Abstract
To determine if green fluorescent protein could be used as a reporter for detecting nitric oxide production, gfp was fused to nnrS from Rhodobacter sphaeroides 2.4.3. nnrS was chosen because its expression requires nitric oxide. The presence of the fusion in R. sphaeroides 2.4.3 resulted in a significant increase in fluorescent intensity of the cells, but only when nitrite reductase was active. Cells lacking nitrite reductase activity and consequently the ability to generate nitric oxide were only weakly fluorescent when grown under denitrification-inducing conditions. One of the R. sphaeroides strains unable to generate nitric oxide endogenously was used as a reporter to detect exogenously produced nitric oxide. Incubation of this strain with sodium nitroprusside, a nitric oxide generator, significantly increased its fluorescence intensity. Mixing of known denitrifiers with the reporter strain also led to significant increases in fluorescence intensity, although the level varied depending on the denitrifier used. The reporter was tested on unknown isolates capable of growing anaerobically in the presence of nitrate, and one of these was able to induce expression of the fusion. Analysis of the 16S rRNA gene sequence of this isolate placed it within the Thauera aromatica subgroup, which is known to contain denitrifiers. These experiments demonstrate that this green fluorescent protein-based assay provides a useful method for assessing the ability of bacteria to produce nitric oxide.
Denitrification is the reduction of nitrate or nitrite to gaseous nitrogen oxides. This pathway occurs mainly in bacteria and is used by most denitrifiers to support respiratory growth under anaerobic conditions (22). The production of nitric oxide (NO) by nitrite reductase is a critical step in denitrification, as NO is the first gaseous intermediate formed in the pathway. Its production from nitrite is also unique to denitrification, and any organism catalyzing this reaction is a denitrifier. Thus, the detection of NO in denitrifying cultures could serve as a useful tool for identifying potential denitrifiers.
Currently, routine methods for detecting NO production by denitrifiers are lacking. The main difficulty is that in order to mitigate its toxicity, denitrifiers maintain steady-state NO levels within the low nanomolar range (6). These levels can only be detected with sensitive gas analyzers, requiring specialized equipment, or NO-binding protein assays, which are difficult to do on a routine basis (6, 7). The development of a simple, sensitive tool for NO detection should hasten the identification of potential denitrifiers.
The goal of this work was to develop a simple technique to screen for potential denitrifiers by detecting the NO produced during denitrification. This was done with an NO-inducible gene from Rhodobacter sphaeroides 2.4.3, a strain carrying genes encoding the complete denitrification pathway. In strain 2.4.3, NnrR, a member of the FNR (fumarate nitrate reduction) regulator family, regulates a number of denitrification genes that respond to the presence of NO (13). The nnrS gene, which lies upstream and is divergently transcribed from the gene encoding NnrR, is a member of the NnrR regulon (2). Its expression requires the presence of NnrR and only occurs when R. sphaeroides is producing NO. Genes in the NnrR regulon have also been shown to be responsive to exogenous sources of NO (13). Studies indicate that a putative NnrR binding site is centered 186 bases upstream of the predicted translational start for nnrS and that deletion of this site prevents expression of an nnrS-lacZ fusion (2).
In this paper, the construction and characterization of an nnrS-gfp fusion is described. The experiments presented demonstrate that this green fluorescent protein (GFP)-based reporter is responsive to the NO levels produced by denitrifiers, is easily utilizable, and is not subject to false-positives resulting from production of other nitrogen oxides. The utility of the GFP-based reporter in screening potential denitrifiers for NO production is also demonstrated.
MATERIALS AND METHODS
Strains and culture conditions.
Escherichia coli strain DH5α was used as the maintenance strain for plasmids. E. coli S17-1 was used as the donor for conjugative transfer of plasmids. R. sphaeroides 2.4.3 (ATCC 17025) and R. sphaeroides 2.4.1 (from S. Kaplan, University of Texas Health Science Center, Houston, Tex.) were used as reporter strains. Strain 11.10 is a nitrite reductase-deficient mutant of 2.4.3 (19). Agrobacterium tumefaciens C58, Achromobacter cycloclastes, Rhodopseudomonas palustris strain CGA009 and Rhizobium “hedysari” strain HCNT1 were used as positive controls for NO production.
E. coli strains were grown in Luria-Bertani (LB) medium. R. sphaeroides and Agrobacterium tumefaciens were grown in Sistrom's medium at 30°C (16). Tetracycline was added to E. coli cultures at 15 μg ml−1. Trimethoprim and tetracycline were added to Rhodobacter cultures at 30 μg ml−1 and 1 μg ml−1, respectively. Rhodopseudomonas palustris was grown on Sistrom's medium supplemented with 5.0 g of tryptone liter−1 and 2.5 g of yeast extract liter−1. Achromobacter cycloclastes was grown on Difco nutrient broth (NB). The HCNT1 strain was grown on TY medium (18). During microaerobic growth, KNO3 was added to cultures of R. sphaeroides, Agrobacterium tumefaciens, and Achromobacter cycloclastes to a final concentration of 11 mM. NaNO2 was added to cultures of Rhodopseudomonas palustris to a final concentration of 290 μM to avoid nitrite toxicity. Nitrogen oxides were not added to the HCNT1 cultures because they are not required for nir expression in this strain (18).
All strains were cultured in 250-ml flasks containing 100 ml of medium. The flasks were sealed with a butyl rubber stopper to prevent oxygen exchange. In this way, cells initially respire oxygen, but as it is consumed, growth becomes microaerobic, permitting expression of the nitrogen oxide reductases (19). Nitrite reductase activity was confirmed with a whole-cell activity assay as previously described (15).
Construction of nnrS-gfp clone.
Initially, a SalI fragment containing the gfp gene from plasmid pVIK165 (11) was introduced into plasmid pRK415, creating pRK415gfp. The SalI fragment in pVIK165 was originally from pTB93F (4), which expresses a stable GFP variant that absorbs at 488 nm. Then, a 928-bp fragment containing the nnrS promoter was amplified with the upstream primer RSSUP (5′-CGCGAATTCAGCAGCAGGTAGAACCGGTC-3′) and the downstream primer 15DK (5′-CGCGGTACCCGATCAGCATGATGAGGAAGG-3′). The underlining indicates KpnI and EcoRI restriction sites added to facilitate cloning. The amplified fragment contained 174 bp upstream of the putative NnrR binding site and 607 bp downstream of the putative translational start of nnrS. This fragment was introduced into pRK415gfp by digesting both the plasmid and fragment with KpnI and EcoRI. A plasmid with both the nnrS promoter and gfp was isolated and designated pMF3. This was transformed into E. coli S17-1 to allow conjugation of pMF3 into R. sphaeroides strains.
Isolation of nitrogen oxide reducers.
Soil samples used for the isolation of nitrogen oxide reducers were obtained from the edge of Beebe Lake in Ithaca, N.Y., or from a Winogradsky column constructed with soil from Ithaca. One gram of each sample was resuspended in 9.0 ml of sterilized water and mixed for 2 min. Samples were diluted 10-fold with sterile water, and 60 μl of each dilution was spread onto solid medium. Plates for isolation of nitrate-respiring strains contained either Sistrom's or NB medium supplemented with 11 mM KNO3. The plates were placed in sealed jars, made anaerobic with BBL Gas-Pak cartridges (Becton Dickinson), and incubated at 30°C until colonies became visible. Standard techniques were used to isolate pure colonies.
Detection of exogenous NO.
Detection of exogenous NO required that the reporter be mixed with an NO generator. In experiments with sodium nitroprusside (SNP), cells of 11.10/pMF3 were grown microaerobically in Sistrom's medium and transferred to 12-ml serum vials after overnight growth. The vials were sealed and incubated for 1 h to reduce oxygen levels. Near the end of this incubation, a 200 mM stock of SNP was prepared in Sistrom's medium. With a syringe, SNP was added to a final concentration of 2 mM, and cultures were incubated for an additional 4 h, at which point cells were removed via syringe for analysis.
To detect exogenous NO production from a bacterial strain, the putative NO producer was grown microaerobically as described above. The cells were harvested and resuspended in culture medium lacking nitrogen oxides. The harvested cells were mixed with pMF3-containing cells that had been cultured microaerobically in medium lacking nitrate before their harvest. In most cases, the 11.10/pMF3 strain was used as the reporter. Typically, the ratio of cell mixtures was 1 volume of the putative NO producer to 9 volumes of the reporter. This ratio was used because previous work demonstrated that only a small amount of the NO producer is necessary for induction of NO-dependent genes (14). The mixed cultures were transferred to 12-ml serum vials, and these were crimp sealed with rubber stoppers and incubated at 30°C with shaking. If needed, vials were supplemented with nitrate or nitrite. The density of cell mixtures was similar to that of the cultures prior to harvest (optical density at 660 nm from 0.7 to 1.0). Following incubation, samples were removed via syringe for microscopic analysis. Cells were placed on slides coated with polylysine to limit cell motility.
Determination of 16S rRNA gene sequence.
Genomic DNA of the NO-producing isolate was prepared with the Wizard genomic DNA kit (Promega). A portion of the 16S rRNA gene was amplified with the 27f [5′-AGAGTTTGATC(C/A)TGGCTCAG-3′] and 1492r [5′-TACGG(C/T)TACCTTGTTACGACTT-3′] universal primers (3, 10). The amplified DNA was gel purified and sequenced with the 1392r [5′-ACGGGCGGTGTGT(A/G)C-3′] oligonucleotide primer (10). Sequencing was carried out at the Cornell BioResource Center DNA sequencing facility with an Applied Biosystems automated 3700 DNA analyzer. A sequence match program at the Ribosomal Database Project (http://rdp.cme.msu.edu/html) was used to compare the 16S rRNA gene sequence of this isolate with known sequence in the database.
Microscopic analysis of samples.
Cells were viewed with an Olympus BX61 epifluorescent microscope equipped with low-power objectives for scanning and a 100× UPlanApo objective, N.A 1.35, for fluorescent imaging and phase contrast microscopy. The microscope is equipped with a fluorescence filter cube for detecting GFP (41017 EndowGFP, Chroma). Images were obtained with a Cooke SensiCam with a Sony Interline chip. The SlideBook software package (Intelligent Imaging Inc.) was used to acquire images and to analyze the intensity of fluorescence. For each image, the background intensity was measured with an area lacking cells, while cell intensity was measured from the internal portion of 25 cells. To determine the average cell intensity for a particular image, the background intensity was subtracted from the measured intensity for each of the 25 cells, and the resulting values were averaged. To produce figures, the average intensity of each image was normalized to the average intensity range for images of the 2.4.3/pMF3 strain. Figures were assembled with Adobe Photoshop version 5.5.
RESULTS
GFP has proven extremely useful in analyzing gene expression and protein localization. However, one limitation of using GFP is that chromophore synthesis requires oxygen, which makes its use in the study of anaerobic processes difficult (21). Denitrification, in particular the nitrite and NO reduction steps of this pathway, are frequently considered anaerobic processes, but in fact their expression does not require strict anaerobic conditions (12). This makes it feasible to determine whether GFP, which remains functional in low-oxygen environments (8), can be used as a reporter for nitrogen oxide respiration. R. sphaeroides 2.4.3 was chosen as a source for a suitable promoter because it preferentially expresses nitrite and NO reductases under microaerobic conditions (15). The promoter for the nnrS gene from R. sphaeroides 2.4.3 was selected because its expression appears to require a single transcriptional regulator, NnrR (2). The plasmid carrying the nnrS::gfp fusion was designated pMF3.
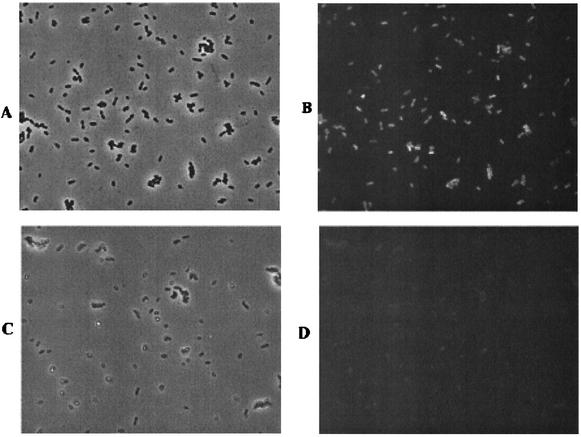
To study GFP production under denitrifying conditions, the pMF3-containing strain of R. sphaeroides 2.4.3 was grown microaerobically in nitrate-amended medium. The 2.4.3/pMF3 cells grown under these conditions were strongly fluorescent (Fig. 1A and B), while the 2.4.3 cells lacking the fusion exhibited only weak fluorescence (not shown). Analysis of the fluorescence intensity indicated that the induced cells were 10 to 15 times more fluorescent than cells lacking the fusion (Table 1). Aerobic growth of 2.4.3/pMF3 resulted in fluorescence similar to the background level observed in cells lacking the fusion (not shown). Under optimal conditions for induction of nnrS, nearly 100% of the cells containing pMF3 showed fluorescence, indicating that there is sufficient oxygen present for chromophore synthesis when the gene is expressed (Fig. 1A and B).
FIG. 1.
Microscopic analysis of strains 2.4.3/pMF3 and 11.10/pMF3. Both strains were grown microaerobically in nitrate-amended medium. (A) Phase contrast micrograph of 2.4.3/pMF3 cells. (B) Fluorescence micrograph of the same field of cells shown in panel A. (C) Phase contrast micrograph of 11.10/pMF3 cells. (D) Fluorescence micrograph of the same field of cells shown in panel C. The intensity of GFP in panel D was normalized to the intensity observed in the 2.4.3/pMF3 cells.
TABLE 1.
Background corrected fluorescence intensity levels of various strainsa
| Strain(s) | Intensity (avg ± SD) |
|---|---|
| 2.4.3 | 36.7 ± 7.9 |
| 2.4.3/pMF3 | 431.8 ± 190.5 |
| 11.10/pMF3 | 89.7 ± 28.4 |
| 11.10/pMF3 + SNP | 533.4 ± 160.2 |
| 11.10/pMF3 + 2.4.3 | 520.1 ± 172.9 |
| 11.10/pMF3 + Achromobacter cycloclastes | 561.4 ± 185.8 |
| 11.10/pMF3 + Agrobacterium tumefaciens | 659.4 ± 221.6 |
| 11.10/pMF3 + Rhodopseudomonas palustris | 276.0 ± 109.3 |
| 11.10/pMF3 + HCNT1 | 288.4 ± 148.6 |
| 11.10/pMF3 + isolate | 583.1 ± 176.2 |
All incubations were done in medium containing either nitrate or nitrite. Intensity values were obtained by averaging the pixel intensity of internal portions of 25 cells in each image. The background intensity of each image was obtained by determining the intensity of an area of the image lacking cells. The background was subtracted from each cell intensity value, and the resulting values were averaged to obtain the final intensity measurements. Values are presented as average ± standard deviation.
To confirm that GFP expression was NO dependent, pMF3 was mobilized into R. sphaeroides 2.4.3 strain 11.10, a nitrite reductase-deficient mutant (19). When grown under inducing conditions, 11.10/pMF3 exhibited weaker fluorescence than seen in the wild-type cells (Fig. 1C and D). The fluorescence intensity of 11.10/pMF3 was greater than that of cells lacking pMF3, consistent with some NO production from the nitrite that accumulates in the absence of nitrite reductase activity (19) (Table 1). Similar results were obtained with R. sphaeroides 2.4.1 containing pMF3 (not shown). R. sphaeroides 2.4.1, like most R. sphaeroides isolates, is naturally deficient in nitrite reductase (9). Autofluorescence levels in 2.4.1 were similar to that observed in experiments with 2.4.3.
Activation of gfp expression by exogenous NO.
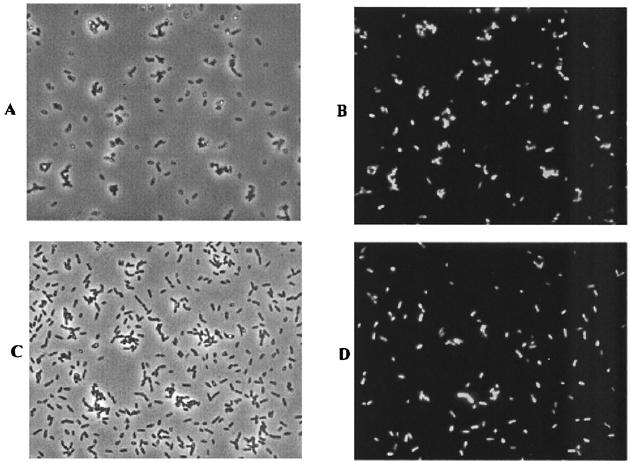
Previous work demonstrated that NO-responsive genes in R. sphaeroides could be activated in the presence of exogenous sources of NO (13). To confirm that the nnrS::gfp fusion could be used to detect exogenous NO, cells of the 11.10/pMF3 reporter strain were mixed with sodium nitroprusside (SNP), an NO donor. Since SNP is toxic, the cells were cultured under microaerobic conditions without SNP, transferred to vials, and then incubated with or without the NO generator. As expected, cells incubated with SNP were visibly fluorescent (Fig. 2A and B), while those incubated without were not (not shown). The background corrected fluorescence intensity of cells induced with SNP was similar to that of strain 2.4.3/pMF3 grown under inducing conditions (Table 1). This experiment demonstrates that the fluorescence observed in pMF3-carrying strains can be induced by an exogenous source of NO.
FIG. 2.
Microscopic analysis of GFP induction from exogenous sources of nitric oxide. (A) Phase contrast micrograph of 11.10/pMF3 cells 3 h after exposure to the NO generator sodium nitroprusside. (B) Fluorescence micrograph of the same field of cells shown in panel A. (C) Phase contrast micrograph of a mixture of 2.4.3 and 11.10/pMF3 cells. For this experiment, 2.4.3 cells were grown microaerobically in nitrate-amended medium and mixed with an equal volume of 11.10/pMF3 cells in a serum vial. The vial was sealed, supplemented with nitrate, and incubated for 2 h before the cells were photographed. (D) Fluorescence micrograph of the same field of cells shown in panel C.
Mixing experiments with R. sphaeroides strains.
Since this assay detected chemically generated NO, an experiment was conducted to assess its ability to detect NO from biological sources. Initial tests involved mixing cells of 11.10/pMF3 with those of wild-type strain 2.4.3 and incubating the mixture under denitrifying conditions. These strains could not be cocultured because addition of tetracycline, which is necessary for maintenance of pMF3 in strain 11.10, inhibits the growth of strain 2.4.3. Instead, cultures of the two strains were grown separately under microaerobic conditions in Sistrom's medium. The 2.4.3 culture was supplemented with nitrate. After overnight growth, cells from the 11.10/pMF3 and 2.4.3 cultures were added to serum vials. Nitrate was added, and each vial was sealed to prevent oxygen exchange. Samples were periodically removed for microscopic assessment of GFP expression.
Incubation of the cell mixtures resulted in visibly detectable fluorescence. In the experiment shown in Fig. 2C and D, equal volumes of the test and reporter strains were used. As shown, about half of the cells present in Fig. 2C were fluorescent in Fig. 2D, which correlates well with a mixture containing half reporter cells. Fluorescence above background was detected within 1 h, but optimal fluorescence typically required about 2 to 3 h of incubation. Interestingly, the fluorescent intensity of cells in this experiment was slightly higher than that of the 2.4.3/pMF3 strain (Table 1). Fluorescence was also observed when 2.4.1/pMF3 was mixed with 2.4.3 using the procedure described above. However, only background levels of fluorescence were observed in mixing experiments with 2.4.1 and 2.4.1/pMF3 (not shown). Strain 11.10/pMF3 was chosen as the reporter for subsequent experiments because it exhibited higher levels of fluorescence in mixing experiments than did strain 2.4.1/pMF3.
Mixing experiments with 11.10/pMF3 and diverse denitrifiers.
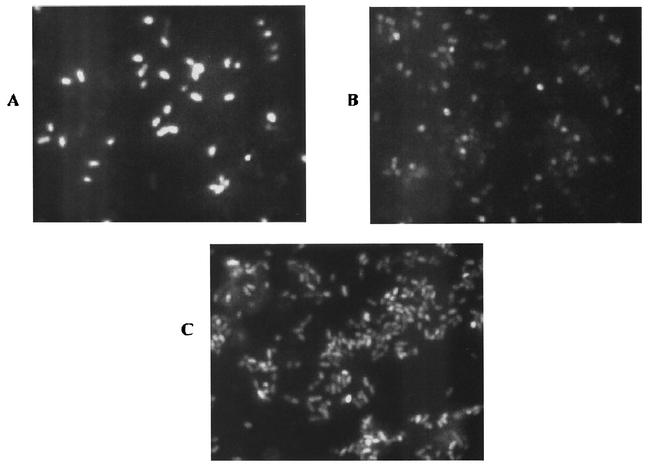
For nnrS::gfp to be a useful tool in detecting NO production, the reporter should respond appropriately in experiments with physiologically diverse bacteria. To assess its utility, the 11.10/pMF3 reporter strain was mixed with a variety of complete and partial denitrifiers. Achromobacter cycloclastes is a complete denitrifier that has been shown to produce steady-state NO levels of 10 to 30 nM during denitrification, which is typical of the concentrations produced by the few denitrifiers for which NO production has been quantitated (6). To determine if 11.10/pMF3 would respond to these NO levels, actively denitrifying cells of Achromobacter cycloclastes were washed to remove residual nitrate and mixed with the reporter strain. The mixture was incubated in sealed vials containing nitrate-supplemented medium. After several hours of incubation, fluorescence was detected from the majority of cells, indicating that the fluorescence was from the pMF3-containing cells (Fig. 3A). The background corrected fluorescence intensity of the reporter strain was similar to that obtained with R. sphaeroides strain 2.4.3 as the NO source (Table 1). This result demonstrates that the low levels of NO produced by Achromobacter cycloclastes were sufficient to induce expression of the reporter. Also, as observed in the experiments with the various R. sphaeroides strains, the NO-producing strain can represent ≤10% of the total mixture and still provide sufficient NO to induce reporter expression in nearly all the cells.
FIG. 3.
Microscopic analysis of fluorescence resulting from mixing of various test strains with 11.10/pMF3. The images in this figure have been magnified two times to clearly show differences in the response of the reporter. Cells in the mixture were (A) 11.10/pMF3 and Achromobacter cycloclastes, (B) 11.10/pMF3 and Rhizobium hedysari HCNT1, (C) 11.10/pMF3 and an environmental isolate. For A and C, the test strain (Achromobacter cycloclastes or an environmental isolate) was cultured microaerobically in medium supplemented with nitrate, harvested, resuspended, and transferred to a serum bottle. Cells of 11.10/pMF3 were added to each vial, and the vials were sealed. Each vial was supplemented with nitrate and incubated for a minimum of 2 h before samples were viewed under a microscope. For B, Rhizobium hedysari HCNT1 was cultured in medium lacking nitrogen oxides and mixed with the reporter strain as described above. In this case, nitrite rather than nitrate was added to the vial prior to incubation.
Similar mixing experiments were done with another complete denitrifier, Agrobacterium tumefaciens (5, 20). This bacterium has been studied extensively, but its capacity for denitrification has been largely overlooked. The fluorescence observed from the reporter when it was mixed with Agrobacterium tumefaciens demonstrated that this bacterium produced NO (not shown). After background correction, the fluorescent intensity of the Agrobacterium tumefaciens-induced reporter was similar to that obtained in mixing experiments with R. sphaeroides 2.4.3 as the NO producer (Table 1).
Genome sequencing of Rhodopseudomonas palustris has shown that it is a partial denitrifier (information found at http://bahama.jgi-psf.org/prod/bin/microbes/rpal/home.rpal.cgi).However, it will not grow anaerobically in the dark with nitrite, and it is much more sensitive to nitrite than related denitrifiers such as R. sphaeroides and Agrobacterium tumefaciens (D. Y. Lee and J. P. Shapleigh, unpublished data). Therefore, this bacterium is unlikely to be identified as a denitrifier in traditional screens. To test if this bacterium will produce NO, cells of Rhodopseudomonas palustris grown microaerobically in nitrite-amended medium were mixed with 11.10/pMF3. As in other experiments, fluorescence of the reporter was observed within about 2 h (not shown). However, the fluorescent intensity of reporter cells incubated with Rhodopseudomonas palustris was significantly lower than that in previous mixing experiments (Table 1).
An extreme example of a truncated denitrification pathway is provided by Rhizobium “hedysari” strain HCNT1. This bacterium cannot reduce nitrate or NO but can reduce nitrite and is known to generate relatively high levels of NO as a result (18). This increased level of NO is not inhibitory to expression of the nnrS::gfp fusion; however, it does seem to limit fluorescence production to some extent (Fig. 3B and Table 1).
As a nondenitrifier control, the 11.10/pMF3 reporter was mixed with E. coli DH5α grown under aerobic conditions. After incubation, the fluorescence level of the reporter showed no increase over background fluorescence (not shown).
Confirmation of NO production in natural isolates.
As an additional test, potential denitrifiers were isolated from soil samples from various locations. Initial isolations were carried out under strict anaerobic conditions, with nitrate as the sole terminal electron acceptor. Several isolates capable of anaerobic growth on nitrate-amended medium were tested for NO production with the GFP reporter. Each isolate was cultured microaerobically in medium containing nitrate and mixed with the 11.10/pMF3 reporter strain. Even though most strains were capable of nitrite reduction, only one of them could induce fluorescence of the reporter (Fig. 3C). This strain was also the only isolate to produce gas bubbles upon incubation with nitrate. The level of fluorescence produced by the reporter in this experiment was similar to that obtained in mixing experiments with strain 2.4.3. The inability of the other isolates to produce gas and induce fluorescence suggests that they are ammonifiers and thus incapable of NO production.
The NO-producing strain was further characterized by partial sequencing of its 16S rRNA gene (not shown). Comparison of the 16S rRNA gene sequence to the small-subunit database indicates that this organism is most closely related to the Thauera aromatica subgroup, which belongs to the beta subclass of proteobacteria. Bacteria in the Thauera cluster are frequently isolated in screens for denitrifiers capable of degrading certain aromatic pollutants (17).
DISCUSSION
By fusing gfp to an NO-inducible promoter derived from the nnrS gene from R. sphaeroides, a method for detecting NO production by monitoring expression of GFP has been developed. As expected from previous work with nnrS, the reporter was only expressed when nitrite respiration was occurring (2). While the reporter is capable of detecting endogenously generated NO, its general applicability lies in the fact that it is equally useful in detecting exogenous production of this denitrification intermediate. This is because NO, like other intermediates produced during denitrification, is freely diffusible throughout the growth medium (7). If a nitrite reductase-deficient strain with the reporter fusion becomes fluorescent in a mixed culture, the NO that caused the nnrS expression must have been produced exogenously. This capacity to detect exogenously produced NO makes it possible to do rapid screening of organisms suspected of producing NO.
One constraint when using GFP is that oxygen is required for production of the GFP chromophore (21). Therefore, incubation of the reporter with NO producers cannot be done under strictly anaerobic conditions. This problem can be avoided, however, by growing the test strains under denitrifying conditions and then mixing the harvested cells with the reporter in a sealed but not anaerobic incubation vessel. In this way, oxygen and nitrogen oxides are present during the incubation, and as oxygen concentrations decrease, respiration of nitrogen oxides will increase. Because the test strains were grown under denitrifying conditions, their nitrogen oxide reductases will already be induced, and denitrification should occur when electron flow to oxygen decreases enough to allow electrons to flow to the nitrogen oxide reductases. Given the results reported here, the shift in oxidants occurs before complete consumption of oxygen. It seems likely that the oxygen concentration at which NO production begins will vary among denitrifiers, but since chromophore synthesis can occur under conditions of low oxygen (8), the production of NO should be detectable in most cases. This assay, however, would be unable to detect a bacterium that requires complete anaerobiosis for denitrification.
In conclusion, the GFP-based assay described here provides a useful method for assessing the ability of bacteria to produce NO. All of the bacteria that tested positive in this study were shown to be denitrifiers, demonstrating that this assay should be effective in confirming if a bacterium is a member of this physiological group. While denitrifiers are likely to represent the majority of NO producers, it is possible that some bacteria may have other means of producing NO, for instance, via NO synthase (1). Given the sensitivity of the nnrS::gfp fusion, it is possible that NO generated by alternative mechanisms might be detected. Therefore, it is important that additional characterization be done to confirm that an NO producer is a denitrifier. There is also no reason this assay should be limited to prokaryotes. As long as the reporter can be incubated in liquid medium with the test strain, NO production should be detected with this sensitive assay.
Acknowledgments
We are grateful to Esther Angert (Cornell University) for use of the fluorescence microscope and imaging equipment as well as invaluable advice. We also thank Joe Flint (Cornell University) for assistance in use of the microscope.
S. Yin was supported by the Natural Science Foundation of China (49971051 and 30170029) and the China Scholarship Council. This work was supported by the Department of Energy (95ER20206).
REFERENCES
- 1.Adak, S., K. S. Aulak, and D. J. Stuehr. 2002. Direct evidence for nitric oxide production by a nitric-oxide synthase-like protein from Bacillus subtilis. J. Biol. Chem. 277:16167-16171. [DOI] [PubMed] [Google Scholar]
- 2.Bartnikas, T. B., Y. Wang, T. Bobo, A. Veselov, C. P. Scholes, and J. P. Shapleigh. 2002. Characterization of a member of the NnrR regulon in Rhodobacter sphaeroides 2.4.3 encoding a heme-copper protein. Microbiol. 148:825-833. [DOI] [PubMed] [Google Scholar]
- 3.Fennell, D. E., A. B. Carroll, J. M. Gossett, and S. H. Zinder. 2001. Assessment of indigenous reductive dechlorinating potential at a TCE-contaminated site with microcosms, polymerase chain reaction analysis, and site data. Environ. Sci. Technol. 35:1830-1839. [DOI] [PubMed] [Google Scholar]
- 4.Gage, D. J., T. Bobo, and S. R. Long. 1996. Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa). J. Bacteriol. 178:7159-7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodner, B., G. Hinkle, S. Gattung, N. Miller, M. Blanchard, B. Qurollo, B. S. Goldman, Y. Cao, M. Askenazi, C. Halling, L. Mullin, K. Houmiel, J. Gordon, M. Vaudin, O. Iartchouk, A. Epp, F. Liu, C. Wollam, M. Allinger, D. Doughty, C. Scott, C. Lappas, B. Markelz, C. Flanagan, C. Crowell, J. Gurson, C. Lomo, C. Sear, G. Strub, C. Cielo, and S. Slater. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323-2328. [DOI] [PubMed] [Google Scholar]
- 6.Goretski, J., and T. Hollocher. 1990. The kinetic and isotopic competence of nitric oxide as an intermediate in denitrification. J. Biol. Chem. 265:889-895. [PubMed] [Google Scholar]
- 7.Goretski, J., and T. C. Hollocher. 1988. Trapping of nitric oxide produced during denitrification by extracellular hemoglobin. J. Biol. Chem. 263:2316-2323. [PubMed] [Google Scholar]
- 8.Hansen, M. C., R. J. J. Palmer, C. Udsen, D. C. White, and S. Molin. 2001. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147:1383-1391. [DOI] [PubMed] [Google Scholar]
- 9.Jain, R., and J. P. Shapleigh. 2001. Characterization of nirV and a gene encoding a novel pseudoazurin in Rhodobacter sphaeroides 2.4.3. Microbiology 147:2505-2515. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, J. L. 1994. Similarity analysis of rRNAs, p. 683-700. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular microbiology. American Society for Microbiology, Washington, D.C.
- 11.Kalogeraki, V. S., and S. C. Winans. 1997. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene 188:69-75. [DOI] [PubMed] [Google Scholar]
- 12.Korner, H., and W. G. Zumft. 1989. Expression of denitrification enzymes in response to the dissolved oxygen and respiratory substrate in continuous culture of Pseudomonas stutzeri. Appl. Environ. Microbiol. 55:1670-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwiatkowski, A., and J. P. Shapleigh. 1996. Requirement of nitric oxide for induction of genes whose products are involved in nitric oxide metabolism in Rhodobacter sphaeroides 2.4.3. J. Biol. Chem. 271:24382-24388. [DOI] [PubMed] [Google Scholar]
- 14.Kwiatkowski, A. V., W. P. Laratta, A. Toffanin, and J. P. Shapleigh. 1997. Analysis of the role of the nnrR gene product in the response of Rhodobacter sphaeroides 2.4.1 to exogenous nitric oxide. J. Bacteriol. 179:5618-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laratta, W. P., P. S. Choi, I. E. Tosques, and J. P. Shapleigh. 2002. Involvement of the PrrB/PrrA two-component system in nitrite respiration in Rhodobacter sphaeroides 2.4.3: evidence for transcriptional regulation. J. Bacteriol. 184:3521-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leuking, D. R., R. T. Fraley, and S. Kaplan. 1978. Intracytoplasmic membrane synthesis in synchronous cell populations of Rhodopseudomonas sphaeroides. J. Biol. Chem. 253:451-457. [PubMed] [Google Scholar]
- 17.Lin, B., H. W. Van Verseveld, and W. F. Roling. 2002. Microbial aspects of anaerobic BTEX degradation. Biomed. Environ. Sci. 15:130-144. [PubMed] [Google Scholar]
- 18.Toffanin, A., Q. Wu, M. Maskus, S. Casella, H. D. Abruna, and J. P. Shapleigh. 1996. Characterization of the gene encoding nitrite reductase and the physiological consequences of its expression in the nondenitrifying Rhizobium “hedysari” strain HCNT1. Appl. Environ. Microbiol. 62:4019-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tosques, I. E., A. V. Kwiatkowski, J. Shi, and J. P. Shapleigh. 1997. Characterization and regulation of the gene encoding nitrite reductase in Rhodobacter sphaeroides 2.4.3. J. Bacteriol. 179:1090-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, J. N. F. Almeida, L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, K. B. Gordon, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 21.Zimmer, M. 2002. Green fluorescent protein (GFP): applications, structure, and related photophysical behavior. Chem. Rev. 102:759-781. [DOI] [PubMed] [Google Scholar]
- 22.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]