Abstract
The fungal population dynamics in soil and in the rhizospheres of two maize cultivars grown in tropical soils were studied by a cultivation-independent analysis of directly extracted DNA to provide baseline data. Soil and rhizosphere samples were taken from six plots 20, 40, and 90 days after planting in two consecutive years. A 1.65-kb fragment of the 18S ribosomal DNA (rDNA) amplified from the total community DNA was analyzed by denaturing gradient gel electrophoresis (DGGE) and by cloning and sequencing. A rhizosphere effect was observed for fungal populations at all stages of plant development. In addition, pronounced changes in the composition of fungal communities during plant growth development were found by DGGE. Similar types of fingerprints were observed in two consecutive growth periods. No major differences were detected in the fungal patterns of the two cultivars. Direct cloning of 18S rDNA fragments amplified from soil or rhizosphere DNA resulted in 75 clones matching 12 dominant DGGE bands. The clones were characterized by their HinfI restriction patterns, and 39 different clones representing each group of restriction patterns were sequenced. The cloning and sequencing approach provided information on the phylogeny of dominant amplifiable fungal populations and allowed us to determine a number of fungal phylotypes that contribute to each of the dominant DGGE bands. Based on the sequence similarity of the 18S rDNA fragment with existing fungal isolates in the database, it was shown that the rhizospheres of young maize plants seemed to select the Ascomycetes order Pleosporales, while different members of the Ascomycetes and basidiomycetic yeast were detected in the rhizospheres of senescent maize plants.
Soil is a complex and dynamic environment in which the biological activity is mostly governed by microorganisms. The beneficial effects of soil microorganisms are manifold and range from nitrogen fixation and organic matter decomposition to breakdown of metabolic by-products and agrochemicals, enhancing the bioavailability of nitrates, sulfates, phosphates, and essential metals. The role of fungi in the soil is an extremely complex one and is fundamental to the soil ecosystem (4). Fungi play an important role in nutrient cycling and plant health and development (4, 23, 39). While some fungi are well known to cause a range of plant diseases and in some cases to devastate agricultural crops (16, 39), others are known to antagonize plant pathogens, decompose plant residues, provide nutrients to plants, and stimulate plant growth. Some fungi (external mycelium of arbuscular mycorrhizae) can also affect the composition of bacterial communities, either directly by changing host plant physiology or indirectly by changing the patterns of root exudation (22, 37, 41). An improved knowledge of the diversity and structure of fungal communities in bulk and rhizosphere soils can lead to a better understanding of their roles in soil ecosystems.
It is estimated that there are 1.5 million fungal species on earth, of which only about 70,000 have been described up to now (11). The cultivation approach has been applied to assess the fungal diversity for several decades, but a problem is the fastidious nature of several fungal species. Thus, fungi such as rust or smut fungi or arbuscular mycorrhizae are difficult or impossible to grow in axenic media under laboratory conditions. However, the use of various cultivation techniques which would be required to properly describe the fungal diversity is rather time-consuming and laborious, thus limiting their use to describe the dynamics, composition, and fungal diversity in rhizosphere and bulk soil samples. The limitations to adequate identification of fungi by morphological techniques have led to the use of molecular methods, which greatly facilitated the identification of fungi (4). Molecular studies with fungal isolates have mainly exploited the rRNA gene cluster consisting of three rRNA subunit genes, internally transcribed spacers and intergenic sequences. The analysis of rRNA genes from DNA obtained directly from environmental samples has expanded our view of microbial diversity in recent years and has proven to be a powerful tool for investigating the microbial diversity in a wide range of environmental samples (1, 29). Molecular fingerprinting techniques such as denaturing gradient gel electrophoresis (DGGE) analysis of ribosomal DNA (rDNA) fragments amplified from total community DNA have been mainly used to analyze the composition of bacterial communities (27). Only very few studies have used this approach to study fungal soil and rhizosphere communities (7, 18, 19, 36, 42). Since this technique can also be combined with cloning and sequencing, this allows us to analyze phylogenetic sequences of bands generated by community members (36). Although the analysis of fungal communities by means of DGGE of rDNA fragments amplified from community DNA is also prone to different kinds of biases (44), it allows the cultivation-independent and parallel analysis of large sample numbers.
The aim of this work was to provide baseline data on fungal population dynamics in the rhizosphere of maize grown in tropical soils. We analyzed the potential effects of two maize cultivars (Nitroflint and Nitrodent) on the fungal community structure in the context of natural variability (e.g., seasonal shifts) during two consecutive planting periods (1999 and 2000). Cloning and sequencing of 18S rDNA fragments PCR amplified from community DNA were used to provide information on the phylogeny of ribotypes in the DGGE patterns. This allowed us also to evaluate the diversity behind bands with identical electrophoretic mobility in the fungal community profiles.
MATERIALS AND METHODS
Field design and sampling.
The field work was performed at Empresa Brasileira de Pesquisa Agropecuária, located in Seropédica, Rio de Janeiro state, Brazil. Maize plants of two cultivars, Nitroflint and Nitrodent, differing in their abilities to utilize N were grown in a field under organic farming practice in Brazil for two growth periods (from 30 September 1999 to 29 November 1999 and from 8 April 2000 to 17 June 2000). The cultivars were planted in three plots (3 by 6 m) in a randomized block design as part of a larger field test. Two independent soil and rhizosphere samples were taken from each plot at 20, 40, and 90 days after sowing. Each of the six replicate rhizosphere samples per sampling comprised total roots with adhering soil from five maize plants. The roots were shaken vigorously to separate soil not tightly adhering to the roots. The soil cores were taken between the maize rows (approximately 50 cm from plants); they were free of roots and homogeneous with depth. Each of the six replicate soil samples per sampling consisted of eight cores (15 cm of top soil) which were mixed by sieving. Samples for DNA extraction were kept frozen at −20°C.
Total community DNA isolation.
DNA extraction was performed with the Ultra Clean Soil DNA kit (MoBio Laboratories, Solana Beach, Calif.). A portion of 0.25 g of bulk soil or root samples was processed according to the protocol provided by the manufacturer with an additional bead-beating step using a cell homogenizer (Braun, Melsungen, Germany) to achieve a harsh cell lysis.
PCR amplification of 18S rRNA gene fragments and DGGE analysis.
PCR was performed with a Tgradient thermal cycler (Biometra GmbH, Göttingen, Germany), and the fungus-specific primers (40) NS1 (5′-GTA GTC ATA TGC TTG TCT C-3′) and FR1 (5′-AIC CAT TCA ATC GGT AIT-3′) were used for amplification of 18S rRNA gene fragments (1,650 bp). The reaction mixture (25 μl) consisted of 1 μl of template DNA (ca. 20 ng), Stoffel buffer (10 mM KCl, 10 mM Tris-HCl [pH 8.3]), 0.2 mM deoxynucleoside triphosphates, 3.75 mM MgCl2, 2% (wt/vol) dimethyl sulfoxide, a 0.2 μM concentration of each primer (NS1 and FR1-GC), and 2 U μl of Taq DNA polymerase (Stoffel fragment; Applied Biosystems, Foster City, Calif.). A GC-rich sequence (indicated as -GC) was attached to primer FR1 to prevent complete melting of PCR products during separation in the denaturating gradient gel. Dimethyl sulfoxide was added to the reaction mixture to improve specificity and facilitate the amplification of GC-rich templates (43). After 8 min of denaturation at 94°C, 35 thermal cycles of 30 s at 94°C, 45 s at 48°C, and 3 min at 72°C were performed, followed by an extension step at 72°C for 10 min.
DGGE analysis was performed as previously described (12, 13) with a denaturing gradient of 18 to 43% denaturant. Aliquots of PCR samples (4 to 6 μl) were applied to the DGGE gel, and DGGE was performed in 1× Tris-acetate-EDTA buffer at 58°C at a constant voltage of 180 V for 18 h. After silver staining of the DGGE gels, they were air dried and scanned as described by Heuer et al. (12).
Cloning and sequencing.
The primers NS1 and FR1 (without a GC clamp) were used for amplification of 18S rRNA gene fragments from DNA extracted from rhizosphere (20 and 90 days) and bulk soil (90 days) samples from both cultivars. The amplified DNA fragments were purified with the UltraClean DNA purification kit (MoBio Laboratories) and then ligated into the pGEM-T vector and transformed into competent cells (Escherichia coli JM109) according to the instructions of the manufacturer (pGEM-T vector system II; Promega, Madison, Wis.). PCR amplification with primers NS1 and FR1-GC was performed directly from the selected white colonies (presumed transformants). These PCR products were analyzed by agarose gel electrophoresis to confirm the presence of the insert in the transformants. Subsequently, PCR products generated from clones with insert were run on a DGGE gel to determine electrophoretic mobility of the insert. Inserts of clones matching dominant bands of the DGGE fungal community pattern were analyzed by amplified rDNA restriction analysis (ARDRA). One clone representing each ARDRA pattern from each DGGE band type was sent for sequencing.
Sequencing of approximately 500 bp of selected clones was done with standard primers SP6 and T7 (IIT GmbH, Bielefeld, Germany). The 18S sequence fragments were analyzed by using the ARB software package (Linux beta version 011107) and the ARB database ssujun02 (Department of Microbiology, Technical University of Munich, Munich, Germany [http://www.arb-home.de]), including additional fungal 18S partial sequences from the EMBL database. The clone sequences were automatically aligned by using the ARB sequence editor with the fast_aligner. The resulting alignment was manually proofread and corrected if necessary. For each clone sequence the similarity to the best database hits was calculated by using a distance matrix with the particular sequence itself as a filter. The initial tree was calculated by using neighbor joining and the full sequences from the database. The resulting tree was later corrected and optimized, and the partial sequences were added by using arb_parsimony and a fungus-specific filter.
ARDRA.
The 18S rDNA fragments of clones with identical DGGE mobility were analyzed by HinfI restriction patterns. A 10-μl aliquot of each PCR product containing approximately 3 μg of DNA was digested with the restriction enzyme HinfI (New England Biolabs, Beverly, Mass.) in a total volume of 30 μl at 37°C for 2.5 h. The digested PCR products were precipitated by addition of 75 μl of ethanol and 3 μl of sodium acetate (3 M, pH 5.2) and kept at −70°C for at least 1 h (31). Separation of the digested PCR fragments was achieved by agarose gel electrophoresis (2.5% Seakem LE agarose; BMA, Rockland, Maine).
Cluster analysis.
Analyses of restriction patterns and fungal community profiles were performed with the software package Gelcompar 4.0 program (Applied Maths, Ghent, Belgium). Background was subtracted by using a rolling-disk method with an intensity of 10 (relative units), and the lanes were normalized. A dendrogram was constructed by using the Pearson correlation index for each pair of lanes within a gel and cluster analysis by the unweighted pair group method using arithmetic averages.
Nucleotide sequence accession numbers.
Sequence accession numbers for sequences submitted to the EMBL nucleotide sequence database are AJ515162 to AJ515172 and AJ515921 to AJ515950.
RESULTS
The total community DNA isolated from bulk and rhizosphere samples was of high molecular weight and of sufficient purity for successful PCR amplification of 18S rDNA fragments. The 18S rDNA fragments were obtained from all DNA samples by direct PCR amplification. Molecular fingerprints of the most dominant fungal populations which were amplifiable under the PCR conditions used were obtained after separation of PCR products by DGGE. With the exception of rhizosphere samples taken 40 days after sowing, the fingerprints showed relatively little variation between different replicates or between the two cultivars. This also suggests good reproducibility of the DNA extraction, the PCR amplification, and the DGGE analysis.
Fungal community shifts during plant growth development.
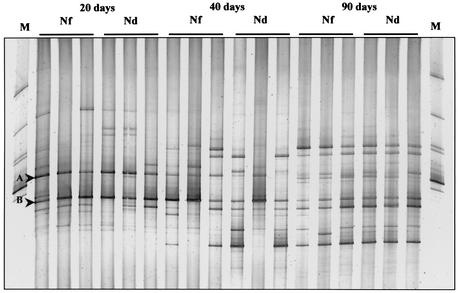
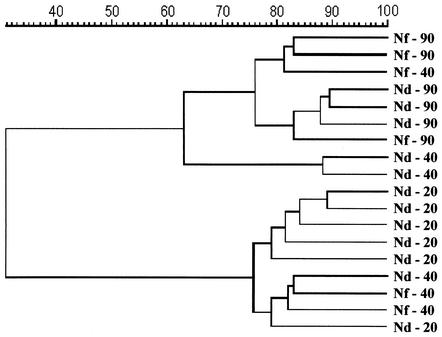
The DGGE profiles revealed that the composition of the fungal community in the rhizosphere of maize was strongly affected by plant age (Fig. 1). The comparison of fungal profiles in the rhizospheres of the cultivars Nitroflint and Nitrodent showed a pronounced shift in the relative abundance of fungal populations during plant growth (20, 40, and 90 days after sowing [R20, R40, and R90, respectively]). The DGGE profiles of young plants (R20) showed the presence of two dominant bands (bands A and B) and some faint bands, indicating the dominance of few populations. Bands with the same electrophoretic mobilities as bands A and B were detectable in the profiles of rhizosphere samples of both cultivars at all sampling times. However, both bands A and B became less dominant in the rhizospheres of samples taken 40 or 90 days after sowing. The DGGE profiles of the R40 samples showed a rather high variability. Cluster analysis based on the unweighted pair group method using arithmetic averages was used to create a dendrogram describing the similarities between fungal community profiles from rhizosphere samples taken at the three time points. The dendrogram clearly indicates two distinct groups with about 75 and 62% similarity within each one (Fig. 2). The grouping supports the notion that the composition of fungal communities in the rhizosphere of young plants was rather different from that in senescent plants. Furthermore, it was confirmed that the fungal rhizosphere community of plants taken 40 days after sowing was in a transition phase and that the DGGE profiles of some of the replicates were more similar to those of young plants while the profiles of others showed a higher similarity to those of mature plants. No relevant differences were found in the fungal community pattern between the cultivars at all time points.
FIG. 1.
DGGE profiles showing the comparison of the fungal rhizosphere communities of the maize cultivars Nitroflint (Nf) and Nitrodent (Nd) at different stages of plant development (20, 40, and 90 days). The fingerprints of fungal communities were generated by separation of 18S rDNA fragments amplified with primers NS1 and FR1-GC. The following fungal species, from top to bottom, were used as standards (lanes M): Colletotrichum sp., Sclerotium tuliparum, Trichoderma harzianum, Myrothecium cinctum, Ustilago nuda, Myrothecium leucotrichum, and Penicillium simplicissimum.
FIG. 2.
Dendrogram constructed with the fungal community fingerprints of the maize cultivars Nitroflint (Nf) and Nitrodent (Nd) at different stages of plant development (20, 40, and 90 days). The differences between the profiles are indicated by percentage of similarity. The dendrogram was based on the Pearson correlation index and cluster analysis by the unweighted pair group method using arithmetic averages.
Rhizosphere effect.
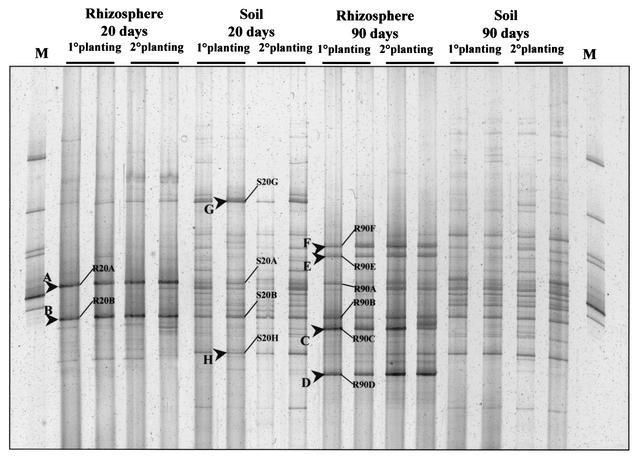
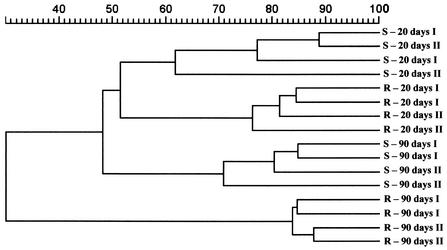
A comparison of the fungal community profiles from the rhizosphere (R20 and R90) with their corresponding bulk soil fingerprints clearly indicated enhancement of the relative abundance of specific fungal populations in the rhizosphere (Fig. 3). The DGGE profiles from bulk soil showed a higher number of roughly equally intense bands at both sampling times. The predominant bands in the R20 fingerprints (bands A and B) were also detectable in the bulk soil pattern (S20 and S90) but with lower intensity. The DGGE profiles from rhizospheres of senescent plants showed an increased number of predominant bands, and in addition to bands R90A and R90B, four new band positions (R90C, R90D, R90E, and R90F) were detected. While the predominant R90 bands C, D, and F were not detected in the corresponding bulk soil pattern, band E appears in the soil pattern but with a lower intensity. Despite an increased number of bands in the DGGE profiles of rhizosphere samples from senescent plants, the corresponding bulk soil presented a different pattern with an even higher number of roughly equally abundant bands. The shift of the fungal community profile in soil and rhizosphere during plant growth development revealed the same trend during two consecutive planting periods (Fig. 3). A cluster analysis of the fungal community profiles from rhizosphere and bulk soil indicated that plant roots indeed affect the fungal diversity in the vicinity of the roots (Fig. 4). Interestingly, the S20 and S90 DGGE profiles formed clearly separate groups, indicating a shift in the relative abundance of fungal populations. While the rhizosphere profiles of samples taken 20 days after sowing still shared about 50% similarity with the DGGE profiles of the bulk soil samples, the DGGE profiles of the rhizosphere samples taken 90 days after sowing grouped separately with 30% similarity. The cluster analysis also did not show any relevant differences between samples from two consecutive planting periods (Fig. 4).
FIG. 3.
DGGE profiles showing the fungal community fingerprints of rhizosphere samples from young (R20) and senescent (R90) plants (Nitrodent) and their corresponding bulk soil fingerprints. Two lanes each represent a typical DGGE profile from the first and second growth periods. The relative band position of the more dominant bands and the cloned DNA fragments are shown. The following fungal species, from top to bottom, were used as standards (lanes M): Colletotrichum sp., Sclerotium tuliparum, Trichoderma harzianum, Myrothecium cinctum, Ustilago nuda, Myrothecium leucotrichum, and Penicillium simplicissimum.
FIG. 4.
Dendrogram constructed with the fungal community fingerprints of rhizosphere (R) samples from young (R20) and senescent (R90) plants (Nitrodent) and their corresponding bulk soil (S20 and S90) fingerprints. Two full growth periods (I and II) were analyzed. The differences between the profiles are indicated by percentage of similarity. The dendrogram was based on the Pearson correlation index and cluster analysis by the unweighted pair group method using arithmetic averages.
Cloning, ARDRA, and sequence analysis.
Cloning of 18S rDNA fragments amplified from R20, R90, and S20 DNA samples resulted in approximately 100 white colonies for each sample type. All white colonies were checked for the presence of inserts by PCR amplification of the 18S rDNA fragment followed by agarose gel electrophoresis. Seventy clones of the S20 clone library, 65 clones of the R20 clone library, and 74 clones of the R90 clone library contained inserts of the correct size. These clones were analyzed by DGGE to identify 18S rDNA fragments matching dominant bands in the corresponding DGGE community profiles. All clones matching a dominant band in the community profiles and sharing the identical electrophoretic mobility were given a code indicating their origin, sampling period, and matched band (e.g., R20A indicates rhizosphere, 20 days, and matching with band position A). The clone codes and their respective band positions are shown in Fig. 3. Although most of the 18S rDNA PCR products amplified from clones comigrated with bands in the DGGE community profiles, only a total of 87 clones matched 12 dominant bands (Fig. 3). The 18S rDNA fragments of these clones were further characterized by their ARDRA patterns, generating different operational taxonomic units (OTU). Inserts representing a unique OTU were selected for sequence analysis. Based on the partial sequence (approximately 500 bp), the 39 selected OTU were assigned to 20 different species (Table 1) with rather high similarity by using the ARB database with approximately 1,897 complete fungal 18S rDNA sequences and 1,035 partial sequences. In addition, the complete 1.65-kb sequence was determined for 13 clones to confirm sequence affiliation based on the partial sequence. However, two of the clones turned out to be chimeric.
TABLE 1.
Results of DGGE, ARDRA, sequence analysis and presumptive phylogenetic affiliations of bands
| Sample and clone type in DGGE | No. of clones matching band type | ARDRAa | Clone code | Closest relative (sequence length, ∼500 bp) | % Similarity | Taxon |
|---|---|---|---|---|---|---|
| Rhizosphere, 20 days | ||||||
| R20A | 3 | 3 | r20-61 | Alternaria cheiranthi | 100 | Ascomycetes-Pleosporales |
| R20B | 29 | 1 | r20-71 | Paraphaeosphaeria quadriseptatab | 99.4 | Ascomycetes-Pleosporales |
| 24 | r20-21 | Paraphaeosphaeria quadriseptatab | 99.5 | Ascomycetes-Pleosporales | ||
| 2 | r20-10 | Setosperia monoceras | 98.8 | Ascomycetes-Pleosporales | ||
| 1 | r20-12 | Setosperia monocerasb | 98.8 | Ascomycetes-Pleosporales | ||
| 1 | r20-111 | Phaeospheria nodorum | 96.9 | Ascomycetes-Pleosporales | ||
| Rhizosphere, 90 days | ||||||
| R90A | 4 | 1 | r90-1 | Gibberella pullcaris | 99.83 | Ascomycetes-Hypocreales |
| 1 | r90-63 | Gibberella pullcaris | 99.65 | Ascomycetes-Hypocreales | ||
| 1 | r90-78 | Gibberella pullcaris | 99.65 | Ascomycetes-Hypocreales | ||
| 1 | r90-85 | Raciborskiomyces longisetosumb | 99.9 | Ascomycetes-Pleosporales | ||
| R90B | 8 | 8 | r90-16 | Paraphaeosphaeria quadriseptatab | 99.2 | Ascomycetes-Pleosporales |
| R90C | 8 | 1 | r90-67 | Chaetomlum globosum | 97.2 | Ascomycetes-Sordariales |
| 1 | r90-74 | Raciborsklomyces longisetosum | 96.0 | Ascomycetes-Pleosporales | ||
| 5 | r90-4 | Raciborsklomyces longisetosumb | 99.3 | Ascomycetes-Pleosporales | ||
| 1 | r90-75 | Raciborsklomyces longisetosumb- Cladosporium cladosporoidesb | 100 | Ascomycetes-Pleosporales | ||
| R90D | 4 | 1 | r90-26 | Sporidiobolus johnsonii-S. salmonicolor | 100 | Basidiomycetes-Sporidiales |
| 1 | r90-5 | Sporidiobolus johnsonii-S. salmonicolor | 100 | Basidiomycetes-Sporidiales | ||
| 1 | r90-71 | Bullera unicab | 97.2 | Basidiomycetes-Filobasidiales | ||
| 1 | r90-95 | Sporidiobolus johnsonii-S. salmonicolor | 98.6 | Basidiomycetes-Sporidiobolaceae | ||
| R90E | 7 | 1 | r90-2 | Cryptococcus magnusb-Filobasidium uniguttulatumb | 99.8 | Basidiomycetes-Filobasidiales |
| 3 | r90-69 | Cryptococcus luteolus | 97.5 | Basidiomycetes-Filobasidiales | ||
| 1 | r90-13 | Cryptococcus luteolus | 99.3 | Basidiomycetes-Filobasidiales | ||
| 1 | r90-29 | Cryptococcus luteolus | 99.3 | Basidiomycetes-Filobasidiales | ||
| 1 | r90-66 | Cryptococcus luteolus | 99.8 | Basidiomycetes-Filobasidiales | ||
| R90F | 7 | 1 | r90-61 | Bullera oryzaeb | 99.7 | Basidiomycetes-Filobasidiales |
| 5 | r90-70 | Bullera hannae | 99.5 | Basidiomycetes-Filobasidiales | ||
| 1 | r90-87 | Bullera hannae | 99.5 | Basidiomycetes-Filobasidiales | ||
| Soil, 20 days | ||||||
| S20A | 5 | 1 | s20-5 | Aspergillus nomius | 99.8 | Ascomycetes-Eurotiales |
| 1 | s20-72 | Aspergillus terreus | 99.7 | Ascomycetes-Eurotiales | ||
| 1 | s20-2 | Blonectria ochroleuca | 99.6 | Ascomycetes-Hypocreales | ||
| 1 | s20-63 | Mortierella wolfil | 99.1 | Zygomycetes-Mucorales | ||
| 1 | s20-105 | Alternaria cheiranthi | 99.83 | Ascomycetes-Pleosporales | ||
| S20B | 4 | 4 | s20-67 | Cucurbita berberidisb | 99.5 | Ascomycetes-Pleosporales |
| S20G | 4 | 1 | s20-8 | Mortierella chlamydospora | 96.87 | Zygomycetes-Mucorales |
| 1 | s20-12 | Mortierella chlamydospora | 99.1 | Zygomycetes-Mucorales | ||
| 1 | s20-19 | Mortierella chlamydospora | 99.1 | Zygomycetes-Mucorales | ||
| 1 | s20-108 | Mortierella chlamydospora | 97.4 | Zygomycetes-Mucorales | ||
| S20H | 4 | 3 | s20-62 | Chaetomium globosum | 99.7 | Ascomycetes-Sordariales |
| 1 | s20-71 | Cladosporium cladosporoidesb | 100 | Ascomycetes-Pleosporales |
Number of clones grouped in each OTU generated by ARDRA analysis.
Sequence length, ∼1,600 bp.
All three clones matching band A had the same ARDRA pattern (clone type R20A), and the sequence determined for one representative indicated 100% similarity to Alternaria cheiranthi. Although bands with the same electrophoretic mobility as bands R20A were detectable in soil (S20A), sequence analysis indicated that different fungal populations contribute to band A. All five S20A clones had different ARDRA patterns, and thus all five S20A clones were sequenced. The S20A clones seem to belong to five different species and four genera (Aspergillus terreus, Aspergillus nomius, Bionectria ochroleuca, Mortirella wolfii, and Alternaria cheiranthi). All 29 R20B clones seem to belong to the Pleosporales. Sequencing of one representative of each ARDRA type revealed that 25 of the 29 R20B clones resembling two different ARDRA types were assigned to Paraphaeosphaeria quadriseptata, three R20B clones showed a high similarity to Setosperia monoceras, and one R20B clone was 97% similar to Phaeospheria nodorum. The sequenced representative of the four S20B clones was 100% similar to Cucurbita berberidis (Pleosporales). The clones matching bands D (R90D), E (R90E), and F (R90F) were assigned to different basidiomycetic yeasts (Filobasidiales, Sporidiales). Three of the R90D clones showed similarity to Sporidiobolus johnsonii (R90D), and two of them even had 100% similarity. One R90D clone was 97% similar to Bullera unica. All four S20G clones showed rather high similarity with Mortierella clamydiospora). Interestingly, band G was not detectable in soil samples taken 90 days after sowing. The cloning and sequencing approach provided information on the phylogeny of most dominant ribotypes in the DGGE patterns (Table 1; Fig. 5). The results showed that with exception of some Mortierella species (Zygomycetes) detected in soil samples, most of the fungal populations present in young roots and their corresponding bulk soil belonged to the Ascomycetes. However, interestingly, when the plants reached the senescent stage, half of the dominant bands resembled basidiomycetic yeasts (Filobasidiales, Sporidiobolaceae).
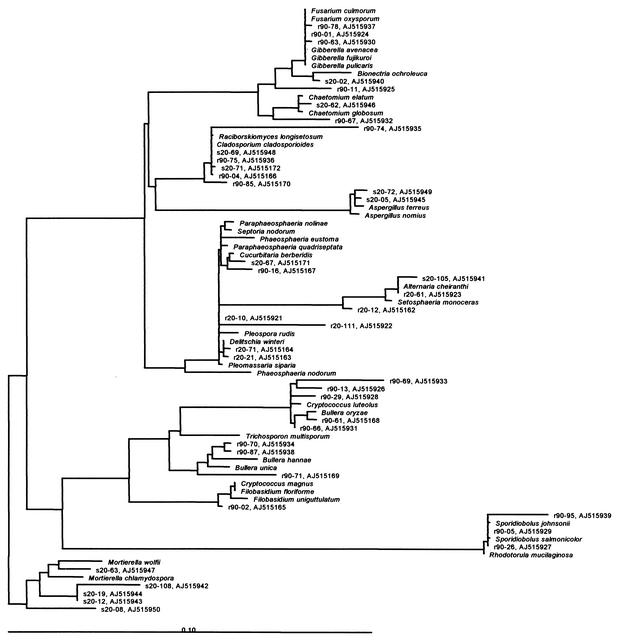
FIG. 5.
Phylogenetic tree for partial sequences of cloned 18S rDNA fragments and the most closely related fungi. The clones are indicated by their code and accession number (National Center for Biotechnology Information), respectively.
DISCUSSION
This is the first report on the composition and dynamics of fungal populations in bulk and maize rhizosphere soils in the tropics analyzed by cultivation-independent, DNA-based approaches. To amplify fungal 18S rDNA fragments from the DNA directly extracted from soil or root samples, we have used primers NS1 and FR1, which have recently been described by Vainio and Hantula (40). In contrast to the case with other primer systems used for fungal community analysis (3, 7, 17, 19, 24, 26, 32, 36, 42), the PCR product analyzed by DGGE is rather large (approximately 1.65 kb). Although it was previously suggested that for DGGE analysis the optimal size of PCR products should be around 500 bp (27), the DGGE profiles obtained with 1.65-kb PCR products amplified from soil or rhizosphere DNA were of surprisingly good quality. Most of the bands were clear and focused. In particular, the absence of single-stranded DNA, which often impairs the evaluation of silver-stained bacterial community fingerprints, improved the quality of the fungal community profiles. The primers used were designed to amplify the 18S rDNAs of all three major phyla of fungi, i.e., Basidiomycota, Ascomycota, and Zygomycota, and our cloning and sequencing results support this specificity. Furthermore, Vainio and Hantula (40) and Pennanen et al. (30) reported that no amplification products were obtained from bacterial and plant DNAs or from oomycota, nematode, or protozoan DNA.
DGGE analysis of 18S rDNA fragments amplified with NS1 and FR1 from DNA directly extracted from soil or root samples allowed us to monitor the shifts in the relative abundance of fungal populations during plant growth development. The DGGE analysis revealed that relative abundance of fungal populations in the rhizosphere of both cultivars, Nitroflint and Nitrodent, strongly shifted during plant growth. All three plant growth stages studied showed distinct profile characteristics. Some bands which were not detected in the profiles of young plants (20 days after sowing) became dominant. Most of the bands appearing in the rhizospheres of 40-day-old plants remained detectable when the plant reached the senescent stage (R90). The DGGE profiles of the rhizospheres of senescent plants (R90) were again relatively stable and had a high number of roughly equally abundant bands. The shifts in the fungal community patterns most likely occurred because of changing root morphology and root exudation patterns during plant development (5, 28). The release of organic substances by plant roots has an interesting ecological aspect, since it influences the nutrient availability in the rhizosphere and indirectly acts on the soil microorganisms that in turn influence plant growth (9, 10, 15, 34, 38, 45, 46). Temporal changes in the bacterial communities in the rhizosphere were indicated by different cultivation-based studies (2, 21, 25). Several other cultivation-independent studies employing molecular fingerprints also have recently shown, for different crops, a plant-dependent bacterial diversity and shifts of the bacterial community composition depending on plant growth developmental stages (8, 14, 20, 33, 35). However, the effects of changing root exudation during plant growth on the fungal community have not yet been demonstrated. Interestingly, the shifts in the relative abundance of the fungal populations were surprisingly similar for two independent growth periods. Furthermore, the analysis of three clone libraries (R20, R90, and S20) revealed that the shifts in the relative abundance of dominant fungal populations predicted on the basis of the comparative evaluation of DGGE profiles are an underestimate, since often different fungal populations contributed to bands with similar electrophoretic mobilities.
Similar to what is observed for bacterial 16S rDNA-based fingerprints (33), different, sometimes phylogenetically nonrelated fungi can have the same electrophoretic mobility in a DGGE run (reference 42 and this study). This could be demonstrated here for clones matching the two dominant DGGE bands (A and B) in the community pattern. While the R20A clones had identical ARDRA patterns and the sequence determined was 100% similar to that of Alternaria cheiranthi, a much greater diversity was reflected by the five clones with an electrophoretic mobility matching that of band A (S20A). The S20A clones were assigned to five different species, based on the partial sequence. Only one of the five S20A clones showed a high similarity to Alternaria cheiranthi. Apparently, Alternaria cheiranthi was enriched in the vicinity of the roots of young maize plants. Thus, the rhizosphere effect, namely, a reduced diversity and an increased relative abundance of a few populations (8, 35), could be observed not only in the DGGE profiles but also for clones matching band A. Although the number of dominant fungal populations in the rhizosphere increased when the plants reached the senescent stage, the rhizosphere profiles were different from the corresponding soil DGGE patterns. Gomes et al. (8) reported also an increased number of equally abundant bacterial populations in the rhizosphere of senescent maize. However, in contrast to the fungal community profiles, the eubacterial rhizosphere patterns largely resembled those of soil. Thus, at all maize development stages analyzed here, a pronounced rhizosphere effect was found, and this could be quantified by computer-assisted analysis. Interestingly, shifts in the relative abundance of fungal populations were even observed for soil. Compared to the shifts detected in the rhizosphere, these changes were less pronounced. While the DGGE analysis of 18S rDNA fragments amplified from community DNA was sensitive enough to detect the rhizosphere effect and shifts in the relative abundance of fungal populations during plant growth development, no differences were detected between the two cultivars. Recently, it was also reported that bacterial DGGE fingerprints did not reveal any differences between these two cultivars (8). However, considering the level of resolution of 18S rDNA-based analysis, it cannot be excluded that at a finer level of resolution, differences in the composition and activity of fungi in the rhizosphere would have become detectable. Thus, Dalmastri et al. (6) reported that the diversity of Burkholderia cepacia in the rhizosphere of maize was affected by the cultivar.
Sequencing of approximately 500 bp of 39 clones, each representing a unique OTU based on the ARDRA analysis, allowed to identify 19 different species belonging to the Ascomycota (Pleosporales, Hypocreales, Sordariales, and Eurotiales), Basidiomycota (Filobasidiales, Sporidiales) and Zygomycota (Mucorales). In several cases different ARDRA types were assigned to the same species (e.g., R20B, R90A, and R90E) (data not shown). These OTU share similar or identical sequences in the approximately 0.5 kb which was sequenced but likely have sequence differences in the remaining 1.1 kb which were detected by ARDRA. However, even sequencing the complete 1.65-kb fragment confirmed the affiliation. The similarity to sequences in the database was surprisingly high. All clones showed a similarity to database entries of more than 95%. Interestingly, the clone library of 18S rDNA amplified with primers EF4 and EF3 from wheat rhizosphere DNA (36) also contained the basidiomycete Bullera and Cryptococcus as well as the zygomycete Mortierella polycephala. In our study, clones which showed a high similarity to Mortierella spp. were only detected in the clone library obtained from soil DNA, while Bullera spp. seemed to belong to the dominant fungal populations in the rhizospheres of senescent maize plants. Populations belonging to the genus Pleospora seemed to be the most dominant fungi in young roots. However, although a similar intensity of bands R20A and R20B in the R20 community profiles indicated that fungal populations with these electrophoretic mobilities seemed to be equally abundant, in the R20 clone library 29 clones matched band B while only three clones matching band A were obtained. Since the same primer pair (except for the GC clamp) was used for 18S rDNA amplification, this might point to a cloning bias, although a PCR bias cannot be ruled out. Another bias which might have affected the DGGE patterns obtained is chimera formation.
In this study we could show that fungal populations in the rhizosphere of maize grown in tropical soils also undergo pronounced changes during the development of the plant. As previously reported only for bacterial communities, an increased relative abundance of some fungal populations in the vicinity of the maize roots was found. The cloning and sequencing approach proved to be crucial to provide information on the phylogeny of dominant bands and to evaluate the ribotype diversity behind the DGGE bands. Based on our data, we strongly suggest that 18S rDNA-based fingerprints of complex fungal communities should be accompanied by use of a clone library for each sample type if a better understanding of the true diversity is to be gained. Furthermore, although it was not done in this study, an attempt to link both cultivation-independent and -dependent approaches should provide better insights into fungal diversity in bulk and rhizosphere soils.
Acknowledgments
This study was financed by a bilateral (German-Brazilian) cooperation in science and technology (grant WTZ 98/005). Support for consumables was provided by Monsanto. O. Fagbola was financed by a fellowship of the Alexander von Humboldt-Stiftung.
We are grateful to G. Deml and M. Götz for carefully reading the manuscript and for discussion.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg, G., N. Roskot, A. Steidle, L. Eberl, A. Zock, and K. Smalla. 2002. Plant-dependent genotypic and phenotypic diversity of antagonistic rhizobacteria isolated from different Verticillium host plants. Appl. Environ. Microbiol. 68:3328-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borneman, J., and R. J. Hartin. 2000. PCR primers that amplify fungal rRNA genes from environmental samples. Appl. Environ. Microbiol. 66:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bridge, P., and B. Spooner. 2001. Soil fungi: diversity and detection. Plant Soil 232:147-154. [Google Scholar]
- 5.Brimecombe, M. J., F. A. De Leij, and J. M. Lynch. 2001. The effect of root exudates on rhizosphere microbial population, p. 95-140. In R. Pinton, Z. Varanini, and P. Nannipieri (ed.), The rhizosphere. Marcel Dekker, Inc., New York, N.Y.
- 6.Dalmastri, C., L. Chiarini, C. Cantale, A. Bevivino, and S. Tabaccioni. 1999. Soil type and maize cultivar affect the genetic diversity of maize root-associated Burkholderia cepacia populations. Microb. Ecol. 38:273-284. [DOI] [PubMed] [Google Scholar]
- 7.Glandorf, D. C. M., P. Verheggen, T. Jansen, J.-W. Jorritsma, E. Smit, P. Leeflang, K. Wernars, L. S. Thomashow, E. Laureijs, J. E. Thomas-Oates, P. A. H. M. Bakker, and L. C. van Loon. 2001. Effect of genetically modified Pseudomonas putida WCS358r on the fungal rhizosphere microflora of field-grown wheat. Appl. Environ. Microbiol. 67:3371-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomes, N. C. M., H. Heuer, J. Schönfeld, R. Costa, L. Hagler-Mendonca, and K. Smalla. 2001. Bacterial diversity of the rhizosphere of maize (Zea mays) grown in tropical soil studied by temperature gradient gel electrophoresis. Plant Soil 232:167-180. [Google Scholar]
- 9.Grayston, S. J., D. Vaughan, and D. Jones. 1997. Rhizosphere carbon flow in trees, in comparison with annual plants: the importance of root exudation and its impact on microbial activity and nutrient availability. Appl. Soil Ecol. 5:29-56. [Google Scholar]
- 10.Grayston, S. J., S. Wang, C. D. Campbell, and A. C. Edwards. 1998. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 30:369-378. [Google Scholar]
- 11.Hawksworth, D. L., and A. Y. Rossman. 1997. Where are all the undescribed fungi? Phytopathology 87:888-891. [DOI] [PubMed] [Google Scholar]
- 12.Heuer, H., G. Wieland, J. Schönfeld, A. Schönwälder, N. C. M. Gomes, and K. Smalla. 2001. Bacterial community profiling using DGGE or TGGE analysis, p. 177-190. In P. Rouchelle (ed.), Environmental molecular microbiology: protocols and applications. Horizon Scientific Press, Wymondham, United Kingdom.
- 13.Heuer, H., K. Hartung, G. Wieland, I. Kramer, and K. Smalla. 1999. Polynucleotide probes that target a hypervariable region of 16S rRNA genes to identify bacterial isolates corresponding to bands of community fingerprints. Appl. Environ. Microbiol. 65:1045-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heuer, H., R. M. Kroppenstedt, J. Lottmann, G. Berg, and K. Smalla. 2002. Effects of T4 lysozyme release from transgenic potato roots on bacterial rhizosphere communities are negligible relative to natural factors. Appl. Environ. Microbiol. 68:1325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaeger, C. H., III, S. E. Lindow, W. Miller, E. Clark, and M. K. Firestone. 1999. Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl. Environ. Microbiol. 65:2685-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarosz, A. M., and A. L. Davelos. 1995. Effects of disease in wild plant populations and the evolution of pathogen aggressiveness. New Phytol. 129:371-387. [Google Scholar]
- 17.Kowalchuk, G. A. 1999. Fungal community analysis using denaturing gradient gel electrophoresis (DGGE), p. 3.4.6.1-16. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 18.Kowalchuk, G. A. 1999. New perspectives in analysing fungal communities in terrestrial ecosystems. Curr. Opin. Biotechnol. 10:247-251. [DOI] [PubMed] [Google Scholar]
- 19.Kowalchuk, G. A., S. Gerards, and J. W. Woldendorp. 1997. Detection and characterization of fungal infections of Ammophila arenaria (marram grass) roots by denaturing gradient gel electrophoresis of specifically amplified 18S ribosomal DNA. Appl. Environ. Microbiol. 63:3858-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lottmann, J., H. Heuer, J. de Vries, A. Mahn, K. Düring, W. Wackernagel, K. Smalla, and G. Berg. 2000. Establishment of introduced antagonistic bacteria in the rhizosphere of transgenic potatoes and their effect on the bacterial community. FEMS Microb. Ecol. 33:41-49. [DOI] [PubMed] [Google Scholar]
- 21.Mahaffee, W. F., and J. W. Kloepper. 1997. Temporal changes in the bacterial communities of soil, rhizosphere, and endorhiza associated with field-grown cucumber (Cucumis sativus L.). Microb. Ecol. 34:210-223. [DOI] [PubMed] [Google Scholar]
- 22.Marschner, P., D. E. Crowley, and R. Lieberei. 2001. Arbuscular mycorrhizal infection changes the bacterial 16S rDNA community composition in the rhizosphere of maize. Mycorrhiza 11:297-302. [DOI] [PubMed] [Google Scholar]
- 23.Martin, F. M., S. Perotto, and P. Bonfante. 2001. Mycorrhizal fungi: a fungal community at the interphase between soil and roots, p. 263-296. In R. Pinton, Z. Varanini, and P. Nannipieri (ed.), The rhizosphere. Marcel Dekker, Inc., New York, N.Y.
- 24.May, L. A., B. Smiley, and M. G. Schmidt. 2001. Comparative denaturing gradient gel electrophoresis analysis of fungal communities associated with whole plant corn silage. Can. J. Microbiol. 47:829-841. [DOI] [PubMed] [Google Scholar]
- 25.Miller, H. J., G. Henken, and J. A. van Veen. 1989. Variation and composition of bacterial populations in the rhizospheres of maize, wheat, and grass cultivars. Can. J. Microbiol. 35:656-660. [Google Scholar]
- 26.Möhlenhoff, P., L. Müller, A. A. Gorbushina, and K. Petersen. 2001. Molecular approach to the characterization of fungal communities: methods for DNA extraction, PCR amplification and DGGE analysis of painted art objects. FEMS Microbiol. Lett. 195:169-173. [DOI] [PubMed] [Google Scholar]
- 27.Muyzer, G., and K. Smalla. 1998. The need for DGGE and TGGE in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 28.Neumann, G., and V. Römheld. 2001. The release of root exudates as affected by the plant's physiological status, p. 41-93. In R. Pinton, Z. Varanini, and P. Nannipieri (ed.), The rhizosphere. Marcel Dekker, Inc., New York, N.Y.
- 29.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 30.Pennanen, T., L. Paavolainen, and J. Hantula. 2001. Rapid PCR-based method for the direct analysis of fungal communities in complex environmental samples. Soil Biol. Biochem. 33:697-699. [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Schabereiter-Gurtner, C., G. Pinar, W. Lubitz, and S. Rölleke. 2001. Analysis of fungal communities on historical church window glass by denaturing gradient gel electrophoresis and phylogenetic 18S rDNA analysis. J. Microbiol. Methods 47:345-354. [DOI] [PubMed] [Google Scholar]
- 33.Schmalenberger, A., and C. C. Tebbe. 2002. Bacterial community composition in the rhizosphere of a transgenic, herbicide-resistant maize (Zea mays) and comparison to its non-transgenic cultivar Bosphore. FEMS Microbiol. Ecol. 40:29-37. [DOI] [PubMed] [Google Scholar]
- 34.Semenov, A. M., A. H. C. van Bruggen, and V. V. Zelenev. 1999. Moving waves of bacterial populations and total organic carbon along roots of wheats. Microbiol. Ecol. 37:116-128. [DOI] [PubMed] [Google Scholar]
- 35.Smalla, K., G. Wieland, A. Buchner, A. Zock, J. Parzy, S. Kaiser, N. Roskot, H. Heuer, and G. Berg. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67:4742-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smit, E., P. Leeflang, B. Glandorf, J. D. van Elsas, and K. Wernars. 1999. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2614-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Söderberg, K. H., P. A. Olsson, and E. Baath. 2002. Structure and activity of the bacterial community in the rhizosphere of different plant species and the effect of arbuscular mycorrhizal colonisation. FEMS Microbiol. Ecol. 40:223-231. [DOI] [PubMed] [Google Scholar]
- 38.Sørensen, J. 1997. The rhizosphere as a habitat for soil microorganisms, p. 21-45. In J. D. van Elsas, J. T. Trevors, and E. M. H. Wellington (ed.), Modern soil microbiology. Marcel Dekker, Inc., New York, N.Y.
- 39.Thorn, G. 1997. The fungi in soil, p. 63-127. In J. D. van Elsas, J. T. Trevors, and E. M. H. Wellington (ed.), Modern soil microbiology. Marcel Dekker, Inc., New York, N.Y.
- 40.Vainio, E. J., and J. Hantula. 2000. Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycol. Res. 104:927-936. [Google Scholar]
- 41.Van der Heijden, M. G. A., J. N. Klironomos, M. Ursic, P. Moutoglis, R. Streitwolf-Engel, T. Boller, A. Wiemken, and I. R. Sanders. 1998. Mycorrhizal fungal diversity determines plant diversity, ecosystem variability and productivity. Nature 396:69-71. [Google Scholar]
- 42.Van Elsas, J. D., G. F. Duarte, A. Keijzer-Wolters, and E. Smit. 2000. Analysis of the dynamics of fungal-specific PCR of soil DNA followed by denaturing gradient gel electrophoresis. J. Microbiol. Methods 43:133-151. [DOI] [PubMed] [Google Scholar]
- 43.Varadaraj, K., and D. M. Skinner. 1994. Denaturants or cosolvents improve the specificity of PCR amplification of a G+C-rich DNA using genetically-engineered DNA-polymerases. Gene 140:1-5. [DOI] [PubMed] [Google Scholar]
- 44.von Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 45.Westover, K. M., A. C. Kennedy, and S. E. Kelley. 1997. Patterns of rhizosphere microbial community structure associated with co-occurring plant species. J. Ecol. 85:863-873. [Google Scholar]
- 46.Yang, C.-H., and D. E. Crowley. 2000. Rhizosphere microbial community structure in relation to root location and plant iron nutritional status. Appl. Environ. Microbiol. 66:345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]