, C. E., C. J. Hurst, and C. P. Gerba. 1995. Survival of the enteric adenoviruses 40 and 41 in tap, sea, and waste water. Water Res. 29:2548-2553. [Google Scholar]
- 8.Enriquez, C. E., M. Abbaszadegan, I. L. Pepper, K. J. Richardson, and C. P. Gerba. 1993. Poliovirus detection in water by cell culture and nucleic acid hybridization. Water Res. 27:1113-1118. [Google Scholar]
- 9.Gantzer, C., A. Maul, J. M. Audic, and L. Schwartzbrod. 1998. Detection of infectious enteroviruses, enterovirus genomes, somatic coliphages, and Bacteroides fragilis phages in treated wastewater. Appl. Environ. Microbiol. 64:4307-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gantzer, C., A. Maul, Y. Levi, and L. Schwartzbrod. 1998. Fate of the genome and infectious units of Coxsackie B3 virus in phosphate buffered saline. Water Res. 32:1329-1333. [Google Scholar]
- 11.Glass, R. I., J. Noel, T. Ando, R. Fankhauser, G. Belliot, A. Mounts, U. D. Parashar, J. S. Bresee, and S. S. Monroe. 2000. The epidemiology of enteric caliciviruses from humans: a reassessment using new diagnostics. J. Infect. Dis. 181:S254-S261. [DOI] [PubMed] [Google Scholar]
- 12.Gratacap-Cavallier, B., O. Genoulaz, I. Brengel-Pesce, H. Soule, P. Innocenti-Francillard, M. Bost, L. Gofti, D. Zmirou, and J. M. Seigneurin. 2000. Detection of human and animal rotavirus sequences in drinking water. Appl. Environ. Microbiol. 66:2690-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green, D. H., and G. D. Lewis. 1999. Comparative detection of enteric viruses in wastewaters, sediments and oysters by reverse transcription-PCR and cell culture. Water Res. 33:1195-1200. [Google Scholar]
- 14.Griffin, D. W., C. J. Gibson, E. K. Lipp, K. Riley, J. H. Paul, and J. B. Rose. 1999. Detection of viral pathogens by reverse transcriptase PCR and of microbial indicators by standard methods in the canals of the Florida Keys. Appl. Environ. Microbiol. 65:4118-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurst, C. J., W. H. Benton, and K. A. McClellan. 1989. Thermal and water source effects upon the stability of enteroviruses in surface freshwaters. Can. J. Microbiol. 35:474-480. [DOI] [PubMed] [Google Scholar]
- 16.Kopecka, H., S. Dubrou, J. Prevot, J. Marechal, and J. M. Lopez-Pila. 1993. Detection of naturally occurring enteroviruses in waters by reverse transcription, polymerase chain reaction, and hybridization. Appl. Environ. Microbiol. 59:1213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Guyader, F., L. Haugarreau, L. Miossec, E. Dubois, and M. Pommepuy. 2000. Three-year study to assess human enteric viruses in shellfish. Appl. Environ. Microbiol. 66:3241-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma, J. F., T. M. Straub, I. L. Pepper, and C. Gerba. 1994. Cell culture and PCR determination of poliovirus inactivation by disinfectants. Appl. Environ. Microbiol. 60:4203-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maul, A. 1991. Aspects statistiques des méthodes de quantification en virologie, p. 143-171. In L. Schwartzbrod (coordinating ed.), Virologie des milieux hydriques. Lavoisier, Paris, France.
- 20.Monpoeho, S., A. Maul, B. Mignotte-Cadiergues, L. Schwartzbrod, S. Billaudel, and V. Ferre. 2001. Best viral elution method available for quantification of enteroviruses in sludge by both cell culture and reverse transcription-PCR. Appl. Environ. Microbiol. 67:2484-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nuanualsuwan, S., and D. O. Cliver. 2003. Capsid functions of inactivated human picornaviruses and feline calicivirus. Appl. Environ. Microbiol. 69:350-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pancorbo, O. C., B. G. Evanshen, W. F. Campbell, S. Lambert, S. K. Curtis, and T. W. Woolley. 1987. Infectivity and antigenicity reduction rates of human Rotavirus strain Wa in fresh waters. Appl. Environ. Microbiol. 53:1803-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puig, M., J. Jofre, F. Lucena, A. Allard, G. Wadell, and R. Girones. 1994. Detection of adenoviruses and enteroviruses in polluted waters by nested PCR amplification. Appl. Environ. Microbiol. 60:2963-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saiki, R. K., S. Scharf, F. Faloona, K. B. Mullis, G. T. Horn, H. A. Erlich, and N. Arnheim. 1985. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230:1350-1354. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Sobsey, M. D., D. A. Battigelli, G. A. Shin, and S. Newland. 1998. RT-PCR amplification detects inactivated viruses in water and wastewater. Water Sci. Technol. 12:91-94. [Google Scholar]
- 27.Toranzo, A. E., J. L. Barja, and F. M. Hetrick. 1983. Mechanism of poliovirus inactivation by cell-free filtrates of marine bacteria. Can. J. Microbiol. 29:1481-1486. [DOI] [PubMed] [Google Scholar]
- 28.Tsai, Y. L., B. Tran, and C. J. Palmer. 1995. Analysis of viral RNA persistence in seawater by reverse transcriptase-PCR. Appl. Environ. Microbiol. 61:363-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward, R. L., D. R. Knowlton, and P. E. Winston. 1986. Mechanism of inactivation of enteric viruses in fresh water. Appl. Environ. Microbiol. 52:450-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yates, M. V., C. P. Gerba, and L. M. Kelley. 1985. Virus persistence in groundwater. Appl. Environ. Microbiol. 49:778-781. [DOI] [PMC free article] [PubMed] [Google Scholar]