Abstract
We describe a nested PCR-restriction fragment length polymorphism (RFLP) method for detecting low densities of Cryptosporidium spp. oocysts in natural mineral waters and drinking waters. Oocysts were recovered from seeded 1-liter volumes of mineral water by filtration through polycarbonate membranes and from drinking waters by filtration, immunomagnetizable separation, and filter entrapment, followed by direct extraction of DNA. The DNA was released from polycarbonate filter-entrapped oocysts by disruption in lysis buffer by using 15 cycles of freeze-thawing (1 min in liquid nitrogen and 1 min at 65°C), followed by proteinase K digestion. Amplicons were readily detected from two to five intact oocysts on ethidium bromide-stained gels. DNA extracted from Cryptosporidium parvum oocysts, C. muris (RN 66), C. baileyi (Belgium strain, LB 19), human-derived C. meleagridis, C. felis (DNA from oocysts isolated from a cat), and C. andersoni was used to demonstrate species identity by PCR-RFLP after simultaneous digestion with the restriction enzymes DraI and VspI. Discrimination between C. andersoni and C. muris isolates was confirmed by a separate, subsequent digestion with DdeI. Of 14 drinking water samples tested, 12 were found to be positive by microscopy, 8 were found to be positive by direct PCR, and 14 were found to be positive by using a nested PCR. The Cryptosporidium species detected in these finished water samples was C. parvum genotype 1. This method consistently and routinely detected >5 oocysts per sample.
Cryptosporidium is a coccidian protozoan parasite that has been implicated in numerous water-borne and food-borne outbreaks of cryptosporidiosis (11, 14, 39). Twelve species of Cryptosporidium are considered to be valid and potentially can be found in the environment: Cryptosporidium hominis predominantly in humans (30), C. parvum in humans and other mammals (46, 48), C. andersoni in cattle (27), C. muris in mice (47), C. felis in cats (2, 19), C. wrairi in guinea pigs (7, 50), C. meleagridis in turkeys (38), C. baileyi in chickens (8), C. saurophilum in lizards (23), C. serpentis in snakes (26), and C. nasorum (18) and C. molnari (1) in fish.
Cryptosporidium oocysts are frequent contaminants of water, with contributions from infected human and nonhuman hosts, livestock and agricultural practices and infected feral and transport hosts (40-42). Recent genetic analyses reveal that more than one species of Cryptosporidium can infect susceptible, immunocompromised (4, 13, 16, 29, 33, 34, 35), and immunocompetent (21, 33, 34, 53) human hosts, but C. parvum remains the most common species infecting humans. Cryptosporidium species reported to have crossed host-specificity barriers and to have been detected in human stools include C. meleagridis, C. felis, C. muris, and C. parvum dog genotype.
The environmental robustness of oocysts permits prolonged persistence in the aquatic environment (11, 45). Oocysts occur at low densities in water (40-42) and sensitive molecular methods, which can determine species and genotype small numbers of organisms reliably and reproducibly from water concentrates, are required. In particular, sensitivity must be maximized for detecting oocysts in natural mineral water sources since the expected level of contamination should be low because mineral water sources are situated in protected sites and exploited under strict regulations. The risk of contamination can derive from increased exploitation of new sites due to steadily increasing product consumption. In the United Kingdom, bottled water consumption has increased from >5 million liters in 1975 (43) to 1,380 million liters in 2000 (www.globaldrinks.com), and in Western Europe the consumption of mineral waters remains high: 91 liters per annum per capita (www.zenithinternational.com). Protozoan contamination of mineral water was detected in three brands of Mexican bottled mineral waters after membrane filtration and in vitro culture. Rivera et al. (37) identified four types of trophic or cystic forms of parasites, including the nonpathogenic (growth in vitro at 28°C but not at 37°C) amoebae Vahlkampfia vahlkampfi, Naegleria gruberi, and Acanthamoeba astronyxis and one flagellate (Bodomorpha minima). In France, a nonpathogenic amoebic species was detected in mineral waters originating from small-capacity, regional plants (9). Recently, Cryptosporidium oocysts were detected by immunofluorescence after membrane filtration (oocyst concentration range, 0.2 to 0.5 liter−1) in 2 of 13 commercial brands of Brazilian noncarbonated mineral waters (12).
Environmental contamination with oocysts of Cryptosporidium species that are not infectious to susceptible human hosts contributes to the difficulties in assessing the risk to public health from water-borne oocysts. The extent of the occurrence of species other than C. parvum in the environment is only now being addressed. Xiao et al. (52) reported the analysis of 29 storm water samples in the United States, which revealed the presence of Cryptosporidium spp. in 27 of them, mainly wildlife Cryptosporidium genotypes. The most common genotypes and species found in surface waters were C. parvum genotypes 1 and 2 and C. andersoni, with C. andersoni reported to be the most commonly found in wastewater (eight samples). However, restriction fragment length polymorphism (RFLP) patterns indicated mixed populations, and sequence analysis of the amplicons indicated that only four genotypes had 100% homology with previously known sequences.
Microscopy is used in both the United Kingdom Regulatory and U.S. standard methods (UK SI no. 1524 and U.S. EPA 1622 and 1623) to identify and differentiate Cryptosporidium spp. oocysts by assessing their morphology and morphometry. For many Cryptosporidium species present in water concentrates, oocyst size and shape are similar, making species identification based on morphometry difficult, if not impossible, due to size overlap.
Here, we describe a method for the detection and simultaneous species identification of small numbers of Cryptosporidium oocysts in United Kingdom finished drinking waters and noncarbonated, natural mineral waters by using epifluorescence microscopy, nested PCR and RFLP with a DNA template prepared directly from oocysts deposited on polycarbonate filters.
MATERIALS AND METHODS
Cryptosporidium oocyst sources.
Cervine-ovine oocysts of the Moredun isolate (genotype 2; MD, Moredun Scientific, Ltd., Edinburgh, United Kingdom) and the bovine Iowa isolate (genotype 2; Pleasant Hill Farm [PHF], Troy, Idaho) were obtained commercially. C. muris (RN 66) and C. baileyi (Belgium strain, LB 19) isolates were donated by K. Webster (Veterinary Laboratories Agency, Weybridge, United Kingdom). Purified human-derived C. meleagridis oocysts were obtained from the Scottish Parasite Diagnostic Laboratory Cryptosporidium oocyst isolate bank and from the Cryptosporidium Reference Unit, Public Health Laboratory Service, Swansea, United Kingdom. Purified DNAs from C. felis (oocysts isolated from a cat) and C. andersoni (oocysts isolated from a cow) were donated by G. Lindegard, Cornell University.
Mineral waters.
Four commercially available, noncarbonated natural mineral waters were selected to include a representative range of total dissolved solids (TDS). These were categorized as mineral waters A (430 mg liter−1), B (280 mg liter−1), C (136 mg liter−1), and D (91 mg liter−1). The TDS values are classified by the Natural Mineral Waters Regulations as follows: very low (<50 mg liter−1), low (50 to 500 mg liter−1), medium (500 to 1,500 mg liter−1), and rich (>1,500 mg liter−1) (5). Mineral waters classified as rich are usually carbonated and were deliberately excluded from the present study. Reverse osmosis (RO) water was used as a reference control. Natural mineral waters were donated by the manufacturers in glass or plastic containers with a 1-, 2.5-, or 5-liter capacity.
Preparation and seeding of mineral water concentrates.
The mineral water with the high (430 mg liter−1) TDS left a white, water-soluble salt deposit when filtered through 1.2-μm-pore-size cellulose nitrate membranes, within which seeded oocysts were contained. To simulate this environment for PCR, we filtered 5 liters of mineral waters A, B, C, and D through 1.2-μm-pore-size, 47-mm-diameter cellulose nitrate membranes and dissolved the membrane-entrapped salts in eluant (aqueous solution of 0.1% Tween 80 and 0.001% Anti-foam A; Sigma, London, United Kingdom). Filters were processed as follows: filters were placed inside a sterile 50-ml plastic tube containing 10 ml of eluant and vortexed for 2 min. The eluant was transferred to a 15-ml plastic tube, and the filter-elution-vortexing procedure was repeated in the 50-ml tube by using a further 5 ml of eluant. Both eluants were combined and then centrifuged at 1,500 × g for 10 min at 4°C, and the eluant was aspirated to leave 5 ml.
Purified C. parvum oocysts of bovine origin (Iowa-PHF; genotype 2) were used for seeding the mineral water concentrates. The density of the oocyst stock suspension used for seeding was determined from the arithmetic mean of three separate hemocytometer counts, and a working dilution of 2 × 104 ml−1 was prepared in RO water. Five microliters of this working dilution, containing ca. 100 oocysts, was seeded in 85-μl aliquots of each mineral water concentrate, and the suspension was buffered with 10 μl of 10× lysis buffer before DNA extraction by freezing and thawing (see below). The extracted DNA and an internal control (IC) were coamplified with the CPB-DIAG primers in direct PCRs to determine whether any PCR inhibitors were present in the mineral water concentrates (32).
Recovery of oocysts seeded into 1-liter volumes of mineral water A by membrane filtration.
For seeding and recovery experiments, fluorescence-activated cell sorting (36) was used to obtain accurate oocyst (MD isolate) seed preparations containing 100 (range, 98 to 101) intact, unstained oocysts in 2 ml of water. From these suspensions 200-μl aliquots, each containing ca. 10 fluorescence-activated cell-sorted oocysts, were seeded into replicates of 1-liter volumes of mineral water A (with the highest TDS). Seeded oocysts were concentrated by filtration through 3-μm-pore-size, 13-mm-diameter polycarbonate filters (Millipore) by using negative pressure. Filter-entrapped oocysts were then fixed, stained, and observed microscopically (see below).
Enhanced morphological detection of oocysts on filters.
MD oocysts trapped on polycarbonate filters by filtration and filter controls, prepared by depositing small numbers (ca. 10) of Iowa-PHF oocysts directly on 3-μm-pore-size, 13-mm-diameter polycarbonate filters by negative pressure, were fixed, stained, and observed directly by microscopy.
The filters containing entrapped oocysts were placed onto microscope slides, air dried at room temperature, methanol fixed, and stained with optimally diluted fluorescein isothiocyanate (FITC)-labeled genus-specific anti-Cryptosporidium sp. monoclonal antibody (FITC-MAb; Crypt-a-Glo; Waterborne, Inc., New Orleans, La.) according to the manufacturer's instructions. The fluorogenic dye DAPI (4′,6′-diamidino-2-phenylindole) was used to visualize sporozoite nuclei according to the method of Grimason et al. (15). A total of 50 μl of DAPI (final concentration of 4 × 10−4 mg ml−1 in 150 mM phosphate-buffered saline [pH 7.2]) was applied onto each sample for 2 min at room temperature. Filters were washed once in RO water to remove excess DAPI, mounted in glycerol-phosphate-buffered saline (60:40 [vol/vol]) containing 2% antifadant 1,4-diazabicyclo(2,2,2)octane (Dabco); a coverslip was then applied, and the filters were examined by epifluorescence microscopy (×400 magnification) on an Olympus BH2 microscope. FITC emissions were viewed by using a blue filter block (480-nm excitation, 520-nm emission), and DAPI emissions were viewed by using a UV filter block (350-nm excitation, 450-nm emission).
DNA extraction from oocysts either in suspension or directly from filters.
A freeze-thaw protocol, which maximizes DNA extraction from oocysts, was followed (31; R. A. B. Nichols and H. V. Smith, unpublished data) and applied either to oocysts in suspension or to filter-entrapped oocysts.
Oocysts in lysis buffer (50 mM Tris-HCl [pH 8.5], 1 mM EDTA [pH 8], 0.5% sodium dodecyl sulfate) were subjected to 15 freeze-thaw cycles (1 min in liquid nitrogen and 1 min at 65°C per cycle). Samples were microcentrifuged at 14,000 × g for 10 s and then incubated with proteinase K (final concentration, 200 μg ml−1) at 55°C for 3 h, while being rotated horizontally in a Techne hybridization oven. Tubes were then incubated for 20 min in a water bath at 90°C to denature proteinase K, chilled on ice for 1 min, and centrifuged at 14,000 × g for 5 min. The supernatant was transferred to a clean tube and either used immediately for PCR amplification or stored at −20°C until used.
Filters were retrieved from slides after microscopic observation by gently immersing the slide vertically in a container of clean RO water to detach the coverslip and then by lifting the filter from the slide by using clean forceps. Each filter was then placed in a 0.5-ml screw-cap microcentrifuge tube and thoroughly wetted in 50 μl of lysis buffer by rolling the capped centrifuge tube between finger and thumb. Oocysts were lysed by using the freeze-thaw protocol described.
PCR protocol.
PCR amplifications were performed in a Perkin-Elmer thermocycler model 9600 in 0.5 ml thin-walled tubes. Reaction volumes of either 50 or 100 μl consisted of premixed reagents containing a 200 mM concentration of each of the four deoxynucleoside triphosphates (Pharmacia), 0.2 μM concentrations of each of primers CPB-DIAGF/R (MWG-Biotech, Milton Keynes, United Kingdom), bovine serum albumin at a final concentration of 400 μg ml−1, 3.5 mM MgCl2, 2% Tween 20 (final concentration), and 2.5 U of Taq polymerase in 1× PCR buffer IV (Advanced Biotechnologies). DNA template did not exceed 10% of the reaction volume.
Nested PCR assay.
To develop a nested PCR assay according to established principles (10, 17), a primer set, which flanked the amplicon defined by the CPB-DIAGF/R primers, was designed. The outer primers were designed to have an annealing temperature significantly higher than the inner primers so that a single-tube nested PCR could also be developed, if required. The GC content was 59.2 and 53.8% forward and reverse primers, respectively, and care was taken to choose a noncomplementary primer set, particularly at the 3′ ends of the two primers, to avoid primer-dimer formation.
A 27-mer forward primer (N-DIAGF2; 5′-CAA TTG GAG GGC AAG TCT GGT GCC AGC-3′) and a 26-mer reverse primer (N-DIAGR2; 5′-CCT TCC TAT GTC TGG ACC TGG TGA GT-3′) were selected as they had homology with all Cryptosporidium species and genotypes listed in Table 1. The expected lengths of amplicons obtained after amplification with N-DIAGF2/R2 vary from 655 to 667 bp depending on the species of Cryptosporidium or C. parvum genotype.
TABLE 1.
Cryptosporidium spp. and genotypes determined by RFLP of the amplicon defined by the CPB-DIAGR/F primers after digestion with the enzymes VspI, DraI, and DdeI
| Cryptosporidium spp. and genotypes (total amplicon length in bp) | Amplicon length(s) (bp) as determined with:
|
GenBank accession no. | ||
|---|---|---|---|---|
| VspI | DraI | DdeI | ||
| C. parvum 1 (438) | 222, 104, 112 | None | 204, 68, 166 | L16997 |
| C. parvum 2 (435) | 219, 104, 112 | None | 201, 68, 166 | L16996, AF161856 |
| C. muris (432) | 320, 112 | None | 42, 224, 166 | AF093498, AF093498, AF093497 |
| C. andersoni (431) | 319, 112 | None | 265, 166 | AF093496, L19069 |
| C. felis (455) | 239, 104, 112 | 50, 405 | 221, 68, 166 | AF087577 |
| C. baileyi (428) | 212, 104, 112 | 84, 344 | 262, 166 | L19068, AF093495 |
| C. meleagridis (434) | 47, 171, 104, 112 | None | 200, 68, 166 | AF112574 |
| C. serpentis (430) | 318, 112 | None | 264, 166 | AF093502a |
| C. wrairi (435) | 219, 104, 112 | None | 201, 68, 166 | AF115378 |
| Cryptosporidium pig (435) | 219, 104, 112 | None | 201, 68, 166 | AF108861 |
| Cryptosporidium desert monitor (432) | 216, 108, 112 | None | 198, 68, 166 | AF112573 |
| Cryptosporidium mouse (439) | 48, 175, 104, 112 | None | 205, 68, 166 | AF108863 |
| Cryptosporidium ferret (438) | 48, 174, 103, 113 | None | 204, 68, 166 | AF112572 |
| Cryptosporidium dog (429) | 213, 104, 112 | None | 195, 68, 166 | AF112576 |
| Cryptosporidium koala (436) | 220, 104, 112 | None | 202, 68, 166 | AF108860 |
| Cryptosporidium kangaroo (436) | 220, 104, 112 | None | 202, 68, 166 | AF112570 |
| Cryptosporidium monkey (436) | 220, 104, 112 | None | 202, 68, 166 | AF112569 |
| Cryptosporidium bear (432) | 216, 104, 112 | None | 87, 111, 68, 166 | AF247535 |
Optimum annealing temperatures for both primer sets (CPB-DIAGF/R and N-DIAGF2/R2) were determined by PCR amplification of DNA equivalent to two oocysts, obtained by serial dilution of DNA extracted from 106 oocysts (Iowa isolate, genotype 2; extracted as described previously). Both outer and inner primers were evaluated by direct PCR assays at the optimum concentration of 200 nM each per reaction tube. The primary and secondary PCR reagent concentrations were as described above. The primary PCR contained 2 μl of DNA in a 50-μl reaction volume, and the secondary PCR reamplified 5 μl of the primary PCR product. The following step cycle protocols were used: primary PCR consisted of initial denaturation at 95°C for 5 min; 25 to 35 cycles at 94°C for 30 s, 68°C for 1 min, and 72°C for 30 s; and extension at 72°C for 10 min. Secondary PCR consisted of initial denaturation at 95°C for 1 min; 25 to 35 cycles at 94°C for 30 s, 60°C for 1 min, and 72°C for 30 s; and extension at 72°C for 10 min.
To minimize cross-contamination pre-PCR, DNA extraction and template addition to reaction tubes and post-PCR manipulations were performed in three separate laboratory areas by using designated micropipettes and filtered tips.
Cryptosporidium oocyst detection in United Kingdom drinking water samples.
Fourteen United Kingdom finished water samples were concentrated and oocysts were purified by immunomagnetizable separation (IMS) according to United Kingdom Standard Operating Protocols identified in part 2 of the protocol for monitoring Cryptosporidium oocysts in water supplies (49) generated to implement the Water Supply (Water Quality) (Amendment) Regulations of 1999 (SI no. 1524). IMS separations from turbid samples were performed in subsamples whenever necessary to have packed pellet volumes equal to 0.5 ml of sediment in a 10-ml sample volume (5% packed pellet volumes). Oocysts recovered from the IMS subsamples were pooled on one polycarbonate membrane filter (13-mm diameter and 1.2-μm pore size) for epifluorescence microscopy and subsequent DNA extraction. The finished water concentrates were analyzed for the presence of Cryptosporidium oocysts by using epifluorescence microscopy, direct PCR, and nested PCR.
Species identification by PCR-RFLP analysis.
Purified oocysts of C. parvum (genotype 2; human isolate), C. muris, C. baileyi, and C. meleagridis and DNA from C. felis and C. andersoni oocysts were used to confirm species identity by PCR-RFLP. Approximately 103 oocysts of each species suspended in 100 μl of PCR buffer were subjected to DNA extraction by freeze-thawing, and 20 μl of the supernatant was used for PCR amplification. Then, 10 μl of the PCR product was digested for 2 h at 37°C with 10 U of each enzyme DraI and VspI (Gibco/Life Technologies) in 35 μl of 1× React-2 buffer (Gibco/Life Technologies/Invitrogen). Undigested controls were run alongside the digested fragments in 2% agarose gels at 100 V for 2 h and then stained with ethidium bromide (0.5 μg ml−1) incorporated into both gel and running buffer. The interpretation of nested PCR-RFLP results was performed as described in Table 1.
RESULTS
Direct PCR detection of oocysts seeded into mock concentrates of natural mineral waters.
Direct PCR coamplification of DNA extracted from 100 oocysts seeded in concentrated mineral waters A, B, C, and D and the IC yielded two bands; a 435-bp band corresponding to amplicons generated by the amplification of 10 oocysts, and a 503-bp band corresponding to the amplification of the IC. No inhibitory effect on the PCR occurred when the mineral waters concentrates were tested (data not shown).
Direct PCR and nested PCR detection of oocysts experimentally entrapped on membrane filters by filtration of seeded United Kingdom mineral water A.
Enumerations of oocysts recovered from replicate filtrations of mineral water A and filter controls by microscopy are presented in Table 2. Direct PCR coamplification of 10 μl of DNA template extracted from each membrane containing entrapped, methanol-fixed, FITC-antibody, and DAPI-labeled oocysts with 10 μl of the IC yielded a product corresponding to the IC (553 bp) from all mineral water A replicate samples tested (data not shown). However, genomic DNA (435 bp) amplicons either were generated at low levels or were undetectable on an agarose gel (Table 2).
TABLE 2.
Comparison of the sensitivity of epifluorescence microscopy, direct PCR, and nested PCR for detecting oocysts retained on membrane filters
| Filter | No. of oocysts counted (FITC labeled/DAPI labeleda) | Sensitivityb as determined by:
|
|
|---|---|---|---|
| Direct PCR | Two-step nested PCR | ||
| F filtersc | |||
| F1 | 11/6 | Negative | ND |
| F2 | 6/5 | 1+ | 4+ |
| F3 | 4/0 | 1+ | 4+ |
| F4 | 8/7 | Negative | 4+ |
| F5 | 7/4 | Negative | 4+ |
| F6 | 8/4 | Negative | 3+ |
| F7 | 7/5 | Negative | 4+ |
| F8 | 4/2 | Negative | 2+ |
| F9 | 6/5 | Negative | 1+ |
| F10 | 12/10 | Negative | 3+ |
| F11 | 10/7 | 1+ | 4+ |
| F12e | 9/2 | Negative | 4+ |
| FC filtersd | |||
| FC1 | 5/2 | Negative | 3+ |
| FC2 | 9/2 | Negative | 4+ |
| FC3 | 9/3 | Negative | 4+ |
| FC4 | 4/1 | Negative | 4+ |
| FC5 | 15/9 | Negative | 3+ |
| FC6 | 2/1 | 1+ | 3+ |
| FC7 | 13/10 | 1+ | 4+ |
| FC8 | 7/2 | Negative | 3+ |
| FC9 | 12/5 | Negative | 3+ |
DAPI labelling refers to oocysts containing one to four nuclei.
Negative, no PCR product detected on gel; 4+, PCR product yielding a strong band; 3+ and 2+, PCR product yielding medium-strength bands; 1+, PCR product yielding a weak band on the gel; ND, not done.
F filters, filters through which seeded mineral water A was passed.
FC filters, filter controls (filters onto which a known number of oocysts were deposited [see text]).
Filtration of 1 liter of RO water.
Using the nested PCR assay with DNA from filter-entrapped oocysts by mineral water A filtrations generated detectable amplicons when 50 cycles of PCR amplification were performed. The sensitivity of the nested PCR assay was two to three oocysts present on each filter with PCR product detected on the gel in sufficient concentration for RFLP analysis (Table 2). No PCR product was ever observed with negative controls.
Detection and specifies identification of Cryptosporidium oocysts concentrated from United Kingdom finished drinking waters. (i) Confirmation of species identification by RFLP.
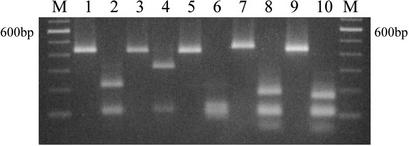
The amplification of C. parvum genotype 2, C. muris, C. baileyi, C. felis, and C. meleagridis oocyst DNA yielded products of 435, 431, 428, 455, and 434 bp, respectively (Table 1 and Fig. 1). On 2% ethidium bromide-stained gels, the double-digest product (with DraI and VspI) produced profiles that confirmed the reported sequence of these cloned genes (Table 1 and Fig. 1). RFLP analysis of the amplicon defined by the diagnostic primers with restriction enzymes used for identifying Cryptosporidium species and C. parvum genotypes is presented in Table 1.
FIG. 1.
Two percent agarose gel electrophoresis analysis of amplicons before and after simultaneous digestion with restriction enzymes DraI and VspI. Lanes 1, 3, 5, 7, and 9, undigested PCR product from C. parvum, C. muris, C. baileyi, C. felis, and C. meleagridis, respectively; lanes 2, 4, 6, 8, and 10, fragmented PCR product from C. parvum (222, 112, and 104 bp), C. muris (319 and 112 bp), C. baileyi (128, 112, 104, and 84 bp), C. felis (189, 112, 104, and 50 bp), and C. meleagridis (171, 112, 104, and 47 bp), respectively. Lanes M, 100-bp DNA ladder. In this figure, bands of sizes 112 and 104 bp comigrate and appear as a single band. This does not affect the interpretation of the RFLP analysis nor the discrimination among the five species.
(ii) Immunofluorescence, direct PCR, and nested PCR detection of oocysts in finished drinking waters.
Oocysts (range, 2 to 37) were detected in 12 of 14 water samples by microscopy on filters (Table 3). Of these, the number of oocysts that contained nuclei (one to four DAPI-stained nuclei per oocyst) ranged from 2 to 30. No oocysts were visible on filters 8 and 11.
TABLE 3.
Detection of Cryptosporidium species and C. parvum genotype by nested PCR-RFLP analysis of filter-extracted oocyst DNA in United Kingdom finished water samples
| Sample (no.) | No. of oocysts 10 liters−1 | IMS vol (ml) | Total no. of labeled oocysts (FITC/DAPI) | Resulta as determined by:
|
RFLP sp.b | |
|---|---|---|---|---|---|---|
| Direct PCR | Nested PCR | |||||
| 1 (1285) | NKc | 5 | 4/4 | 2+ | 4+ | C. parvum |
| 2 (1285) | NK | 18 | 37/30 | 2+ | 4+ | C. parvum† |
| 3 (1304) | 0.07 | 10 | 3/3 | 1+ | 4+ | C. parvum† |
| 4 (1305) | 0.1 | 5 | 12/10 | 2+ | 4+ | C. parvum† |
| 5 (1291) | 0.5 | 5 | 10/8 | 2+ | 4+ | C. parvum† |
| 6 (1293) | 0.9 | 5 | 19/15 | 2+ | 4+ | C. parvum† |
| 7 (1297) | 0.6 | 5 | 27/24 | 1+ | 4+ | C. parvum† |
| 8 (1315) | 0.02 | 5 | 0/0 | Negative | 4+ | C. parvum |
| 9 (1372) | 0.015 | 5 | 2/2 | Negative | 4+ | C. parvum |
| 10 (1389) | 0.07 | 5 | 2/2 | 1+ | 4+ | C. parvum |
| 11 (1468) | 0.02 | 5 | 0/0 | Negative | 2+* | NI |
| 12 (1419) | 0.05 | 5 | 2/1 | Negative | 2+* | NI |
| 13 (1418) | 0.09 | 5 | 5/5 | Negative | 4+* | C. parvum |
| 14 (1416) | 0.1 | 5 | 7/6 | Negative | 4+ | C. parvum |
Of the 14 DNA samples that were amplified by direct PCR, 8 were found to be positive when 10 μl of lysate was amplified (Table 3). No inhibition of test sample occurred when 5 μl of lysate was coamplified with 5 μl of the IC (10−9 dilution), since IC amplicons were observed with all samples (data not shown). A total of 11 samples were determined to be positive (10 strong and 1 weak, sample 13 [1418]) with the nested PCR assay after 50 cycles of nested amplification (samples 11, 12, and 13, Table 3). Increasing the number of amplification cycles to 70 (35 cycles for both the primary and secondary PCRs) yielded amplicons from the three previously negative samples (Table 3). Samples 11 and 12 generated amplicons that were longer (ca. 450 bp) than expected for C. parvum (435 bp), and less PCR product was detected visually compared to samples 13 and 14 (data not shown).
(iii) Species and genotype identification of oocysts recovered from finished drinking waters.
RFLP analysis of the amplicons by simultaneous digestion with the enzymes VspI and DraI revealed a profile compatible with C. parvum species in 12 of 14 samples (Table 3). Samples 11 and 12 gave identical profiles; these profiles differed from those expected with known species or genotypes in that three fragments were obtained corresponding to ca. 240, 104, and 50 bp. In an attempt to clarify the digestion pattern of these samples, separate restrictions with DraI and SspI were performed, but no digestion product was generated with these enzymes (data not shown). Of the drinking water concentrates tested, six were genotyped with the single-tube nested COWP assay (17), and RFLP analysis of the PCR product indicated that C. parvum genotype 1 was the Cryptosporidium genotype detected (Table 3).
DISCUSSION
Both species identification and detection of small numbers of oocysts are prerequisites for a successful PCR-based method for detecting Cryptosporidium oocysts in natural mineral waters and in drinking waters. PCR amplification of multicopy genes is a useful approach to sensitive molecular detection. Le Blancq et al. (24) determined that 20 copies of the Cryptosporidium ribosomal DNA gene are present per oocyst. Since the complete sequence of the small-subunit (SSU) rRNA gene from oocysts originating from numerous hosts and from different Cryptosporidium species is currently available from the GenBank database, species identification by PCR-RFLP analysis of this Cryptosporidium gene has been used by many authors (3, 22, 25, 28, 51).
Leng et al. (25) devised primers that amplify the entire 18S rRNA gene based on sequences deposited in the GenBank by Pieniazek et al. (N. J. Pieniazek, M. J. Arrowood, B. L. Blagburn, H. M. Mathews, and S. B. Slemenda, unpublished data [GenBank accession no. L16996]) and confirmed the feasibility of distinguishing among C. parvum, C. muris, and C. baileyi by PCR-RFLP by double digestion with the enzymes DraI and VspI. These primers, aimed at conserved sequences of the gene, also amplify Eimeria neischultzi and E. papillata DNA (44). In contrast, the diagnostic primers used in the present study are suitable for species identification by PCR-RFLP (Table 1). To our knowledge this is the first report to confirm the GenBank sequences by simultaneous restriction digestion with the enzymes DraI and VspI using PCR products obtained with the diagnostic primers CPB-DIAGF and CPB-DIGR of Johnson et al. (20).
Champliaud et al. (6) compared the performance of eight primer pairs described in the literature for their ability to distinguish among C. parvum, C. muris, C. baileyi, and C. meleagridis. These authors showed that all primer pairs amplified DNA from C. meleagridis and C. parvum, including the two genus-specific primer pairs directed to the 18S rRNA gene (3, 20) that amplified DNA from all four species. Digestion of the 18S rRNA gene product, obtained with the primers developed by Awad-El-Kariem et al. (3), with MaeI yielded incomplete digestion, as expected. However, these authors did not digest the PCR products obtained with the primers of Johnson et al. (20) with the enzymes DraI and VspI used in the present study.
Complete sequences of the SSU rRNA gene from different isolates of C. parvum, C. muris, C. baileyi, and C. serpentis have been deposited in GenBank (51). A phylogenetic study by sequence analysis of this gene locus confirmed these species as distinct taxonomic groups. A nested PCR-RFLP assay for interspecies discrimination was devised by digesting the PCR product with SspI, and intraspecies variation of C. parvum from human or animal genotypes was tested by digestion with VspI (51). The sensitivity of this test with purified oocysts was reported to be of a single oocyst; however, the sensitivity and specificity of detection in environmental samples are yet to be evaluated fully.
In the present study we have shown that species identification can be accomplished by nested PCR-RFLP from the amplification of five or fewer oocysts entrapped on polycarbonate membranes and isolated from noncarbonated, natural mineral waters and from drinking waters. When optimized PCR and DNA extraction protocols are used, these primers are sensitive and suitable for use in direct PCR and nested PCR assays. The five Cryptosporidium species simultaneously detected because of the polymorphism of this region of the 18S rRNA, defined by the CPB-DIAGF and CPB-DIGR primers, are relevant to a detection method for natural mineral and drinking waters since these species can be present in the United Kingdom environment. Differentiation between C. andersoni and C. muris is possible by further digesting the PCR product with the restriction enzyme DdeI (data not shown).
In addition, filter concentrates from noncarbonated mineral water do not interfere with the PCR and the DNA extracted from methanol-fixed and FITC-MAb and DAPI-stained oocysts entrapped on polycarbonate membranes does not interfere with nested PCR amplification, given the current sensitivity of two to five oocysts. Since only 20% of the extracted DNA is used for the first amplification, other PCR tests, including C. parvum genotyping, can also be performed on the same sample by amplifying other polymorphic loci with sensitive and specific primers (17).
The identification of C. parvum genotype 1 (renamed C. hominis [30]) DNA in all finished drinking water concentrates is compatible with the fact that the water samples were collected from an oocyst-contaminated drinking water strongly associated with a C. parvum genotype 1 outbreak. Thus, our nested PCR appears to be more sensitive than microscopy for detecting oocysts in these water concentrates. In concentrates that had no identifiable oocysts present, occluding debris may have obscured oocyst observation by microscopy; however, given that the water samples were collected from a Cryptosporidium-positive water source, the presence of amplifiable, naked DNA in the water concentrate remains a plausible explanation for these results. We identified the same Cryptosporidium species and C. parvum genotype both in outbreak-associated human stools and in the oocysts present in the finished drinking water supplying that outbreak area. Sequencing the PCR products obtained from samples 11 and 12 may assist in elucidating the Cryptosporidium species-C. parvum genotype signature of the amplifiable Cryptosporidium DNA isolated.
REFERENCES
- 1.Alvarez-Pellitero, P., and A. Sitjà-Bobadilla. 2002. Cryptosporidium molnari n. sp. (Apicomplexa: Cryptosporidiidae) infecting two marine fish species, Sparus aurata L. and Dicentrarchus labrax L. Int. J. Parasitol. 32:1007-1021. [DOI] [PubMed] [Google Scholar]
- 2.Asahi, H., T. Koyama, H. Arai, Y. Funakoshi, H. Yamamura, R. Shirasaka, and K. Okutomi. 1991. Biological nature of Cryptosporidium sp. isolated from a cat. Parasitol. Res. 77:237-240. [DOI] [PubMed] [Google Scholar]
- 3.Awad-El-Kariem, F. M., D. C. Warhurst, and V. McDonald. 1994. Detection and species identification of Cryptosporidium oocysts using a system based on PCR and endonuclease restriction. Parasitology 109:19-22. [DOI] [PubMed] [Google Scholar]
- 4.Cacciò, S., E. Pinter, R. Fantini, I. Mezzaroma, and E. Pozio. 2002. Human infection with Cryptosporidium felis: case report and literature review. Emerg. Infect. Dis. 8:85-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cahill, J. 1994. Bottled water: do the public know what they are getting? Commun. Dis. Environ. Health Scotland Wkly. Rep. 28:94/51; 94/52. [Google Scholar]
- 6.Champliaud, D., P. Gobet, M. Naciri, O. Vagner, J. Lopez, J. C. Buisson, I. Varga, G. Harly, R. Mancassola, and A. Bonnin. 1998. Failure to differentiate Cryptosporidium parvum from C. meleagridis based on PCR amplification of eight DNA sequences. Appl. Environ. Microbiol. 64:1454-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chrisp, C. E., W. C. Reid, G. D. Rush, M. A. Suckow, A. Bush, and M. J. Thomann. 1990. Cryptosporidiosis in guinea pigs: an animal model. Infect. Immun. 56:674-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Current, W. L., S. J. Upton, and T. B. Haynes. 1986. Life cycle of Cryptosporidium baileyi n. sp. (Apicomplexa: Cryptosporididae) infecting chickens. J. Protozool. 33:289-296. [DOI] [PubMed] [Google Scholar]
- 9.Dive, D., J. P. Picard, and H. Leclerc. 1979. Les amibes dans les eaux d'alimentation: evaluation des risques. Ann. Microbiol. Inst. Pasteur 130A:487-498. [PubMed] [Google Scholar]
- 10.Erlich, H. A., D. Gelfand, and J. J. Sninsky. 1991. Recent advances in the polymerase chain reaction. Science 252:1643-1651. [DOI] [PubMed] [Google Scholar]
- 11.Fayer, R., U. Morgan, and S. J. Upton. 2000. Epidemiology of Cryptosporidium: transmission, detection and identification. Int. J. Parasitol. 30:1305-1322. [DOI] [PubMed] [Google Scholar]
- 12.Franco, R. M. B., and R. Cantusio Neto. 2002. Occurrence of cryptosporidial oocysts and Giardia cysts in bottled mineral water commercialized in the City of Campinas, State of São Paulo, Brazil. Mem. Inst. Oswaldo Cruz 97:205-207. [DOI] [PubMed] [Google Scholar]
- 13.Gatei, W., R. W. Ashford, N. J. Beeching, S. K. Kamwati, J. Greensill, and C. A. Hart. 2002. Cryptosporidium muris infection in an HIV-infected adult in Kenya. Emerg. Infect. Dis. 8:204-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girdwood, R. W. A., and H. V. Smith. 1999. Cryptosporidium, p. 487-497. In R. Robinson, C. Batt, and P. Patel (ed.), Encyclopaedia of food microbiology, vol. 1. Academic Press, Ltd., London, United Kingdom.
- 15.Grimason, A. M., H. V. Smith, J. F. W. Parker, Z. Bukhari, A. T. Campbell, and L. J. Robertson. 1994. Application of DAPI and immunofluorescence for enhanced identification of Cryptosporidium spp. oocysts in water samples. Water Res. 28:733-736. [Google Scholar]
- 16.Guyot, K., A. Follet-Dumoulin, E. Lelievre, C. Sarfati, M. Rabodonirina, G. Nevez, J. C. Cailliez, D. Camus, and E. Dei-Cas. 2001. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J. Clin. Microbiol. 39:3472-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Homan, W., T. van Gorkom, Y. Y. Kan, and J. Hepener. 1999. Characterization of Cryptosporidium parvum in human and animal feces by single-tube nested polymerase chain reaction and restriction analysis. Parasitol. Res. 85:707-712. [DOI] [PubMed] [Google Scholar]
- 18.Hoover, D. M., F. J. Hoerr, and W. W. Carlton. 1981. Enteric cryptosporidiosis in a naso tang, Naso lituratus Bloch and Schneider. J. Fish Dis. 4:425-428. [Google Scholar]
- 19.Iseki, M. 1979. Cryptosporidium felis sp. n. (Protozoa: Eimeriorina) from the domestic cat. Jpn. J. Parasitol. 28:285-307. [Google Scholar]
- 20.Johnson, D. W., N. J. Pieniazek, D. W. Griffin, L. Misener, and J. B. Rose. 1995. Development of a PCR protocol for sensitive detection of Cryptosporidium in water samples. Appl. Environ. Microbiol. 61:3849-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katsumata, T., D. Hosea, I. G. Ranuh, S. Uga, T. Yanagi, and S. Kohono. 2000. Short report: possible Cryptosporidium muris infections in humans. Am. J. Trop. Med. Hyg. 62:70-72. [DOI] [PubMed] [Google Scholar]
- 22.Kimbell, L. M., III, D. L. Miller, W. Chavez, and N. Altman. 1999. Molecular analysis of the 18S rRNA gene of Cryptosporidium serpentis in a wild-caught corn snake (Elaphe guttata) and a five-species restriction fragment length polymorphism-based assay that can additionally discern C. parvum from C. wrairi. Appl. Environ. Microbiol. 65:5345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koudela, B., and D. Modry. 1998. New species of Cryptosporidium (Apicomplexa: Cryptosporidiidae) from lizards. Folia Parasitol. 45:93-100. [Google Scholar]
- 24.LeBlancq, S. M., N. V. Khramtsov, F. Zamani, S. J. Upton, and T. W. Wu. 1997. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol. Biochem. Parasitol. 90:463-478. [DOI] [PubMed] [Google Scholar]
- 25.Leng, X., D. A., Mosier, and R. D. Oberst. 1996. Differentiation of Cryptosporidium parvum, C. muris, and C. baileyi by PCR-RFLP analysis of the 18sRNA gene. Vet. Parasitol. 62:1-7. [DOI] [PubMed] [Google Scholar]
- 26.Levine, N. D. 1980. Some corrections of coccidian (Apicomplexa: Protozoa) nomenclature. J. Parasitol. 66:830-834. [PubMed] [Google Scholar]
- 27.Lindsay, D. S., S. J. Upton, D. S. Owens, U. M. Morgan, J. R. Mead, and B. L. Blagburn. 2000. Cryptosporidium andersoni n. sp. (Apicomplexa: Cryptosporiidae) from cattle, Bos taurus. J. Eukaryot. Microbiol. 47:91-95. [DOI] [PubMed] [Google Scholar]
- 28.Lowery, C. J., J. E. Moore, B. C. Millar, D. P. Burke, K. A. J. McCorry, E. Crothers, and J. S. G. Dooley. 2000. Detection and speciation of Cryptosporidium spp. in environmental water samples by immunomagnetic separation, PCR and endonuclease restriction. J. Med. Microbiol. 49:779-785. [DOI] [PubMed] [Google Scholar]
- 29.Morgan, U. M., R. Weber, L. Xiao, I. Sulaiman, R. C. A. Thompson, W. Ndiritu, A. Lal, A. Moore, and P. Deplazes. 2000. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J. Clin. Microbiol. 38:1180-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan-Ryan, U. M., A. Fall, L. A. Ward, N. Hijjawi, I. Sulaiman, R. Fayer, R. C. A. Thompson, M. Olson, A. Lal, and L. Xiao. 2002. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol. 49:433-440. [DOI] [PubMed]
- 31.Nichols, R. A. B. 2002. Ph.D. thesis. University of Strathclyde, Glasgow, United Kingdom.
- 32.Nichols, R. A. B., C. A. Paton, B. M. Campbell, J. Wastling, and H. V. Smith. 2002. A sensitive, semiquantitative direct PCR-RFLP assay for simultaneous detection of five Cryptosporidium species in treated drinking waters and mineral waters. Water Sci. Technol. Water Supply 2:9-15. [Google Scholar]
- 33.Pedraza-Diaz, S., C. Amar, A. M. Iversen, P. J. Stanley, and J. McLauchlin. 2001. Unusual Cryptosporidium species recovered from human faeces: first description of Cryptosporidium felis and Cryptosporidium dog type from patients in England. J. Med. Microbiol. 50:293-296. [DOI] [PubMed] [Google Scholar]
- 34.Pedraza-Diaz, S., C. Amar, J. McLauchlin, G. L. Nichols, K. M. Cotton, P. Godwin, A. M. Iversen, L. Milne, J. R. Mulla, K. Nye, H. Panigrahl, S. R. Venn, R. Wiggins, M. Williams, and E. R. Youngs. 2001. Cryptosporidium meleagridis from humans: molecular analysis and description of affected patients. J. Infect. 42:243-250. [DOI] [PubMed] [Google Scholar]
- 35.Pieniazek, N. J., F. J. Bornay-Llinares, S. B. Slemenda, A. J. daSilva, I. N. S. Moura, M. J. Arrowood, O. Ditrich, and D. G. Addiss. 1999. New Cryptosporidium genotypes in HIV-infected persons. Emerg. Infect. Dis. 5:444-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reynolds, D. T., R. B. Slade, N. J. Sykes, A. Jonas, and C. R. Fricker. 1999. Detection of Cryptosporidium oocysts in water: techniques for generating precise recovery data. J. Appl. Microbiol. 87:804-813. [DOI] [PubMed] [Google Scholar]
- 37.Rivera, F., M. Galvan, E. Robles, P. Leal, L. Gonzalez, and A. M. Lacy. 1981. Bottled mineral waters polluted by protozoa in Mexico. J. Protozool. 28:54-56. [DOI] [PubMed] [Google Scholar]
- 38.Slavin, D. 1955. Cryptosporidium meleagridis (sp. nov.). J. Comp. Pathol. Ther. 65:262-266. [DOI] [PubMed] [Google Scholar]
- 39.Slifko, T. R., H. V. Smith, and J. R. Rose. 2000. Emerging parasite zoonoses associated with water and food. Int. J. Parasitol. 30:1379-1393. [DOI] [PubMed] [Google Scholar]
- 40.Smith, H. V. 1995. Intestinal protozoa, p. 79-117. In S. H. Gillespie and P. M. Hawkey (ed.), Medical parasitology: a practical approach. IRL Press/Oxford University Press, Oxford, United Kingdom.
- 41.Smith, H. V., and J. B. Rose. 1990. Waterborne cryptosporidiosis. Parasitol. Today 6:8-12. [DOI] [PubMed] [Google Scholar]
- 42.Smith, H. V., and J. B. Rose. 1998. Waterborne cryptosporidiosis: current status. Parasitol. Today 14:14-22. [DOI] [PubMed] [Google Scholar]
- 43.Stickler, D. J. 1989. The microbiology of bottled natural mineral waters. J. R. Soc. Health 109:118-124. [DOI] [PubMed] [Google Scholar]
- 44.Sulaiman, I. M., L. Xiao, and A. A. Lal. 1999. Evaluation of Cryptosporidium parvum genotyping techniques. Appl. Environ. Microbiol. 65:4431-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamburrini, A., and E. Pozio. 1999. Long-term survival of Cryptosporidium parvum oocysts in seawater and in experimentally infected mussels (Mytilis galloprovincialis). Int. J. Parasitol. 29:711-715. [DOI] [PubMed] [Google Scholar]
- 46.Tyzzer, E. E. 1907. A sporozoan found in the peptic gland of the common mouse. Proc. Soc. Exp. Biol. Med. 5:12-13. [Google Scholar]
- 47.Tyzzer, E. E. 1910. An extracellular coccidium Cryptosporidium muris of the gastric glands of the common mouse. J. Med. Res. 23:487-509. [PMC free article] [PubMed] [Google Scholar]
- 48.Tyzzer, E. E. 1912. Cryptosporidium parvum (sp. nov.): a coccidian found in the small intestine of the common mouse. Arch. Protistenkd. 26:394-412. [Google Scholar]
- 49.United Kingdom Drinking Water Inspectorate. 2000. The Water Supply (Water Quality) (Amendment) Regulations 1999. Cryptosporidium in water supplies: laboratory analysis, part 2, revision 3 (November 2000). Protocol containing standard operating protocols (SOPs) for monitoring Cryptosporidium oocysts in water supplies. United Kingdom Drinking Water Inspectorate, London, United Kingdom. [Online.] www.dwi.detr.gov.uk.
- 50.Vetterling, J. M., H. R. Jervis, T. G. Merrill, and H. Sprinz. 1971. Cryptosporidium wrairi sp. n. from the guinea pig, Cavia porcellus, with an emendation of the genus. J. Protozool. 18:243-247. [DOI] [PubMed] [Google Scholar]
- 51.Xiao, L., L. Escalante, C. Yang, I. Sulaiman, A. A. Escalante, J. R. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao, L., J. Limor, K. Alderisio, A. Singh, T. Graczyk, S. Gradus, M. Royer, and A. A. Lal. 2000. Genotyping Cryptosporidium oocysts in water samples as a tool for the identification of contamination sources. In 2000 Water Quality Technology Conference proceedings. American Water Works Association, Washington, D.C.
- 53.Xiao, L., C. Bern, J. Limor, I. Sulaiman, J. Roberts, W. Checkley, L. Cabrera, R. H. Gilman, and A. A. Lal. 2001. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis. 183:492-497. [DOI] [PubMed] [Google Scholar]