Abstract
A bacterium exhibiting activities of several inorganic polyphosphate [poly(P)]- and ATP-dependent kinases, including glucokinase, NAD kinase, mannokinase, and fructokinase, was isolated, determined to belong to the genus Arthrobacter, and designated Arthrobacter sp. strain KM. Among the kinases, a novel enzyme responsible for the poly(P)- and ATP-dependent mannokinase activities was purified 2,200-fold to homogeneity from a cell extract of the bacterium. The purified enzyme was a monomer with a molecular mass of 30 kDa. This enzyme phosphorylated glucose and mannose with a high affinity for glucose, utilizing poly(P) as well as ATP, and was designated poly(P)/ATP-glucomannokinase. The Km values of the enzyme for glucose, mannose, ATP, and hexametaphosphate were determined to be 0.50, 15, 0.20, and 0.02 mM, respectively. The catalytic sites for poly(P)-dependent phosphorylation and ATP-dependent phosphorylation of the enzyme were found to be shared, and the poly(P)-utilizing mechanism of the enzyme was shown to be nonprocessive. The gene encoding the poly(P)/ATP-glucomannokinase was cloned from Arthrobacter sp. strain KM, and its nucleotide sequence was determined. This gene contained an open reading frame consisting of 804 bp coding for a putative polypeptide with a calculated molecular mass of 29,480 Da. The deduced amino acid sequence of the polypeptide exhibited homology to the amino acid sequences of the poly(P)/ATP-glucokinase of Mycobacterium tuberculosis H37Rv (level of homology, 45%), ATP-dependent glucokinases of Corynebacterium glutamicum (45%), Renibacterium salmoninarum (45%), and Bacillus subtilis (35%), and proteins of bacteria belonging to the order Actinomyces whose functions are not known. Alignment of these homologous proteins revealed seven conserved regions. The mannose and poly(P) binding sites of poly(P)/ATP-glucomannokinase are discussed.
Inorganic polyphosphate [poly(P)] is an energy- and phosphorus-rich biopolymer that is present in a variety of organisms. The energy contained in phosphodiester bonds in poly(P) is thermodynamically equivalent to the energy of ATP and can be utilized indirectly and directly for the phosphorylation of cellular molecules (13). The presence of various poly(P)-dependent enzymes responsible for the phosphorylation reaction in microbes has been reported previously. These enzymes include poly(P) kinase, glucokinase, NAD kinase, AMP phosphotransferase (3), and 1,3-diphosphoglycerate phosphotransferase (14).
Among these enzymes, poly(P) kinase, which is found only in prokaryotic cells and catalyzes the formation of poly(P) through addition of the terminal phosphate of ATP to a growing poly(P) chain, has been studied extensively and has been well characterized (13). In addition to poly(P) kinase, poly(P)/ATP-glucokinase and poly(P)/ATP-NAD kinase have also been well studied and have been shown to catalyze the phosphorylation of glucose and NAD, respectively, by use of poly(P) and ATP. Poly(P)/ATP-glucokinase was first found by Szymona and Ostrowski in Mycobacterium phlei (27) and was subsequently purified from Propionibacterium shermanii (20) and Mycobacterium tuberculosis H37Ra (9), and the gene for the enzyme was cloned from M. tuberculosis H37Rv (10). Poly(P)/ATP-NAD kinase was first found by Murata et al. (18) in bacteria belonging to order Actinomyces, and the enzyme and the gene encoding the enzyme were recently isolated from M. tuberculosis H37Rv and Micrococcus flavus (12).
The presence of these poly(P)-dependent enzymes, including poly(P) kinase, poly(P)/ATP-glucokinase, and poly(P)/ATP-NAD kinase, suggests that poly(P) functions as a physiological energy carrier in certain microbes. Furthermore, poly(P)/ATP-glucokinase and poly(P)/ATP-NAD kinase are presumed to be evolutionarily intermediate enzymes between an ancient poly(P)-specific kinase and a present-day ATP-specific kinase, since poly(P) can be formed and can participate in ATP synthesis under ancient prebiotic conditions (28, 29). Therefore, studies on poly(P)- and ATP-dependent kinases are important not only for revealing the physiological role of poly(P) but also for understanding the presumed evolutionary process for kinases.
However, except for poly(P)/ATP-glucokinase and poly(P)/ATP-NAD kinase, poly(P)-dependent kinases that utilize both poly(P) and ATP as phosphoryl donors have not been purified, nor have the genes encoding the enzymes been isolated. The lack of information concerning poly(P)- and ATP-dependent kinases prevents us from understanding the physiological roles of poly(P) and the presumed evolutionary process for kinases. Thus, we attempted to find novel poly(P)- and ATP-dependent kinases, and we describe here purification and characterization of a novel poly(P)/ATP-glucomannokinase.
MATERIALS AND METHODS
Materials.
Metaphosphate, sodium metaphosphate (molecular weight, 2,000), hexametaphosphate (molecular weight, 1,630), and poly(P) were obtained from Wako Pure Chemical Industries, Osaka, Japan. Nucleoside phosphates, glucose-6-phosphate dehydrogenase, mannose-6-phosphate isomerase, glucose-6-phosphate isomerase, pyruvate kinase, lactate dehydrogenase, and isocitrate dehydrogenase were obtained from Sigma-Aldrich Japan, Tokyo, Japan. DEAE-cellulose and hydroxylapatite were purchased from Nacalai Tesque, Kyoto, Japan. Phosphocellulose (P-11) was obtained from Whatman International, Maidstone, England. Sephadex G-150 was obtained from Amersham Pharmacia Biotech, Uppsala, Sweden. Silica Gel 60/Kieselguhr F254 thin-layer chromatography (TLC) plates were obtained from E. Merck, Darmstadt, Germany. A Gene Clean kit was purchased from Bio 101, Vista, Calif. pUC118 was obtained from Takara Biomedicals, Kyoto, Japan.
Screening of microorganisms.
To isolate microorganisms exhibiting poly(P)-dependent kinase activities, soil samples were directly spread on agar (1.5%) plates containing a medium (pH 7.0) consisting of 0.5% glucose, 0.1% (NH4)2SO4, 0.1% MgSO4 · 7H2O, 0.3% yeast extract, 1.5 M inorganic phosphate (Pi), and 2% metaphosphate. One of the large colonies appearing on the plates after 24 h of incubation at 30°C was isolated. Identification of the bacterium was performed at NCIMB Japan, Shizuoka, Japan.
Enzyme assays.
Cells of Arthrobacter sp. strain KM grown aerobically for 24 h at 30°C in Luria-Bertani (LB) medium (1) were suspended in 10 mM potassium phosphate (pH 7.0) containing 0.1 mM EDTA (buffer I) and then ultrasonically disrupted at 9 kHz (Insonator model 200 M; Kubota, Tokyo, Japan) and 0°C for 20 min. The cell homogenate was centrifuged at 20,000 × g and 4°C for 20 min, and the resultant clear solution (cell extract) was used for reactions. Unless otherwise specified, the reaction was carried out at 30°C, and poly(P) represented metaphosphate. For all enzymes 1 U of activity was defined as 1.0 μmol of product formed in 1 min at 30°C, and the specific activity was expressed in units per milligram of protein. Km values were determined by using Lineweaver-Burk plots (7). Protein contents were determined by the method of Bradford (4) with bovine serum albumin as the standard.
(i) Glucokinase.
Glucokinase activity was determined by monitoring the formation of NADPH spectrophotometrically at A340 (17). The assay mixture (1.0 ml) comprised 5.0 mM glucose, 0.5 mM NADP, a phosphoryl donor [5.0 mM ATP or 2.0 mg of poly(P) per ml], 5.0 mM MgCl2, 0.5 U of glucose-6-phosphate dehydrogenase, and 100 mM Tris-HCl (pH 7.0). The reaction was initiated by addition of the enzyme preparation. When the effects of pH, temperature, metal ions, and other factors on glucokinase activity were examined, NADP and glucose-6-phosphate dehydrogenase were omitted from the assay mixture described above. The reaction was terminated by immersing the test tube in boiling water for 3 min, and then the amount of glucose 6-phosphate in the reaction mixture was enzymatically determined (17). The activity of glucose-6-phosphate dehydrogenase was confirmed not to be inhibited by the factors whose effects were examined.
(ii) Mannokinase.
Mannokinase activity was determined by monitoring the formation of NADPH spectrophotometrically at A340 (24). The assay mixture (1.0 ml) comprised 50 mM mannose, 0.5 mM NADP, a phosphoryl donor [5.0 mM ATP or 2.0 mg of poly(P) per ml], 5.0 mM MgCl2, 0.5 U of mannose-6-phosphate isomerase, 0.6 U of glucose-6-phosphate isomerase, 0.5 U of glucose-6-phosphate dehydrogenase, and 100 mM Tris-HCl (pH 7.0). The reaction was initiated by addition of the enzyme preparation. When the effects of pH, temperature, metal ions, and other factors on mannokinase activity were assayed, NADP and the enzymes required for conversion of the reaction products were omitted from the assay mixture. After the reaction, the mixture was treated as described above, and then the amount of mannose 6-phosphate in it was enzymatically determined (24). The activities of mannose-6-phosphate isomerase, glucose-6-phosphate isomerase, and glucose-6-phosphate dehydrogenase were confirmed not to be affected by the factors whose effects were examined.
(iii) Fructokinase.
Fructokinase activity was determined by monitoring the formation of NADPH spectrophotometrically at A340 (24). The assay mixture (1.0 ml) comprised 50 mM fructose, 0.5 mM NADP, a phosphoryl donor [5.0 mM ATP or 2.0 mg of poly(P) per ml], 5.0 mM MgCl2, 0.5 U of glucose-6-phosphate isomerase, 0.5 U of glucose-6-phosphate dehydrogenase, and 100 mM Tris-HCl (pH 7.0). The reaction was initiated by addition of the enzyme preparation.
(iv) NAD kinase.
NAD kinase activity was determined as described previously (12). Briefly, the formation of NADP was enzymatically determined with isocitrate dehydrogenase in an assay mixture (1.0 ml) comprising 5.0 mM NAD, a phosphoryl donor [5.0 mM ATP or 2.0 mg of poly(P) per ml], 5.0 mM MgCl2, and 100 mM Tris-HCl (pH 7.0).
(v) Other enzymes.
Poly(P)- and ATP-dependent activities for phosphorylation of glucosamine, galactose, and 2-deoxyglucose were assayed at 30°C in a reaction mixture comprising a sugar at a concentration of 5.0 mM, a phosphoryl donor [5.0 mM ATP or 2.0 mg of poly(P) per ml], 5.0 mM MgCl2, and 100 mM Tris-HCl (pH 7.0). The reaction was initiated by addition of the enzyme preparation. After incubation for 10 min, the reaction was terminated by immersing the test tube in boiling water for 3 min, and then the reaction products in the clear solution were analyzed. When ATP was used as the phosphoryl donor, the amount of ADP in the solution was enzymatically determined by using a pyruvate kinase and lactate dehydrogenase system (11). When poly(P) was used, an appropriate amount of the solution was spotted onto a TLC plate, and then the reaction products were separated by TLC by using a solvent system consisting of isobutyric acid and a 0.5 N ammonia solution (5:3, vol/vol) and were visualized by heating the TLC plate at 130°C for 5 min after it was sprayed with 10% (vol/vol) sulfuric acid in ethanol.
Purification of poly(P)/ATP-glucomannokinase.
All purification procedures were performed at 4°C by using centrifugation at 20,000 × g for 20 min. After each purification step, the activities with both ATP and poly(P) were determined. Cells (130 g, wet weight) of Arthrobacter sp. strain KM grown aerobically for 40 h at 30°C in LB medium were suspended in buffer I and then ultrasonically disrupted as described above. The clear cell extract (9,792 mg of protein) obtained after centrifugation was saturated with ammonium sulfate (30%) and then kept for 9 h. After the precipitate was removed by centrifugation, the supernatant was dialyzed against buffer I overnight and then applied to a DEAE-cellulose column (4.5 by 30 cm) previously equilibrated with buffer I. The enzyme was eluted with a linear gradient of KCl (0 to 600 mM) in buffer I (3,000 ml) and was collected in 20-ml fractions at 10-min intervals. The active fractions containing poly(P)- and ATP-dependent mannokinase activities, which were eluted with approximately 200 mM KCl, were pooled, dialyzed against buffer I overnight, and then applied to a phosphocellulose (P-11) column (2.6 by 13 cm) previously equilibrated with buffer I. The enzyme was eluted with a linear 10 to 600 mM potassium phosphate (pH 7.0) gradient (1,200 ml) containing 0.1 mM EDTA and was collected in 4-ml fractions at 3-min intervals. The active fractions containing poly(P)- and ATP-dependent mannokinase activities, which were eluted with 500 to 600 mM potassium phosphate (pH 7.0), were combined. After ammonium sulfate was added to the combined fractions to obtain 30% saturation, the combined fractions were applied to a Butyl-Toyopearl 650 M column (2.6 by 19 cm) previously equilibrated with ammonium sulfate (30%) in buffer I. The enzyme was eluted with a linear gradient of ammonium sulfate (30 to 0%) in buffer I (300 ml) and was collected in 2.5-ml fractions at 2-min intervals. The active fractions containing poly(P)- and ATP-dependent mannokinase activities, which were eluted with 10 and 0% ammonium sulfate in buffer I, were pooled and then concentrated by ultrafiltration with a CENTRIPREP (Amicon, Beverly, Mass.). The concentrate was loaded onto a Sephadex G-150 column (2.5 by 66 cm) previously equilibrated with buffer I. The enzyme was eluted with the same buffer in 2.2-ml fractions at 4-min intervals. The active fractions (fractions 68 to 80) containing poly(P)- and ATP-dependent mannokinase activities were collected and used as the purified poly(P)/ATP-glucomannokinase.
Gel electrophoresis and gel filtration chromatography.
The subunit molecular mass of the enzyme was determined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (15). The molecular mass of the enzyme was determined by gel filtration chromatography on a Sephadex G-150 column as described above and by native gradient PAGE with Multigel 2/15 (Daiichi Pure Chemicals, Tokyo, Japan) as recommended by the manufacturer. The average chain length of poly(P) was determined by PAGE by using a 15% polyacrylamide gel as described previously (19), except that the gel was 28 by 37 by 0.35 mm, and was estimated as described by Clark and Wood (6).
Reverse reactions.
The phosphorylation of ADP and poly(P) (reverse reaction) by the purified poly(P)/ATP-glucomannokinase in the presence of glucose 6-phophate was examined by using a reaction mixture (1.0 ml) comprising 5.0 mM glucose 6-phosphate, a phosphoryl acceptor [5.0 mM ADP or 2.0 mg of poly(P) per ml], 5.0 mM MgCl2, 100 mM Tris-HCl (pH 7.0), and 0.10 μg of purified poly(P)/ATP-glucomannokinase. As a control, a reaction mixture containing boiled enzyme was also prepared. The residual amounts of glucose 6-phosphate in the mixtures after incubation at 30°C for 1, 3, 6, and 12 h were determined as described above.
Competition plot.
A competition plot for poly(P)/ATP-glucomannokinase against poly(P) and ATP was constructed as described previously (5). The glucokinase activities (reaction rates) of poly(P)/ATP-glucomannokinase toward poly(P) and ATP were determined as described above by using the purified enzyme (0.10 μg). Poly(P) and ATP were used at millimolar concentrations of [poly(P)]0 · p and [ATP]0 · (1 − p), respectively, where p varies from 0 to 1. [Poly(P)]0 and [ATP]0 were chosen so that the reaction rates were identical when p was 0 and 1. Hexametaphosphate was used as poly(P). The molecular weight of hexametaphosphate was assumed to be 1,630 based on the results of a PAGE analysis.
Poly(P)-utilizing mechanism.
The poly(P)-dependent glucokinase reaction was performed in the mixture (1.0 ml) used for the glucokinase activity assay, except that 1.0 μg of purified poly(P)/ATP-glucomannokinase (0.03 μM) and 2.0 mg of sodium metaphosphate per ml (1.0 mM) were used. The average molecular weight of sodium metaphosphate was assumed to be 2,000 based on the results of a PAGE analysis. After incubation for 0, 5, 10, and 20 min, an aliquot of the mixture (0.05 ml) was withdrawn, treated by the method of Pepin and Wood (19), and analyzed by PAGE as described above. As a control, a reaction mixture with boiled enzyme was prepared and treated in the same manner.
N-terminal amino acid sequence.
The N-terminal amino acid sequence was determined by Edman degradation by using a Procise 492 protein sequencing system (Applied Biosystems Div., Perkin-Elmer, Foster City, Calif.).
Construction of a genomic DNA library.
Genomic DNA of Arthrobacter sp. strain KM was isolated as described previously (1), partially digested with Sau3AI, and then separated by 0.80% agarose gel electrophoresis. The partially digested DNA fragments (4.0 to 8.0 kb) were isolated with a Gene Clean kit and ligated to pUC118 digested with BamHI, which resulted in recombinant DNA clones. Cells of Escherichia coli DH5α were transformed with these DNA clones and then used as the genomic DNA library of Arthrobacter sp. strain KM.
Cloning and analysis of the gene encoding poly(P)/ATP-glucomannokinase.
The genomic DNA library of Arthrobacter sp. strain KM (about 25,000 clones) was screened by the colony hybridization method (1) by using a 32P-labeled probe (5′ AT[A/C/T]GG[A/C/G/T]AT[A/C/T]GA[C/T]AT[A/C/T]GG 3′) designed based on the N-terminal amino acid sequence of poly(P)/ATP-glucomannokinase. Plasmid DNA was isolated from one of the positive clones obtained, and the nucleotide sequence of the insert DNA was determined by the dideoxy chain termination method with an automated DNA sequencer (model 377; Applied Biosystems Div., Perkin-Elmer). The DNA sequence was determined with the GENETYX program (Software Development, Tokyo, Japan). Homology, alignment, and phylogenetic analyses of amino acid sequences were performed with the BLAST and CLUSTAL W programs on the DDBJ server (http://www.ddbj.nig.ac.jp). Sequence data were obtained from GenBank (http://www.ncbi.nlm.nih.gov).
Nucleotide sequence accession number.
The sequence data determined in this study have been deposited in the DDBJ and GenBank databases under accession no. AB096174.
RESULTS
Isolation and identification of Arthrobacter sp. strain KM.
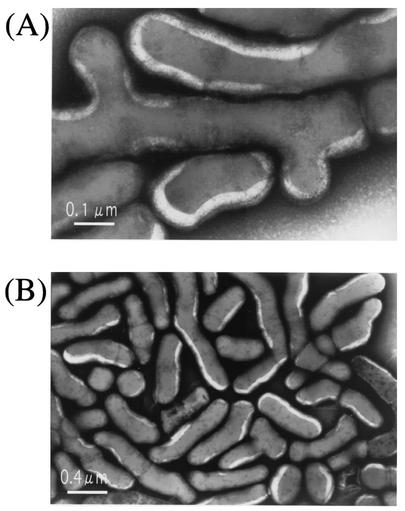
As the first step in research involving novel poly(P)- and ATP-dependent kinases, we attempted to isolate a bacterium that grows well with a high concentration of Pi, since such a bacterium is expected to contain various Pi-metabolizing enzymes, including some poly(P)- and ATP-dependent kinases. As described in Materials and Methods, a bacterium resistant to a high concentration of Pi (1.5 M) was obtained. This bacterium was a gram-positive, nonmotile, non-spore-forming, irregular rod (Fig. 1). The cell wall of the bacterium contained l-lysine. The main cellular fatty acids were anteiso-C15:0 (12-methyltetradecanoic acid), iso-C15:0 (13-methyltetradecanoic acid), and anteiso-C17:1 (14-methylhexadecanoic acid). The colonies were smooth, round, and lustrous. The organism grew at 30 and 37°C, and it did not grow anaerobically at 42°C. It was pyrazinamidase, prolidonylarylamidase, β-glucuronidase, N-acetyl-β-glucosaminidase, urease, and oxidase positive. It was negative in the oxidation-fermentation test and for nitrate reduction, alkaline phosphatase, β-galactosidae, β-glucosidase, glucosidase, catalase, and gelatin liquefaction activities. It did not ferment glucose, ribose, xylose, mannitol, maltose, lactate, glycogen, starch hydrolysate, or sucrose. Based on its biochemical and taxonomic properties, in addition to the results of an analysis of its 16S rRNA nucleotide sequence, the bacterium was determined to belong to the genus Arthrobacter, and it was designated Arthrobacter sp. strain KM.
FIG. 1.
Electron micrographs of Arthrobacter sp. strain KM.
A cell extract of Arthrobacter sp. strain KM grown for 24 h at 30°C in LB medium exhibited several poly(P)- and ATP-dependent kinase activities, including poly(P)-dependent mannokinase and poly(P)-dependent fructokinase activities (Table 1). Furthermore, DEAE-cellulose column chromatography of the cell extract revealed that the active peaks of the poly(P)-dependent mannokinase and the poly(P)-dependent fructokinase could be superimposed on those of an ATP-dependent mannokinase and an ATP-dependent fructokinase (data not shown), respectively, suggesting the bacterium isolated contains a poly(P)- and ATP-dependent mannokinase and a poly(P)- and ATP-dependent fructokinase.
TABLE 1.
Poly(P)- and ATP-dependent kinase activities in a cell extract of Arthrobacter sp. strain KM
| Enzyme | Sp act (U/mg of protein)a
|
|
|---|---|---|
| Poly(P) | ATP | |
| Glucokinase | 0.167 | 0.257 |
| NAD kinase | 0.0068 | 0.026 |
| Mannokinase | 0.064 | 0.077 |
| Fructokinase | 0.013 | 0.020 |
The poly(P)- and ATP-dependent activities in cell extracts were assayed in the presence of poly(P) and ATP, as described in Materials and Methods.
Purification and properties of poly(P)/ATP-glucomannokinase.
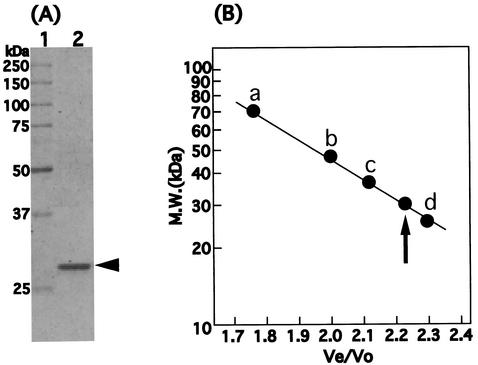
The poly(P)/ATP-mannokinase was purified approximately 2,200-fold from a cell extract of Arthrobacter sp. strain KM with an approximately constant ratio of ATP activity to poly(P) activity (Table 2). The purified enzyme, obtained as a single peak corresponding to a calculated molecular mass of 30 kDa on a Sephadex G-150 column (Fig. 2B), migrated as a single band corresponding to a molecular mass of 30 kDa on an SDS-PAGE gel (Fig. 2A) and a native gradient PAGE gel (data not shown). These results indicated that the purified enzyme was monomeric and responsible for both the poly(P)- and ATP-dependent mannokinase activities. The properties of the purified enzyme were as follows.
TABLE 2.
Purification of poly(P)/ATP-glucomannokinase from Arthrobacter sp. strain KM
| Stepa | Total protein (mg) | Sp act (U/mg of protein)
|
Total activity (U)
|
Purification (fold)
|
Yield (%)
|
Ratio of sp actb | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Poly(P) | ATP | Poly(P) | ATP | Poly(P) | ATP | Poly(P) | ATP | |||
| Cell extract | 9,792 | 0.10 | 0.08 | 979 | 783 | 1 | 1 | 100 | 100 | 0.80 |
| (NH4)2SO4 | 3,583 | 0.20 | 0.15 | 717 | 537 | 2.0 | 1.9 | 73.2 | 68.6 | 0.75 |
| DEAE-cellulose | 1,168 | 0.36 | 0.30 | 421 | 350 | 3.6 | 3.8 | 43.0 | 44.7 | 0.83 |
| Phosphocellulose P-11 | 22.3 | 13.2 | 10.9 | 294 | 243 | 132 | 136 | 30.0 | 31.0 | 0.83 |
| Butyl-Toyopearl 650M | 1.50 | 145.9 | 76.8 | 219 | 115 | 1,459 | 960 | 22.4 | 14.7 | 0.53 |
| Sephadex G-150 | 0.61 | 220 | 110 | 134 | 67 | 2,200 | 1,375 | 13.7 | 8.6 | 0.50 |
The purification procedures are described in Materials and Methods.
Ratio of the ATP-dependent specific activity to the poly(P)-dependent specific activity.
FIG. 2.
Determination of the molecular mass of purified poly(P)/ATP-glucomannokinase. (A) SDS-PAGE of poly(P)/ATP-glucomannokinase. Lane 1, protein markers (Bio-Rad Laboratories, Hercules, Calif.); lane 2, purified poly(P)/ATP-glucomannokinase (0.5 μg). (B) Gel filtration chromatography of poly(P)/ATP-glucomannokinase. The standard proteins used were bovine serum albumin (67 kDa) (a), ovalbumin (43 kDa) (b), alginate lyase A1-III (37 kDa) (c), and chymotrypsinogen A (25 kDa) (d). Molecular mass (M.W.) was plotted versus elution volume/void volume. The void volume was determined with blue dextran 2000 (2,000 kDa). The elution position of the purified poly(P)/ATP-glucomannokinase is indicated by an arrow.
(i) Substrate specificity.
The poly(P)- and ATP-dependent mannokinase phosphorylated not only mannose but also glucose by utilizing poly(P) as well as ATP, and it had a high affinity for glucose. The glucokinase-to-mannokinase activity ratios for the purified enzyme were 3:2 and 3:1 for the poly(P)- and ATP-dependent activities, respectively. Fructose, galactose, glucosamine, and 2-deoxyglucose were inert as phosphoryl acceptors irrespective of the phosphoryl donor [poly(P) or ATP]. Thus, we designated the purified enzyme poly(P)/ATP-glucomannokinase.
Purified poly(P)/ATP-glucomannokinase utilized both nucleoside triphosphates and poly(P) for the phosphorylation of mannose and glucose. Acetylphosphate, phosphoenolpyruvate, phosphocreatine, carbamylphosphate, and phosphoramidate were not utilized for the phosphorylation of mannose and glucose. The relative mannokinase and glucokinase activities of the enzyme in the presence phosphoryl donors at a concentration of 10 mM were as follows (the activities in the presence of 10 mM ATP were defined as 100%): ATP, 100 and 100%, respectively; GTP, 50 and 38%, respectively; CTP, 50 and 67%, respectively; UTP, 25 and 14%, respectively; ADP, 0 and 0%, respectively; AMP, 0 and 0%, respectively; pyrophosphate, 0 and 0%, respectively; tripolyphosphate, 0 and 0%, respectively; trimetaphosphate, 0 and 0%, respectively; and tetrapolyphosphate, 20 and 14%, respectively. The relative mannokinase and glucokinase activities of the enzyme in the presence of poly(P)s at a concentration of 2 mg per ml were as follows (the activities in the presence of 2 mg of metaphosphate per ml were defined as 100%): metaphosphate, 100 and 100%, respectively; poly(P), 110 and 112%, respectively; and hexametaphosphate, 120 and 177%, respectively.
The Km values of the poly(P)/ATP-glucomannokinase for glucose, mannose, ATP, and poly(P) (hexametaphosphate) were 0.50, 15, 0.20, and 0.02 mM, respectively.
The reverse reactions [i.e., phosphorylation of ADP and poly(P) by use of glucose 6-phosphate] were assessed as described in Materials and Methods. The amount of glucose 6-phosphate in the reaction mixture was equal to the amount in the control mixture with boiled enzyme during the reaction (data not shown), and therefore, we concluded that the reverse reaction was not catalyzed by poly(P)/ATP-glucomannokinase.
(ii) pH and temperature.
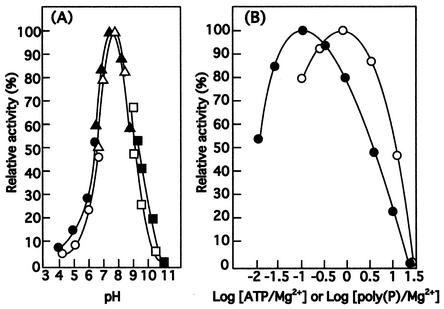
The poly(P)- and ATP-dependent mannokinase activities, as well as the poly(P)- and ATP-dependent glucokinase activities, of the purified poly(P)/ATP-glucomannokinase were maximum in Tris-HCl at pH 7.5 and 45°C (Fig. 3A), and one-half of the activity was lost after treatment at 40°C for 5 min.
FIG. 3.
Effects of pH and the ATP/Mg2+ ratio on the glucokinase activity of poly(P)/ATP-glucomannokinase. (A) Effect of pH. Poly(P)-dependent (solid symbols) and ATP-dependent (open symbols) glucokinase activities were assayed as described in Materials and Methods with the following buffers (100 mM): sodium acetate (○ and •), Tris-HCl (▵ and ▴), and glycine-KOH (□ and ▪). The poly(P)- and ATP-dependent glucokinase activities at pH 7.5 in Tris-HCl were defined as 100%. (B) Effects of the ATP/Mg2+ and poly(P)/Mg2+ ratios. ATP-dependent (○) and poly(P)-dependent (•) glucokinase activities were assayed as described in Materials and Methods with various concentrations of Mg2+. The activities when the ATP/Mg2+ and poly(P)/Mg2+ ratios were 1:1 and 1:10, respectively, were defined as 100%.
(iii) Metal ions and other reagents.
Irrespective of the phosphoryl donor, the poly(P)/ATP-glucomannokinase required a bivalent metal ion for glucokinase and mannokinase activities. The relative poly(P)-dependent and ATP-dependent glucokinase activities of the enzyme in the presence of metal ions at a concentration of 5.0 mM were as follows: Mg2+, 100 and 100%, respectively; Mn2+, 82 and 88%, respectively; Zn2+, 80 and 19%, respectively; Co2+, 86 and 30%, respectively; Cu2+, 11 and 0%, respectively; Fe2+, 0 and 15%, respectively; and Hg2+, 0 and 0%, respectively. The relative poly(P)- and ATP-dependent mannokinase activities had metal ion requirements similar to those of the glucokinase activities. The maximum ATP- and poly(P)-dependent glucokinase activities of the poly(P)/ATP-glucomannokinase were observed when the ATP/Mg2+ and poly(P)/Mg2+ ratios were 1:1 and 1:10, respectively (Fig. 3B). Other reagents, including EDTA, glutathione (reduced), 2-mercaptoethanol, dithiothreitol, and iodoacetamide, had no significant effects at a concentration of 1.0 mM on either the poly(P)- and ATP-dependent glucokinase or poly(P)- and ATP-dependent mannokinase activities of the poly(P)/ATP-glucomannokinase.
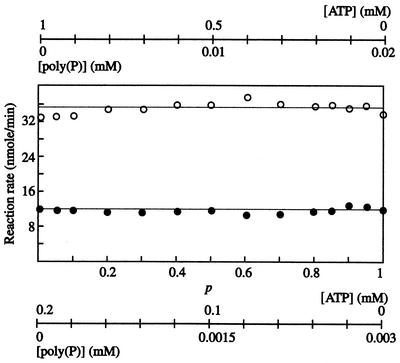
(iv) Competition plot.
A competition plot for poly(P)/ATP-glucomannokinase against poly(P) and ATP was constructed (Fig. 4). This plot shows that the reaction rates were independent of the concentrations of poly(P) and ATP with the two sets of variations (Fig. 4) and indicates that the catalytic sites for poly(P)- and ATP-dependent phosphorylation by the enzyme were shared.
FIG. 4.
Competition plot. A competition plot for poly(P)/ATP-glucomannokinase against poly(P) and ATP was constructed as described in Materials and Methods. Two combinations of [poly(P)]0 and [ATP]0 were used, as follows: [poly(P)]0 of 0.003 mM and [ATP]0 of 0.20 mM (lower line), and [poly(P)]0 of 0.020 mM and [ATP]0 of 1.0 mM (upper line).
(v) Mechanism of poly(P) utilization.
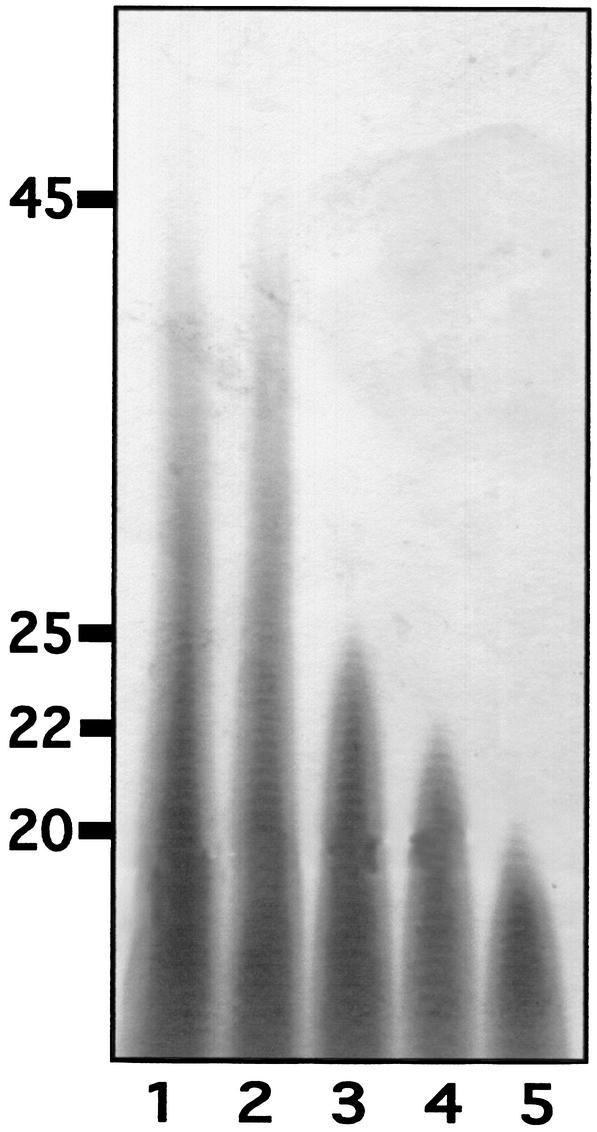
Whether poly(P)/ATP-glucomannokinase utilizes poly(P) processively or nonprocessively was investigated by monitoring poly(P) during the poly(P)-dependent glucokinase reaction. As the reaction proceeded, the length of poly(P) decreased gradually (Fig. 5, lanes 2 to 5), while no decrease in length was observed when the boiled enzyme was present in the reaction mixture (Fig. 5, lane 1). The observed poly(P) utilization pattern of the enzyme indicated that poly(P) was utilized nonprocessively and not hydrolyzed through nonenzymatic reactions. The possibility that excess enzyme acted on poly(P) progressively could be ruled out, since the amount of the enzyme (0.03 μM) was significantly lower than the amount of poly(P) (1.0 mM).
FIG. 5.
Mechanism of poly(P) utilization. The poly(P)-dependent glucokinase reaction catalyzed by poly(P)/ATP-glucomannokinase was monitored as described in Materials and Methods. Lane 1, control reaction mixture with boiled enzyme incubated for 20 min; lanes 2 to 5, reaction mixtures incubated for 0, 5, 10, and 20 min, respectively. The numbers on the left indicate the average chain lengths of poly(P).
(vi) N-Terminal amino acid sequence.
The N-terminal amino acid sequence of the purified enzyme was determined to be 1AKKDEKSHNAPLIGIDIGG19.
Cloning of the gene for poly(P)/ATP-glucomannokinase.
The nucleotide sequence (1,200 bp) of the insert DNA in the positive clone was determined as described in Materials and Methods. In the sequence, an open reading frame (ORF) consisting of 804 bp was found. The ORF starts at TTG and is preceded 8 bp upstream by a possible Shine-Dalgarno sequence (GGAAA). The ORF encodes a polypeptide consisting of 268 amino acid residues, and the predicted molecular mass and N-terminal amino acid sequence of this polypeptide are 29,480 Da and 2AKKDEKSHNAPLIGIDIGG20, respectively. The predicted molecular mass and N-terminal amino acid sequence are comparable to and identical to those of the purified poly(P)/ATP-glucomannokinase, respectively. Thus, we concluded that the ORF encoded the poly(P)/ATP-glucomannokinase. Possible promoter and terminator regions were not found.
Homology analysis.
Homology analysis of the primary structure of the poly(P)/ATP-glucomannokinase was performed with BLAST, and the primary structure was found to exhibit homology with the primary structures of the poly(P)/ATP-glucokinase from M. tuberculosis H37Rv (10) (45% identity over 101 amino acids), ATP-dependent glucokinases from Corynebacterium glutamicum (26) (45% identity over 100 amino acids), Renibacterium salmoninarum (16) (45% identity over 29 amino acids), and Bacillus subtilis (22) (35% identity over 36 amino acids), and proteins having unknown functions from Streptomyces coelicolor (GenBank accession no. AL590942) (50% identity over 112 amino acids), Mycobacterium leprae (AL583904) (44% identity over 98 amino acids), and Rhodoccocus fascians (AJ308422) (43% identity over 96 amino acids).
However, the primary structures of the ATP-dependent glucokinases from E. coli (U22490) and Zymomonas mobilis (2) exhibited little homology with the primary structure of the poly(P)/ATP-glucomannokinase, as determined by BLAST.
DISCUSSION
In this study, a bacterium exhibiting poly(P)-dependent kinase activities was isolated, and a novel poly(P)/ATP-glucomannokinase and the gene encoding the enzyme were isolated for the first time from the bacterium. The bacterium was determined to belong to the genus Arthrobacter and was designated Arthrobacter sp. strain KM. Arthrobacter is classified in the order Actinomyces. This order includes the genera Micrococcus, Mycobacterium, Corynebacterium, and Brevibacterium, all of which have been shown to possess poly(P)-dependent kinases (9, 10, 12, 18, 21, 27).
ATP-dependent mannokinases have been partially purified so far from E. coli (25) and Streptomyces violaceoruber (23), although their molecular masses have not been determined. These two mannokinases phosphorylate glucose in addition to mannose by using ATP, and the affinity for glucose is extremely low. Therefore, the name ATP-dependent mannokinase seems to be appropriate for these two enzymes. An ATP-dependent glucomannokinase and an ATP-dependent fructomannokinase have been purified from Prevotella bryantii (8) and Leuconostoc mesenteroides (24), respectively. These two enzymes phosphorylate glucose or fructose in addition to mannose by using ATP, and the affinities of the enzymes for glucose or fructose are almost the same as the affinities for mannose. The poly(P)/ATP-glucomannokinase purified in this study also phosphorylates glucose and mannose, and the affinity for glucose is high; therefore, the designation glucomannokinase is considered appropriate. Furthermore, since the enzyme utilizes poly(P) as well as ATP, it is thought to be a novel glucomannokinase.
Poly(P)/ATP-glucomannokinase is a monomer with a molecular mass of 30 kDa. The subunit molecular mass of the enzyme is similar to the subunit molecular masses of the poly(P)/ATP-glucokinases of P. shermanii (30 kDa) (20) and M. tuberculosis H37Ra (33 kDa) (9) and the ATP-dependent glucomannokinase of P. bryantii (34 kDa) (8) but different from the subunit molecular mass of the ATP-dependent fructomannokinase of L. mesenteroides (47 kDa) (24). The monomer structure of the poly(P)/ATP-glucomannokinase is different from the structures of the poly(P)/ATP-glucokinases of P. shermanii (dimer) and M. tuberculosis H37Ra (dimer) and the ATP-dependent glucomannokinase of P. bryantii (dimer). However, the catalysis modes of poly(P)/ATP-glucomannokinase, including the shared catalytic sites for poly(P)- and ATP-dependentphosphorylation, and the nonprocessive poly(P)-utilizing mechanism are similar to those of the poly(P)/ATP-glucokinases of P. shermanii and M. tuberculosis H37Ra (21).
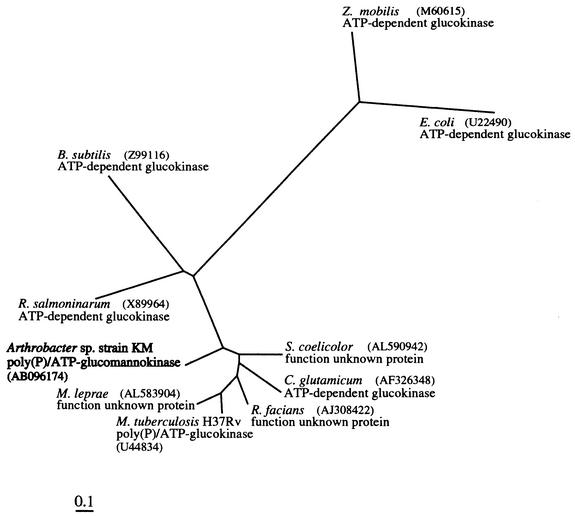
The primary structure of the poly(P)/ATP-glucomannokinase was determined. This is the first report of the primary structure of a glucomannokinase, since the primary structure of an ATP-dependent glucomannokinase has not been reported previously (8). The primary structure of the poly(P)/ATP-glucomannokinase exhibited homology with the primary structures of several proteins, including the poly(P)/ATP-glucokinase from M. tuberculosis H37Rv. However, the primary structure of the poly(P)/ATP-glucomannokinase exhibited little homology with the primary structures of some ATP-dependent glucokinases. Based on the primary structures of these homologous and nonhomologous proteins, a phylogenetic tree was constructed (Fig. 6). The tree showed that the primary structure of the poly(P)/ATP-glucokinase from M. tuberculosis H37Rv was closely related to the primary structures of the ATP-dependent glucokinases from C. glutamicum and proteins of unknown function from S. coelicolor, M. leprae, and R. fascians. Bacteria possessing these closely related proteins belong to the order Actinomyces. Taking into account the fact that several poly(P)-dependent enzymes have frequently been found in bacteria belonging to the order Actinomyces, these closely related proteins are thought to be poly(P)/ATP-glucokinases. Our phylogenetic tree, however, revealed that the primary structure of the poly(P)/ATP-glucomannokinase was slightly different from the primary structures of these poly(P)/ATP-glucokinase homologs, suggesting that the poly(P)/ATP-glucomannokinase evolved in a slightly different way than these poly(P)/ATP-glucokinase homologs. The poly(P)/ATP-glucomannokinase utilizes mannose as well as glucose, and this may be why the primary structure of the poly(P)/ATP-glucomannokinase is not closely related to the primary structures of poly(P)/ATP-glucokinases.
FIG. 6.
Phylogenetic tree constructed from the primary structures of poly(P)/ATP-glucomannokinase and other glucokinases. The GenBank accession numbers are indicated in parentheses.
Multiple-sequence alignment of the primary structures of the poly(P)/ATP-glucomannokinase and other poly(P)/ATP-glucokinase homologs was performed. As a result of the alignment, seven regions were found to be highly conserved (Fig. 7). Four of these regions, the phosphate-1, phosphate-2, connect-1, and connect-2 regions, were thought to be ATP binding sites, and the adenosine, glucose, and phosphate-3 regions were presumed to be adenosine, glucose, and poly(P) binding sites, respectively (21). In the glucose region, only the heptapeptide PEAPAAG was inserted in the primary structure of poly(P)/ATP-glucomannokinase, and this heptapeptide may be responsible for the mannose-phosphorylating ability of the poly(P)/ATP-glucomannokinase. In the phosphate-3 region of the primary structure of the poly(P)/ATP-glucokinase from M. tuberculosis H37Rv, 193Trp, 198Trp, and several charged groups (188Lys, 189Glu, 190Lys, 192Asp, 197Lys, and 200Lys) have been reported to be important for the binding of poly(P) (21). However, similarity was recognized in the phosphate-3 region of the primary structures of the poly(P)/ATP-glucomannokinase and other homologous proteins, but two residues, 193Trp and 198Trp, were not completely conserved in the poly(P)/ATP-glucomannokinase and other poly(P)/ATP-glucokinase homologs. Therefore, it is not known whether these two tryptophan residues in these poly(P)-dependent kinases interact with poly(P). At present, the residues that interact with poly(P) cannot be determined based on the primary structure information alone.
FIG. 7.
Multiple-sequence alignment of the primary structure of poly(P)/ATP-glucomannokinase with the primary structures of homologous proteins. The primary structures of the proteins shown in Fig. 6 except the ATP-dependent glucokinases from E. coli, Z. mobilis, R. salmoninarum, and B. subtilis are aligned. Seven conserved regions, the phosphate-1, phosphate-2, phosphate-3, connect-1, connect-2, glucose, and adenosine regions, are enclosed in boxes. Identical and similar amino acid residues are indicated by asterisks and dots, respectively. The heptapeptide in the glucose region of the poly(P)/ATP-glucomannokinase from Arthrobacter sp. strain KM is underlined. 193Trp and 198Trp in the phosphate-3 region of the poly(P)/ATP-glucokinase from M. tuberculosis H37Rv are underlined and double underlined, respectively. The amino acid residues aligned with 193Trp and 198Trp are shaded.
In any case, comparison of the three-dimensional structure of the poly(P)/ATP-glucomannokinase with the three-dimensional structures of poly(P)/ATP-dependent glucokinases and other ATP-dependent glucokinases should facilitate precise determination of the poly(P) binding site and should provide some clues for understanding the presumed kinase evolution from a poly(P)-specific kinase to an ATP-specific kinase. Furthermore, structural comparison of these enzymes should help reveal the process of acquisition of the mannose-phosphorylating activity of glucokinase. The disclosure of such a process may facilitate understanding the evolution from a glucomannokinase to a eukaryotic hexokinase that phosphorylates several sugars.
REFERENCES
- 1.Ausubel, F. M., R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 2.Barnell, W. O., K. C. Yi, and T. Conway. 1990. Sequence and genetic organization of Zymomonas mobilis gene cluster that encodes several enzymes of glucose metabolism. J. Bacteriol. 172:7227-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonting, C. F. C., G. J. J. Kortstee, and A. J. B. Zehnder. 1991. Properties of polyphosphate: AMP phosphotransferase of Acinetobacter strain 210A. J. Bacteriol. 173:6484-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye-binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Chevillard, C., M. L. Cardenas, and A. Cornish-bowden. 1993. The competition plot: a simple test of whether two reactions occur at the same active site. Biochem. J. 289:599-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark, J. E., and H. G. Wood. 1987. Preparation of standards and determination of sizes of long-chain polyphosphates by gel electrophoresis. Anal. Biochem. 161:280-290. [DOI] [PubMed] [Google Scholar]
- 7.Conn, E. E., P. K. Stumpe., G. Bruening., and R. H. Doi. 1987. Enzymes, p. 115-163. In E. E. Conn, P. K. Stumpe, G. Bruening, and R. H. Doi (ed.), Outlines of biochemistry. John Wiley and Sons, New York, N.Y.
- 8.Fields, M. W., and J. B. Russell. 2001. The glucomannokinase of Prebvotella bryantii B14 and its potential role in regulating β-glucanase expression. Microbiology 147:1035-1043. [DOI] [PubMed] [Google Scholar]
- 9.Hsieh, P. C., B. C. Shenoy, J. E. Jentoft, and N. F. B. Phillips. 1993. Purification of polyphosphate and ATP glucose phosphotransferase from Mycobacterium tuberculosis H37Ra: evidence that poly(P) and ATP glucokinase activities are catalyzed by the same enzyme. Protein Expr. Purif. 4:76-84. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh, P. C., B. C. Shenoy, D. Samols, and N. F. B. Phillips. 1996. Cloning, expression, and characterization of polyphosphate glucokinase from Mycobacterium tuberculosis. J. Biol. Chem. 271:4909-4915. [DOI] [PubMed] [Google Scholar]
- 11.Jaworek, D., and J. Welsch. 1985. Adenosine 5′-diphosphate and adenosine 5′-monophosphate, p. 365-370. In H. U. Bergmeyer, J. Bergmeyer, and M. Graßl (ed.), Methods of enzymatic analysis, vol. 7. Verlagsgesellschaft mbH, Weinheim, Germany.
- 12.Kawai, S., S. Mori, T. Mukai, S. Suzuki, T. Yamada, W. Hashimoto, and K. Murata. 2000. Inorganic polyphosphate/ATP-NAD kinase of Micrococcus flavus and Mycobacterium tuberculosis H37Rv. Biochem. Biophys. Res. Commun. 276:57-63. [DOI] [PubMed] [Google Scholar]
- 13.Kornberg, A. 1999. Inorganic polyphosphate: a molecule of many functions, p. 1-16. In H. C. Schröder and W. E. G. Müller (ed.), Inorganic polyphosphates, vol. 23. Springer-Verlag, New York, N.Y.
- 14.Kulaev, I. S., and M. A. Bobyk. 1971. The detection of a new enzyme, 1,3-phosphoglycerate-polyphosphate phosphotransferase, in Neurospora crassa. Biokhimiya 36:426-429. [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Margarita, I. C., and L. Gloria. 2000. Cloning, functional expression and partial characterization of the glucose kinase from Renibacterium salmoninarum. FEMS Microbiol. Lett. 186:97-101. [DOI] [PubMed] [Google Scholar]
- 17.Murata, K., J. Kato, and I. Chibata. 1978. Properties of polyphosphate glucokinase in Acromobacter butyri. Agric. Biol. Chem. 42:2221-2226. [Google Scholar]
- 18.Murata, K., T. Uchida, K. Tani, J. Kato, and I. Chibata. 1980. Metaphosphate: a new phosphoryl donor for NAD phosphorylation. Agric. Biol. Chem. 44:61-68. [Google Scholar]
- 19.Pepin, C. A., and H. G. Wood. 1986. Polyphosphate glucokinase from Propionibacterium shermanii. J. Biol. Chem. 261:4476-4480. [PubMed] [Google Scholar]
- 20.Phillip, N. F. B., P. J. Horn, and H. G. Wood. 1993. The polyphosphate and ATP-dependent glucokinase from Propionibacterium shermanii: both activities are catalyzed by the same protein. Arch. Biochem. Biophys. 300:309-319. [DOI] [PubMed] [Google Scholar]
- 21.Phillip, N. F. B., P. C. Hsieh, and T. H. Kowalczyk. 1999. Polyphosphate glucokinase, p. 101-122. In H. C. Schröder and W. E. G. Müller (ed.), Inorganic polyphosphates, vol. 23. Springer-Verlag, New York, N.Y.
- 22.Pierre, S., and D. Michael. 1998. The glucose kinase of Bacillus subtilis. J. Bacteriol. 180:3222-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabater, B., J. Sebastian, and C. Asensio. 1972. Identification and properties of an inducible mannokinase from Streptomyces violaceoruber. Biochim. Biophys. Acta 284:406-413. [DOI] [PubMed] [Google Scholar]
- 24.Sapico, V., and R. L. Anderson. 1967. An adenosine 5′-triphosphate:hexose-6-phosphotransferase specific for d-mannose and d-fructose from Leuconostoc mesenteroides. J. Biol. Chem. 242:5086-5091. [PubMed] [Google Scholar]
- 25.Sebastian, J., and C. Asenosio. 1967. Identification of a mannokinase in Escherichia coli. Biochem. Biophys. Res. Commun. 28:197-202. [DOI] [PubMed] [Google Scholar]
- 26.Sun, Y. P., K. K. Hyung, K. Y. Seung, K. O. Tae, and K. L. Jung. 2000. Characterization of glk coding for glucose kinase of Corynebacterium glutamicum. FEMS Microbiol. Lett. 188:209-215. [DOI] [PubMed] [Google Scholar]
- 27.Szymona, M., and W. Ostrowski. 1964. Inorganic polyphosphate glucokinase of Mycobacterium phlei. Biochim. Biophys. Acta 85:283-295. [DOI] [PubMed] [Google Scholar]
- 28.Waehneldt, T. V., and S. Fox. 1967. Phosphorylation of nucleosides with polyphosphoric acid. Biochim. Biophys. Acta 134:9-16. [Google Scholar]
- 29.Yamagata, Y., H. Watanabe, M. Saitoh, and T. Namba. 1991. Volcanic production of polyphosphates and its relevance to prebiotic evolution. Nature 352:516-519. [DOI] [PubMed] [Google Scholar]