Abstract
The TATA box-binding activity of transcription factor IID (TFIID) is autoinhibited by the N-terminal domain of the Drosophila TATA box-binding protein- (TBP) associated factor 230/yeast TBP-associated factor 145 subunit, which binds to the TATA box-binding domain of TBP by mimicking the TATA box structure. Here, we propose a mechanism of transcriptional activation that involves antirepression of this autoinhibitory activity by transcriptional activators. Like the autoinhibitory domain of TFIID, various acidic activators interact with the TATA box-binding domain of TBP. Moreover, the autoinhibitory domain of TFIID, which is known to interact with only the TATA box-binding domain of TBP, acts as an activation domain when fused to the GAL4 DNA-binding domain, indicating that interaction with the TATA-binding domain of TBP is crucial for activation of transcription. In a reciprocal fashion, the acidic activation domains can function as the autoinhibitory domain when the latter is replaced by the former within TFIID. These results indicate that activation domains and the autoinhibitory domain of TFIID are interchangeable, supporting a role for transcriptional activators as antirepressors of the autoinhibitory activity of the TATA box binding of TFIID.
Transcriptional initiation of eukaryotic protein-encoding genes requires at least six general transcription initiation factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) in addition to RNA polymerase II (reviewed in ref. 1). Furthermore, biochemical analyses with individual factors have defined a sequence of steps that, in vitro, lead to the formation of a preinitiation complex on a TATA box-containing promoter (reviewed in ref. 1). Such stepwise model has been challenged by the discovery of holoenzyme complexes containing RNA polymerase II, a subset of general transcription factors, SRB/MED mediators, and other factors (reviewed in ref. 2). An alternative model postulates that the holoenzyme enters the preinitiation complex as a preassembled unit (2, 3). Although the nature of the assembly pathway most relevant to the in vivo context remains unclear, access of TFIID to the core promoter is likely to be a critical step in any pathway, given that only TFIID binds to the promoter in a sequence-specific manner. In support of this view, in vitro recruitment experiments using immobilized promoters demonstrated that the holoenzyme is not recruited independently of TFIID and TFIIA (4).
TFIID is a multimeric protein complex consisting of the TATA box-binding protein (TBP) and >10 distinct TBP-associated factors (TAFs), which are conserved from yeast to man (reviewed in ref. 5). In vitro transcriptional experiments in reconstituted systems demonstrated that, whereas TFIID can support both basal and activated transcription, TBP mediates only basal transcription (reviewed in ref. 5). Consistent with the view that TAFs are important cofactors for activated transcription, several TAFs have been shown to provide interaction sites for distinct activators (reviewed in ref. 6). Moreover, studies using reconstituted recombinant TFIID complexes showed a correlation of activator function and the ability of individual TAFs to selectively interact with various activation domains in vitro (reviewed in ref. 6). However, this proposal has been challenged by studies in yeast demonstrating that the conditional knockout of yeast (y)TAF145 or yTAF90 has no significant effect on global gene expression (reviewed in ref. 7). More recently, histone-like TAFs were shown to be required for transcription more broadly than other TAFs but still not universally (reviewed in ref. 7). Consistent with these observations in vivo, TAF-independent activation was demonstrated in transcriptional experiments in vitro, using TFIID-depleted HeLa nuclear extracts, which could possess activator targets other than TAFs (8), or even with a highly purified cell-free transcription system (9, 10). Furthermore, transcription activation is reconstituted by supplementing purified yeast holoenzyme with only TBP (11, 12). These observations suggest that activators contribute to activated transcription by interacting with multiple targets (e.g., basal factors, TAFs, SRB mediators, or other unknown cofactors).
Although TAFs are well recognized as positive cofactors, in our view, at least certain TAFs are negative cofactors. Importantly, highly purified TFIID manifests lower transcriptional activity than TBP alone (13) and binds to the core promoter poorly (reviewed in ref. 14). This inhibitory activity for promoter binding in TFIID is suppressed by limited proteolysis of TFIID or by TFIIA (14, 15). These results strongly suggest that this inhibitory activity could be intrinsic to TFIID and derived from TAF(s) and is sensitive to proteases. Consistent with this idea, Drosophila (d)TAF230, or the homologous yTAF145, inhibits TBP binding to the TATA box when these TAFs are mixed with TBP in vitro (16–18). Mutational analyses of dTAF230 indicate that the N-terminal 156 residues inhibit TATA box binding through direct interaction with TBP (19, 20). This N-terminal domain (designated as TAND; TAF N-terminal domain), has been dissected into subdomain 1 (dTAND-1; residues 11–77) and subdomain 2 (dTAND-2; residues 82–156), which bind to the concave undersurface and the convex upper surface of TBP in a competitive fashion with the VP16 activation domain and TFIIA, respectively (20, 21). Importantly, NMR structural analyses revealed that the dTAND-1 interacts with the TATA-binding undersurface of TBP by mimicking the minor groove surface of a partially unwound TATA box in the TBP-TATA complex (22).
Here, we show the functional conservation between acidic activation domains and the yTAF145 subdomain 1 (yTAND-1). When fused to the DNA-binding domain, yTAND-1 functions as an activation domain in yeast. Conversely, activation domains function as yTAND-1 when yTAND-1 is replaced with acidic activation domains within TFIID. Furthermore, we show that the interaction with the concave surface of TBP is crucial for the functions of both activators and TAND-1. These results indicate that transcriptional activation steps involves suppression of the inhibitory activity of TAND-1 by transcriptional activators.
Experimental Procedures
Phenotypic Analyses.
Standard techniques were used for the growth and transformation of the yeast strains (23). Yeast strains for phenotypic analyses are derived from Y22.1 (18). To determine the abilities of the ytaf145 mutants to support yeast cell growth at elevated temperatures, a plasmid shuffle technique was used (24). 5-Fluoroorotic acid-resistant colonies harboring wild-type or mutant yTAF145 genes on plasmids derived from pRS314 (TRP1) were incubated on yeast extract/peptone/dextrose plates for 3–4 days at 30°C or 37°C to compare their growth properties.
Plasmids.
To make constructs encoding the activation domain-yTAF145 chimeras, yTAND-1 in pM11 (the yTAF145 gene in pRS314) and pM734 (the yTAF145 gene encoding residues 6–96 in pUC19) (21) were replaced by DNA fragments encoding VP16 (457–490), VP16 (470–490), GAL4 (842–874), the Epstein–Barr virus nuclear protein 2 (426–462), or Sp1 (82–263), which were prepared by amplifying corresponding genes by PCR as BamHI–BssHII fragments. yTAND-2 was removed from the resulting plasmids by digesting with BssHII and MluI and recircularizing with DNA ligase, as required. For bacterial expression of the activation domain-yTAND-2 chimeric proteins, the BamHI–EcoRI fragments of pM734 derivatives were subcloned into pGEX2T (Amersham Pharmacia Biotech). To prepare yeast expression of GAL4 fusion constructs, corresponding regions shown in Fig. 3 were amplified by PCR as EcoRI–BamHI fragments and were subcloned into pGBT9 (CLONTECH).
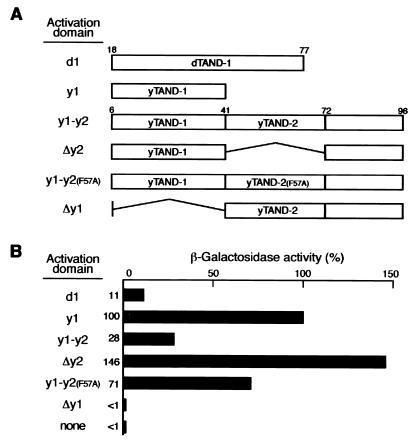
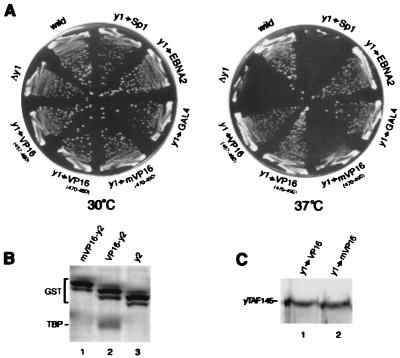
Figure 3.
TAND-1 functions as an activator when fused to a DNA-binding domain. (A) Overall structures of GAL4 fusions tested in this experiment. Fragments shown here were fused with GAL4 DNA-binding domain (residues 1–147) for expression in yeast. Numbers on the bars indicate the amino acid positions in the respective factors. (B) GAL4-dependent transcriptional activation in yeast. Expression vectors described in A were transformed into yeast, and transcription activity was determined by measuring the lacZ-reporter activity. The relative β-galactosidase activity of authentic activation domains measured under the same conditions was as follows: VP16(457–490), 430%; VP16(470–490), 220%; GAL4, 300%; EBNA2, 270%; and Sp1, <1%.
Alanine-Scanning Mutation Analysis.
Site-directed mutagenesis (25) was performed to introduce alanine-scanning substitutions into yTAND-1 of the yTAF145 gene, using pM11 [the yTAF145 gene in pRS314] and pM5 [the yTAF145 gene encoding residues 6–96 in pBluescript II (Stratagene)] (21) as templates.
Preparation of Recombinant Proteins and Protein–Protein Interaction Experiments.
The N-terminal region of yTAF145 and its derivatives were expressed in Escherichia coli (DH5α) as glutathione S-transferase (GST) fusion proteins. Yeast TBP was expressed as a histidine-tagged protein using a pET system (Novagen). The preparation of these proteins and GST pulldown experiments were conducted as described previously (18).
β-Galactosidase Assay.
GAL4 fusions were expressed in yeast strain SFY526, which contains an integrated lacZ reporter driven by the GAL1 promoter. Yeast strains were grown in selective medium and assayed for β-galactosidase activity according to the manufacturer's protocol (CLONTECH).
Electrophoretic Mobility-Shift Assay.
Electrophoretic mobility-shift assay was performed as described previously (16) with affinity purified GST-activation domain-TAND derivatives and yeast TBP. Adenovirus major late promoter (−119 to +61 of the transcription initiation site) was used as a probe.
Results
Acidic Activators Bind to the TATA-Binding Domain of TBP.
A previous report showed that the mutation L114K in yTBP, which maps to the concave surface of TBP, which interacts with the TATA box, impairs interaction of TBP with the VP16 activation domain in vitro (26). Consistent with these results on VP16 interactions, this TBP mutant cannot mediate VP16-induced activation in vitro, although it can still provide a basal level of transcription (26). To obtain further evidence that the concave face of TBP is a bona fide target of activators, we performed the following experiments. The N terminus of yTAF145 possesses two domains that interact with TBP, yTAND-1 and yTAND-2, which interact with the concave and convex surfaces of TBP, respectively (18, 21). Consistent with previous observations (18, 21), neither yTAND-1 nor yTAND-2 alone forms a stoichiometric complex with TBP (Fig. 1 A and B, lanes 2 and 3). On the other hand, when yTAND-1 and yTAND-2 are fused, a stoichiometric complex is formed with TBP (lane 1) (21). Given that the VP16 activation domain and TAND-1 have similar TBP-binding characteristics (20), we considered the possibility that the VP16 activation domain forms a stoichiometric complex with TBP when fused to TAND-2. Consistent with previous reports (27–29), the VP16 activation domain (residues 457∼490 or 470∼490) per se interacts very weakly with TBP. Although TBP bound to glutathione S-transferase-VP16 (GST-VP16) can be detected by immunoblotting (Fig. 1B, lanes 5 and 7), this interaction is far from stoichiometric and barely detectable by Coomassie blue staining (Fig. 1A, lanes 5 and 7). Importantly, as predicted, the VP16 activation domain forms a stoichiometric complex with TBP, which is detectable with Coomassie blue staining, when fused to TAND-2 (lanes 4 and 6). These results strengthen our view that the VP16 activation domain interacts with the concave surface of TBP, and this interaction is stabilized by fusing the VP16 activation domain to TAND-2, which interacts with the convex surface of TBP.
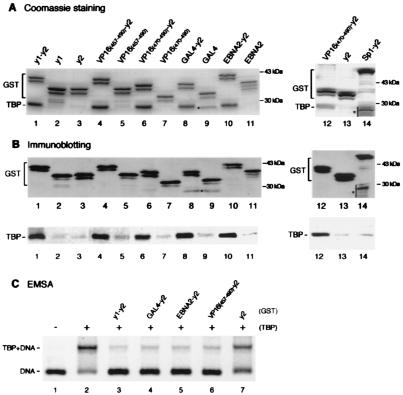
Figure 1.
Acidic activation domains form stable complexes with TBP and inhibit TBP binding to the TATA box when fused to TAND-2. (A and B) VP16 (residues 457–490 or 470–490; lanes 4, 6, and 12), GAL4 (lane 8), EBNA2 (lane 10), and Sp1 (lane 14) activation domains as well as yTAND-1 (lanes 1) were fused to yTAND-2 (y2) and expressed in E. coli as GST-tagged proteins. These activation domains (lanes 5, 7, 9, and 11), yTAND-1 (lane 2), and yTAND-2 (lanes 3 and 13) also were expressed as GST-tagged proteins. GST fusion proteins were incubated with an equimolar amount of yTBP and purified by glutathione Sepharose. The purified materials were analyzed by SDS-PAGE followed by Coomassie blue staining (A) or immunoblotting with anti-GST and anti-TBP antibodies (B). The positions of GST fusions and copurified TBP are indicated. GST fusions appeared as multiple bands due to the protease-hypersensitive sites near the C terminus. Although the bands marked by asterisks in lanes 9 and 14 migrated near TBP, these bands derived from GST fusion proteins according to immunoblotting shown in B. (C) Inhibition of TBP binding to the TATA box by GST-activation domain-TAND-2 chimeras. Gel retardation assays were performed with equimolar amounts of TBP and GST-fusion derivatives that were affinity purified. The positions of TBP-DNA complex and free probe were indicated at the left. Much larger amounts of proteins were required to see the inhibitory effects for activation domains alone (data not shown).
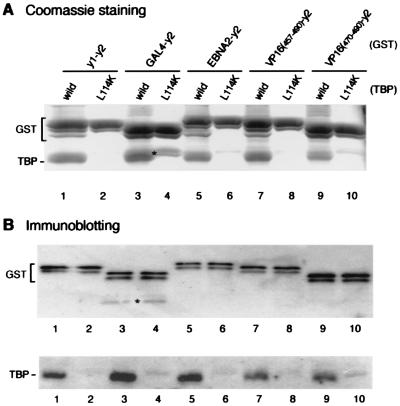
To determine whether the concave surface of TBP is a target for various acidic activators, GAL4 (30) and EBNA2 (31) activation domains were tested in a similar way. Like the VP16 activation domain, the GAL4 or EBNA2 activation domain interacts weakly with TBP, the interaction being barely detectable by Coomassie blue staining (Fig. 1, lanes 9 and 11). However, both GAL4 and EBNA2 activation domains form stoichiometric complexes with TBP when connected with TAND-2 (lanes 8 and 10). These results demonstrate that various acidic activators, including the VP16, GAL4, and EBNA2 activation domains, bind to the concave face of TBP in a manner similar to TAND-1. In support of these results, the L114K mutation in the concave TBP surface almost completely abolishes the interaction with these fusions of activation domains when fused to TAND-2 (Fig. 2A and B, even lanes). In addition, these activation domains can inhibit TBP binding to TATA box just like TAND-1 when connected with TAND-2 (Fig. 1C).
Figure 2.
Acidic activation domains interact with the concave surface of TBP. yTAND-1 (lanes 1 and 2) and various activation domains including GAL4 (lanes 3 and 4), EBNA2 (lanes 5 and 6), VP16 (residues 457–490 or 470–490; lanes 7–10) were fused to yTAND-2 (y2), and expressed as GST fusion proteins. These GST proteins were incubated with equimolar amounts of wild-type (odd lanes) or L114K mutant TBP (even lanes). After GST precipitation, the purified materials were analyzed by Coomassie blue staining (A) or immunoblotting with anti-GST and anti-TBP antibodies (B) as described in the Fig. 1 legend. Degraded GST proteins that closely migrated with TBP are indicated by asterisks.
To understand whether the concave surface of TBP is a common target for various types of activators, we tested the glutamine-rich activation domain of Sp1. Although Drosophila TAF110 or human TAF130 are known to be targets for Sp1 (32), there is a report showing interaction between Sp1 and TBP (33). Thus, the Sp1 activation domain was fused to TAND-2 and tested for interaction with TBP (Fig. 1, lane 14). Almost no interaction between TBP and the Sp1-TAND-2 fusion was detected. Thus, these results indicate that the concave surface of TBP is a target for various acidic activators, but not for all activators.
Interaction with the TATA-Binding Domain of TBP Is Important for Transcriptional Activation.
Because both TAND-1 and the acidic activators interact with the concave surface of TBP in a similar manner, we felt that TAND-1 might act as an activation domain when fused to a DNA-binding domain. To test this possibility, TAND-1 was fused to the GAL4 DNA-binding domain (residues 1–147) and transfected into yeast harboring the integrated GAL1–LacZ-reporter gene to test for GAL4-dependent activation in vivo (Fig. 3A, B). Importantly, the dTAF230 subdomain 1 (dTAND-1) functions as an activation domain. As demonstrated by NMR structural analysis (22), dTAND-1 binds to the concave surface of TBP by mimicking the TATA box structure unwound by TBP (22). Thus, these results strongly suggest that interaction with the concave surface of TBP is important for transcriptional activation in vivo, at least in the GAL1 promoter.
The yTAF145 subdomain 1 (yTAND-1) binds to the concave surface of TBP less stably than does dTAND-1 (18, 20). Regardless of the weaker interaction, yTAND-1 functions as a better activator in yeast than does dTAND-1 (Fig. 3B). These results suggest that an unstable interaction with the concave surface of TBP might be an advantage for activating transcription (see Discussion). In support of this view, the affinity of yTAND-1 for TBP is similar to that of the VP16 activation domain by in vitro interaction experiments (data not shown).
The yTAF145 fragment containing both yTAND-1 and yTAND-2 (y1-y2), which provides stable interaction with TBP, functions as a weaker activation domain than does yTAND-1 alone (y1) (Fig. 3 A and B). Internal deletion (Δy2) or point mutation (y1-y2 F57A) within yTAND-2 increases its potential as an activator. Moreover, yTAND-2 itself (Δy1) does not function as an activation domain, although the affinity of the yTAND-2 for TBP is similar to that of yTAND-1 (21). These results indicate that TAND-2 not only has no potential as an activation domain but also has an inhibitory effect when fused to TAND-1. This effect might be due to inhibition of TFIIA binding to TBP because TFIIA and TAND-2 competitively bind to TBP (18). Alternatively, the TAND-1/TAND-2 fusion might bind TBP too tightly for effective activation as suggested above.
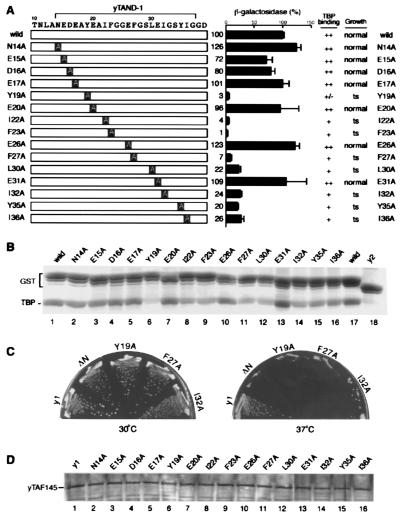
Next, we tested a series of alanine-scanning mutants within yTAND-1 to determine the correlation between binding to the concave face of TBP and the potential of TAND-1 as an activator (Fig. 4). Alanine-scanning mutations were introduced into yTAND-1 and expressed as fusion constructs with the GAL4 DNA-binding domain in yeast to test for activation. To test the TBP-binding activity in vitro, yTAND-1 mutants were connected to yTAND-2 and expressed in E. coli as GST fusions. Importantly, we found a general correlation between potential as an activator and the TBP-binding ability. The yTAND-1 mutants with deficiency in TBP-binding activity also completely (Y19A, I22A, and F23A) or partially (F27A, L30A, I32A, Y35A, and I36A) lose their potential as activators. Taken together with the results shown in Fig. 3, we thus conclude that interaction with the concave surface of TBP is a crucial aspect of transcriptional activation.
Figure 4.
The interaction of yTAND-1 with the concave surface of TBP is essential for its functions. (A) Alanine-scanning substitution mutants of yTAND-1 used in this study are schematically presented. These mutants were used for TBP-binding, growth, and transcription assays, and the results are summarized. For transcription analysis, indicated fragments were expressed in yeast as fusions with the GAL4 DNA-binding domain and tested for GAL4-dependent transcription, as described in the Fig. 3 legend. Note that although the point mutations within the yTAF145 gene caused only subtle effects, more obvious, but correlated, effects were observed when the point mutations were introduced into the yTAF145 gene in which TAND-2 (43–71 aa) had been internally deleted. The results with the gene background lacking TAND-2 are represented in this figure. (B) TBP-binding activity of the yTAND-1 mutants. The yTAND-1 mutants were fused to yTAND-2 and expressed in E. coli as GST-tagged proteins. Interaction of these fusions with TBP was determined as described in the Fig. 1 legend, and a Coomassie blue-stained gel is represented. (C) Growth phenotype of yeast carrying a defined mutation in yTAF145 gene. The yTAF145 genes harboring alanine-scanning mutations were introduced into yeast, replacing the wild-type gene by plasmid shuffling. After plasmid shuffling, the resulting strains were grown on yeast extract/peptone/dextrose plates at 30°C (Left) and 37°C (Right) for 3 days. Results are summarized in A, and photographs of yeast plates represent the selected mutants. Strains ΔN and y1 express yTAF145 proteins lacking the whole TAND (6–96 aa) and TAND-2 (43–71 aa), respectively (D). Expression level of the yTAF145 mutants in yeast. Yeast strains harboring yTAF145 mutants were grown at 25°C and then shifted to 37°C. Cells were harvested 24 h after the temperature shift and tested for expression level of yTAF145 mutants by immunoblotting.
Interaction with the TATA-Binding Domain of TBP Is Essential for Function of TAND-1.
We previously demonstrated that deletion of TAND-1 within the yTAF145 gene leads to a growth defect at 37°C (21). Thus we next investigated whether recognition of the concave surface of TBP by TAND-1 is important for growth at 37°C. To test this correlation, the alanine-scanning mutations shown in Fig. 4 were introduced into the yTAF145 gene. The yTAF145 genes with mutations within TAND-1 were introduced into yeast, replacing the wild-type gene by plasmid shuffling. After plasmid shuffling, the resulting strains were subjected to growth assay at 37°C. When we tested these mutations in strains harboring both TAND-1 and TAND-2, they showed smaller effects on growth (data not shown). To produce larger effects, we also tested them in strains that express yTAF145 gene of which the whole TAND-2 region was deleted because the growth of such strains depends much more strongly on the TAND-1 function (21) (Fig. 4 A and C). Importantly, we found good correlation between growth at 37°C and the TBP-binding ability of TAND-1. The TAND-1 mutants with deficiency in TBP-binding activity also fail to support normal growth at 37°C. We confirmed that all yTAF145 mutant genes are expressed at a level comparable to that of the wild-type gene (Fig. 4D). We thus conclude that interaction with the concave surface of TBP is essential for the function of yTAND-1.
Acidic Activation Domains Can Function as TAND-1 When Integrated Within TFIID.
As demonstrated above, TAND-1 domains function as activation domains when fused to a DNA-binding domain. Next we performed reciprocal experiments to determine whether activation domains can function as TAND-1 when integrated within the TFIID complex. For these experiments, yTAND-1 was replaced with various activation domains, including VP16, GAL4, and EBNA2, that interact with the concave face of TBP. These chimeric constructs completely support yeast growth at 37°C when they replace yTAND-1 (Fig. 5A). In contrast to these acidic activation domains, the Sp1 activation domain, which does not target the concave face of TBP (see Fig. 1), did not support yeast growth at 37°C (Fig. 5A). It was confirmed by immunoblotting that the expression of this chimeric protein is comparable to that of wild-type yTAF145 (data not shown). Next, we tested whether a VP16 mutant (F475S, M478T, F479S), which has no potential as an activation domain (29), can function as yTAND-1 when integrated into the TFIID complex. This triple point mutation within the VP16 segment (residues 470∼490) completely abolishes both transcription ability (data not shown) and interaction activity with TBP (Fig. 5B), supporting our view that interaction with the concave TBP face is crucial for transcriptional activation (see Discussion). Importantly, while the wild-type VP16 segment can completely substitute for yTAND-1, the VP16 mutant cannot, judging from yeast growth at 37°C (Fig. 5A). Expression levels of these chimeric yTAF145 proteins are comparable (Fig. 5C). Thus, we conclude that the VP16 mutant, lacking the ability to interact with the concave surface of TBP, functions neither as activation domain nor as TAND-1.
Figure 5.
Acidic activation domains function as yTAND-1 when integrated into TFIID. (A) Suppression analysis of growth defect by deleting yTAND-1 from the yTAF145 gene. yTAND-1 was deleted from the yTAF145 gene (Δy1) or replaced with various activation domains. Activation domains used are as follows: Sp1, residues 82–263; EBNA2, residues 426–462; GAL4, residues 842–874; VP16, residues 457–490 or 470–490, mutant VP16 (referred to as mVP16); triple mutant (F475S, M478T, F479S) in residues 470–490. The yTAF145 genes with these substitutions were transformed into yeast, replacing the wild-type gene by plasmid shuffling. After plasmid shuffling, the resulting strains were grown on yeast extract/peptone/dextrose plates at 30°C (Left) and 37°C (Right) for 3 days. (B) TBP-binding activity of the VP16 mutant. Wild-type (lane 2) and the triple mutant (F475S, M478T, F479S) (lane 1) of VP16 activation domain (residues 470–490) were fused to yTAND-2 and expressed in E. coli as GST fusions. TBP-binding activity was determined as described in the Fig. 1 legend, and a Coomassie blue stained gel was represented. (C) Expression level of the VP16-yTAF145 fusions. Expression level of the VP16-yTAF145 chimeric proteins was detected as described in the legend to Fig. 4. Growth of these strains is represented in A.
Discussion
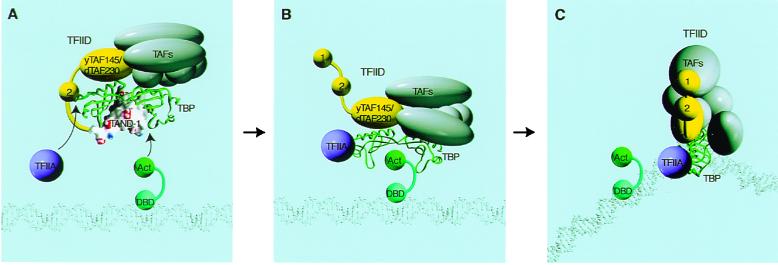
Two-Step “Hand Off” Model for Transcriptional Activation.
In this paper, we demonstrate that TAND-1 and certain activation domains are comparable and functionally interchangeable in yeast. Consistent with this interchangeability, the affinity of TAND-1 for TBP is similar to that of the VP16 activation domain (unpublished observations). Therefore, activation domains could counteract the negative regulation by TAND-1 by competition in yeast. Unlike yeast TFIID, the affinity of human and Drosophila TAND-1 for TBP is much higher than that of the activation domains. However, the observations that TFIIA can facilitate TFIID binding to core promoters dramatically (14, 15) may indicate that binding of TFIIA to the convex surface of TBP weakens the interaction between TAND-1 and the concave surface of TBP, allowing access of TFIID to core promoters. Therefore, even though affinity of TAND-1 for TBP is much higher than that of activators in higher eukaryotes, it is likely that certain activation domains, in conjunction with TFIIA, counteract the inhibitory interactions of TAND, allowing formation of an intermediate complex as illustrated in Fig. 6B. This intermediate complex could be essential for such activators, since interaction between the activation domain and the concave surface of TBP appears to be crucial for transcriptional activation (Fig. 4).
Figure 6.
A two-step “hand off” model for reversal of the autoinhibitory effect of TFIID. When activators are absent, TAND-1 and -2 bind to the concave and convex surface of TBP, respectively, and keep TFIID in a latent state as shown in A. Acidic activators and TAND-1 competitively bind to the concave surface of TBP, whereas TFIIA and TAND-2 competitively bind to the convex surface of TBP. Competitive binding by activation domains and TFIIA could cause a conformational alteration of TFIID that may result in formation of an intermediate state as illustrated in B. Importantly, several lines of evidence shown in this paper indicate that this intermediate stage is essential for activation of transcription. The activation domain still prevents TBP from binding to the TATA box in the intermediate stage. However, activators and TFIIA may induce conformational alteration of TFIID, allowing stabilized interaction of TFIID with sequences near and downstream from the transcriptional initiation site. Through these interactions, TBP is handed from activation domains to TATA box as shown in C.
In view of the close apposition of the concave surface of TBP and the TATA box in the co-crystal structure, it is unlikely that TBP and the activator form a ternary complex with the TATA box. Therefore, we believe that TFIID must dissociate from the activation domain for TATA box binding (Fig. 6C). To initiate this transition, certain TAFs in TFIID may bind near or downstream of the transcription initiation site, and subsequently TBP is handed from the activation domain to the TATA box. Consistent with this idea, TFIID recognizes not only the TATA sequence but also nucleotides near or downstream of the initiation site (15, 34). Thus, TFIID could bind to core promoters by virtue of TAF-DNA interactions even when TBP is tethered to an activation domain.
Previous studies demonstrated that activators, in conjunction with TFIIA, induce conformational alterations in TFIID, allowing more stabilized interaction of TFIID with sequences near and downstream from the transcriptional initiation site (reviewed in ref. 35). Importantly, this conformational change of TFIID appears to be essential for transcriptional activation. Therefore, formation of the proposed intermediate state (Fig. 6B) may contribute to the alteration of TFIID conformation. This two-step “hand off” model (36) is further supported by our observation showing that dTAND-1 is a weaker activator than yTAND-1 (Fig. 3). Given that the affinity of dTAND-1 for TBP is much stronger than that of yTAND-1 or the VP16 activation domain, the transition of TFIID from the intermediate state to core promoter binding could be blocked when dTAND-1 is used as an activation domain.
Functional Targets for Activators.
Despite extensive effort, it is still controversial which factors are functional targets for activators in physiological contexts. For instance, the most thoroughly studied activator, VP16, has been shown to bind to multiple factors in vitro, including TFIIA, TBP, dTAF40, TFIIB, TFIIF, TFIIH, PC4, p52/p75, and RNA polymerase II holoenzyme (reviewed in ref. 37). Although it is likely that activators stimulate transcription through multiple pathways by communicating with distinct targets, the molecular functions of activators are obviously complex. We demonstrate here that the dTAND-1, which is known to interact with the concave surface of TBP (22) functions as an activation domain, when fused to the GAL4–DNA-binding domain. The relevance of this interaction is underscored by mutational analysis of yTAND-1 (Fig. 4). Significantly, yTAND-1 mutants that lack the ability for interaction with the concave surface of TBP also cannot act as an activation domain. This strongly supports the view that TAND-1, when fused to the DNA-binding domain, activates transcription via interaction with the concave face of TBP.
The activator bypass experiments indicate that artificial recruitment of RNA polymerase II transcriptional machinery on promoters would be sufficient for transcriptional activation (3, 38, 39). These experiments have been done using various components of the RNA polymerase II transcriptional machinery including GAL11, TBP, TAFs, TFIIB, Sin4, and Srbs (reviewed in ref. 38). When these factors are fused to a DNA binding domain and recruited on promoters, they activate transcription by bypassing the activation domain. However, importantly, the bypass experiments do not work nearly as well in mammalian cells as in yeast (39). TAND-1 of hTAF250 or dTAF230 has higher affinity for TBP than that of yTAF145 (20), and therefore, this strong inhibition may not be counteracted by simple recruitment of the RNA polymerase II transcriptional machinery in mammalian cells.
Acknowledgments
We thank Drs. T. Miyake and K. Kasahara for helpful discussions, Y. Tsukihashi for antibodies, and A. Kobayashi, T. Ichimiya and Y. Shindoh for technical assistance. We also thank Dr. S. D. Hayward for providing the EBNA2 clone and Drs. N. Tanese and R. Tjian for the Gal-Sp1 plasmid. This study was supported by grants from the Ministry of Education, Science, and Culture of Japan, and from the CREST Japan Science and Technology Corporation, the Uehara Memorial Foundation, the Inamori Foundation, and the Yamada Science Foundation (to T.K.), and a grant from the Medical Research Council of Canada (to M.I.). M.I. is a Howard Hughes Medical Institute International Research Scholar and a Medical Research Council (Canada) Scientist. Editorial assistance was provided by Dr. Birgit An der Lan (BAAR Biomed, Bethesda, MD).
Abbreviations
- TFIID
transcription factor IID
- TBP
TATA box-binding protein
- TAF
TBP-associated factor
- TAND
TAF N-terminal domain
- GST
glutathione S-transferase
- EBNA
Epstein–Barr virus nuclear protein 2
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.120074297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.120074297
References
- 1.Orphanides G, Lagrange T, Reinberg D. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 2.Myer V E, Young R A. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- 3.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 4.Ranish J A, Yudkovsky N, Hahn S. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burley S K, Roeder R G. Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 6.Verrijzer C P, Tjian R. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 7.Hahn S. Cell. 1998;95:579–582. doi: 10.1016/s0092-8674(00)81625-6. [DOI] [PubMed] [Google Scholar]
- 8.Oelgeschlager T, Tao Y, Kang Y K, Roeder R G. Mol Cell. 1998;1:925–931. doi: 10.1016/s1097-2765(00)80092-1. [DOI] [PubMed] [Google Scholar]
- 9.Wu S Y, Kershnar E, Chiang C M. EMBO J. 1998;17:4478–4490. doi: 10.1093/emboj/17.15.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fondell J D, Guermah M, Malik S, Roeder R G. Proc Natl Acad Sci USA. 1999;96:1959–1964. doi: 10.1073/pnas.96.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y-J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 12.Koleske A J, Young R A. Nature (London) 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 13.Guermah M, Malik S, Roeder R G. Mol Cell Biol. 1998;18:3234–3244. doi: 10.1128/mcb.18.6.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozer J, Mitsouras K, Zerby D, Carey M, Lieberman P M. J Biol Chem. 1998;273:14293–14300. doi: 10.1074/jbc.273.23.14293. [DOI] [PubMed] [Google Scholar]
- 15.Emami K H, Jain A, Smale S T. Genes Dev. 1997;11:3007–3019. doi: 10.1101/gad.11.22.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kokubo T, Gong D-W, Yamashita S, Horikoshi M, Roeder R G, Nakatani Y. Genes Dev. 1993;7:1033–1046. doi: 10.1101/gad.7.6.1033. [DOI] [PubMed] [Google Scholar]
- 17.Bai Y, Perez G M, Beechem J M, Weil P A. Mol Cell Biol. 1997;17:3081–3093. doi: 10.1128/mcb.17.6.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kokubo T, Swanson M J, Nishikawa J I, Hinnebusch A G, Nakatani Y. Mol Cell Biol. 1998;18:1003–1012. doi: 10.1128/mcb.18.2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kokubo K, Yamashita S, Horikoshi M, Roeder R G, Nakatani Y. Proc Natl Acad Sci USA. 1994;91:3520–3524. doi: 10.1073/pnas.91.9.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishikawa J, Kokubo T, Horikoshi M, Roeder R G, Nakatani Y. Proc Natl Acad Sci USA. 1997;94:85–90. doi: 10.1073/pnas.94.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotani T, Miyake T, Tsukihashi Y, Hinnebusch A G, Nakatani Y, Kawaichi M, Kokubo T. J Biol Chem. 1998;273:32254–32264. doi: 10.1074/jbc.273.48.32254. [DOI] [PubMed] [Google Scholar]
- 22.Liu D, Ishima R, Tong K I, Bagby S, Kokubo T, Muhandiram D R, Kay L E, Nakatani Y, Ikura M. Cell. 1998;94:573–583. doi: 10.1016/s0092-8674(00)81599-8. [DOI] [PubMed] [Google Scholar]
- 23.Guthrie C, Fink G R. Methods Enzymol. 1991;194:3–21. [Google Scholar]
- 24.Boeke J D, Trueheart J, Natsoulis G, Fink G R. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 25.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 26.Kim T K, Hashimoto S, Kelleher R J, Flanagan P M, Kornberg R D, Horikoshi M, Roeder R G. Nature (London) 1994;369:252–255. doi: 10.1038/369252a0. [DOI] [PubMed] [Google Scholar]
- 27.Stringer K F, Ingles C J, Greenblatt J. Nature (London) 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 28.Tansey W P, Herr W. Proc Natl Acad Sci USA. 1995;92:10550–10554. doi: 10.1073/pnas.92.23.10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi N, Horn P J, Sullivan S M, Triezenberg S J, Boyer T G, Berk A J. Mol Cell Biol. 1998;18:4023–4031. doi: 10.1128/mcb.18.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melcher K, Johnston S A. Mol Cell Biol. 1995;15:2839–2848. doi: 10.1128/mcb.15.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen J I. Proc Natl Acad Sci USA. 1992;89:8030–8034. doi: 10.1073/pnas.89.17.8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoey T, Weinzierl R O, Gill G, Chen J L, Dynlacht B D, Tjian R. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 33.Emili A, Greenblatt J, Ingles C J. Mol Cell Biol. 1994;14:1582–1593. doi: 10.1128/mcb.14.3.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burke T W, Kadonaga J T. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chi T, Carey M. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- 36.Burley S K, Roeder R G. Cell. 1998;94:551–553. doi: 10.1016/s0092-8674(00)81596-2. [DOI] [PubMed] [Google Scholar]
- 37.Sullivan S M, Horn P J, Olson V A, Koop A H, Niu W, Ebright R H, Triezenberg S J. Nucleic Acids Res. 1998;26:4487–4496. doi: 10.1093/nar/26.19.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaudreau L, Keaveney M, Nevado J, Zaman Z, Bryant G O, Struhl K, Ptashne M. Proc Natl Acad Sci USA. 1999;96:2668–2673. doi: 10.1073/pnas.96.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nevado J, Gaudreau L, Adam M, Ptashne M. Proc Natl Acad Sci USA. 1999;96:2674–2677. doi: 10.1073/pnas.96.6.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]