Abstract
Representative encapsulated strains of Vibrio vulnificus from market oysters and oyster-associated primary septicemia cases (25 isolates each) were tested in a blinded fashion for potential virulence markers that may distinguish strains from these two sources. These isolates were analyzed for plasmid content, for the presence of a 460-bp amplicon by randomly amplified polymorphic DNA PCR, and for virulence in subcutaneously (s.c.) inoculated, iron-dextran-treated mice. Similar percentages of market oyster and clinical isolates possessed detectable plasmids (24 and 36%, respectively), produced the 460-bp amplicon (45 and 50%, respectively), and were judged to be virulent in the mouse s.c. inoculation-iron-dextran model (88% for each). Therefore, it appears that nearly all V. vulnificus strains in oysters are virulent and that genetic tests for plasmids and specific PCR size amplicons cannot distinguish between fully virulent and less virulent strains or between clinical and environmental isolates. The inability of these methods to distinguish food and clinical V. vulnificus isolates demonstrates the need for alternative subtyping approaches and virulence assays.
In the United States, Vibrio vulnificus is the leading cause of death associated with consumption of seafood (12, 15). Consumption of raw Gulf Coast oysters from April to November is responsible for nearly all of the cases. Although V. vulnificus is abundant in oysters during that time of year, cases are rare even in the high-risk group (i.e., those with preexisting liver disease or who are immunocompromised) (7). A major obstacle in developing effective control strategies is the inability to identify in the oysters V. vulnificus strains that are capable of causing human illness.
Two major research needs identified at a 1994 V. vulnificus workshop sponsored by the Food and Drug Administration (FDA) were to develop methods to distinguish virulent V. vulnificus strains from avirulent strains and to determine the infectious dose (22). Since human volunteer studies with V. vulnificus are not ethical, a consensus approach was proposed to determine the infectious dose by relating disease frequency with exposure. It was also suggested that a collection of strains from oysters and human septicemia cases associated with oyster consumption should be characterized in various assays in an attempt to determine traits that may be linked to virulence. The Centers for Disease Control and Prevention (CDC), FDA, and various state departments of health collected approximately 75 well-characterized clinical strains from human septicemia cases with known sources of oysters consumed, patient histories, etc. A recent study of the abundance of V. parahaemolyticus and V. vulnificus in retail oysters by the Interstate Shellfish Sanitation Conference and the FDA generated a large collection of V. vulnificus cultures that is seasonally and geographically diverse and well defined (3). Molecular characterization and virulence assays of representative V. vulnificus isolates from these two collections might reveal the importance of various traits for human infection and help determine the significance of total V. vulnificus numbers in oysters in terms of human illness.
Several putative virulence factors, such as the cytolysin-hemolysin, lipopolysaccharide, capsule, and siderophores, have been identified in V. vulnificus (10, 11, 18). The frequencies of occurrence of these factors are similar among clinical and environmental isolates (11); however, few isolates have been tested, and they usually have not been well defined. On the other hand, strains can easily be discriminated with various molecular techniques, such as pulsed-field gel electrophoresis and ribotyping. However, most strains examined have shown different genotypes, and so far the fingerprints generated by these techniques have not been useful in virulence prediction (2, 8, 19, 21).
Recent studies that have revealed the ability to discriminate between environmental and clinical strains present promising new approaches. Plasmids are associated with virulence of many bacterial species. A more sensitive plasmid detection technique for V. vulnificus, described by Danish researchers, was used to find plasmids that were not detected in clinical strains by standard plasmid isolation protocols (L. Høi, J. A. Gooch, A. Dalsgaard, and A. DePaola, Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999, abstr. Q-316, p. 594, 1999). A randomly amplified polymorphic DNA (RAPD) PCR procedure was found to produce an extra DNA band (178 to 200 bp) for all 31 clinical V. vulnificus isolates tested but for only 2 of 39 environmental isolates (21). However, no particular gene was associated with this DNA band. University of Florida researchers recently reported a 1,000-fold difference in the doses of virulent and selected naturally attenuated V. vulnificus required to cause disease in subcutaneously (s.c.) inoculated, iron-dextran-treated mice (16). In each of these studies, either few isolates were tested or the source of the isolates and their association with oysters were not reported.
The objective of this study was to test the hypothesis that proposed molecular markers of virulence (RAPD-PCR amplicons or plasmids) or virulence (s.c.-inoculated, iron-dextran-treated mouse model) are more prevalent in clinical isolates of V. vulnificus than in isolates from market oysters.
MATERIALS AND METHODS
V. vulnificus isolates.
Culture designations, sources, and other information for 50 representative V. vulnificus isolates are shown in Table 1 (oyster isolates) and Table 2 (clinical isolates). Half of the isolates were from human primary septicemia cases linked to raw-oyster consumption, and half were from a nationwide market survey of shell oysters (3). All of the isolates were judged to be encapsulated on the basis of their opaque colony morphology after overnight incubation at 35°C on tryptic soy agar (Difco, Sparks, Md.). Most of the clinical isolates had been previously tested for lipopolysaccharide type (23). The isolates were coded to conceal their source from the investigators. The investigators were Anders Dalsgaard of the Royal Veterinary and Agricultural University of Denmark (for plasmid analysis), James Oliver of the University of North Carolina—Charlotte (for RAPD-PCR typing), and Paul A. Gulig of the University of Florida (for virulence testing with s.c.-inoculated mice).
TABLE 1.
Source information on V. vulnificus strains isolated from market oysters
| Culture identification no. | Harvest state | Harvest date (mo/day/yr) | MPN/ga |
|---|---|---|---|
| 99-624 DP-C10 | Texas | 1/5/99 | 17 |
| 99-779 DP-D2 | Louisiana | 4/16/99 | 490 |
| 99-736 DP-C7 | Florida | 4/5/99 | 1,100 |
| 99-645 DP-C4 | Texas | 5/1/99 | 4,900 |
| 98-624 DP-C9 | Louisiana | 8/18/98 | 130,000 |
| 99-581 DP-C7 | Louisiana | 12/8/98 | 1,300 |
| 99-796 DP-E7 | Florida | 4/6/99 | <240 |
| 99-584 DP-B12 | Texas | 3/31/99 | 1,300 |
| 98-640 DP-E9 | Louisiana | 8/23/98 | 170,000 |
| 99-743 DP-B6 | Texas | 5/8/99 | 9,500 |
| 98-783 DP-A1 | Louisiana | 5/1/99 | 790 |
| 99-780 DP-E1 | Louisiana | 4/14/99 | 7.8 |
| 99-625 DP-D8 | Texas | 1/5/99 | 4.9 |
| 99-738 DP-B5 | Florida | 4/19/99 | 700 |
| 99-537 DP-G7 | Maryland | 11/9/98 | 13 |
| 99-540 DP-B6 | Texas | 11/21/98 | 4,600 |
| 99-742 DP-A9 | Mississippi | 5/11/99 | 330 |
| 99-578 DP-B1 | Louisiana | 11/5/98 | 13,000 |
| 99-623 DP-F5 | Florida | 12/2/98 | >1,600 |
| 99-520 DP-B8 | Rhode Island | 12/29/98 | 0.2 |
| 99-505 DP-C8 | Texas | 11/9/98 | 3,400 |
| 99-609 DP-A4 | Oregon | 2/1/99 | <0.18 |
| 98-641 DP-G8 | Louisiana | 8/23/98 | 23,000 |
| 99-622 DP-E4 | Texas | 12/9/98 | >1,600 |
| 99-509 DP-A6 | Texas | 1/8/99 | 220 |
Most probable numbers (MPN) of V. vulnificus were determined as described by Cook et al. (3).
TABLE 2.
Source information on V. vulnificus strains from primary septicemia cases
| Culture identification no. | LPSa | Harvest state of oysters consumed | Harvest date (mo/day/yr) | Death |
|---|---|---|---|---|
| NSV 5829 (CDC 9149-95) | unk | Florida/Louisiana | 5/23/95 | No |
| NSV 5736 (CDC 9349-95) | 1/5 | Alabama | 7/16/95 | Yes |
| DAL 6-5000 (CDC 9345-95) | unk | Louisiana | 9/30/95 | Yes |
| ATL 9579 | unk | Texas | 8/23/94 | No |
| ATL 71503 (CDC 9075-96) | 2 | Florida | 10/24/96 | Yes |
| ATL-9824 | 1/5 | Texas | 11/6/94 | No |
| DAL 79040 (CDC 9070-96) | 4 | Texas | 10/3/96 | No |
| NSV 5830 (CDC 9348-95) | 3 | Florida | 5/23/95 | No |
| ATL 9823 (CDC 9352-94) | 1/5 | Louisiana | 10/23/94 | Yes |
| ATL 71504 (CDC 9076-96) | 1/5 | Louisiana | 10/29/96 | No |
| DAL 7-9087 (CDC 9005-97) | unk | Louisiana | 5/14/97 | No |
| ATL-9572 | unk | Florida | 6/30/94 | No |
| ORL 8324 (CDC 9340-95) | 1/5 | Florida/Louisiana | 7/26/95 or 8/7/95 | Yes |
| ATL-9580 | 1/5 | Texas/Louisiana | 9/2/94 | Yes |
| DAL 7-9002 (CDC 9060-96) | 3 | Texas | 8/28/96 | Yes |
| FLA 9509 (CDC 9003-97) | 1/5 | Louisiana | 4/29/97 | Yes |
| LOS 7343 (CDC 9062-96) | 1/5 | Louisiana | 5/18/96 | Yes |
| FLA 8869 (CDC 9053-96) | 1/5 | Texas | 8/16/96 | No |
| LOS 6966 (CDC 9342-95) | 3 | Texas/Louisiana | 8/2/95 or 7/24/95 | No |
| ATL 6-1306 (CDC 9031-96) | unk | Florida | 4/30/96 | No |
| ORL 1506 (CDC 9030-95) | 3 | Florida | 5/95-8/95 | No |
| LOS 7318 (CDC 9038-96) | unk | Texas | 4/27/96 | Yes |
| DAL 7-9000 (CDC 9067-96) | 1/5 | Texas | 9/23/96 | Yes |
| ATL 71491 (CDC 9074-96) | 1/5 | Texas/Louisiana | 10/9/96 | Yes |
| ORL 8074 (CDC 9032-95) | 1/5 | Texas | 5/13/95 | No |
Lipopolysaccharide (LPS) determined previously by Zuppardo et al. (23). unk, unknown.
Plasmid extraction and detection.
Isolates were grown in Luria broth (Difco) supplemented with 1% (wt/vol) NaCl at 37°C for 18 to 24 h. Control strains included V. vulnificus strain CDC 9344-95 (containing an 11.5-kb plasmid); V. cholerae O1 strain 1075/25 (150-kb plasmid); and the molecular size marker E. coli strains V517 (54-, 7.4-, 5.6-, 5.1-, 4.4-, 3.0-, 2.7-, and 2.1-kb plasmids) and 39R861 (147-, 63-, 36-, and 7-kb plasmids). Plasmid extraction was by the method of Birnboim and Doly (1) as modified by Høi et al. (Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999). Briefly, plasmids were extracted using SET buffer (50 mM Tris [pH 8], 50 mM EDTA, 0.58 M saccharose), lysozyme, and RNase enzyme. Following alkaline lysis at 56°C, potassium acetate buffer (3 M CH3COOK, 1 mM EDTA, 2 M CH3COOH) was added to precipitate protein, and plasmids were extracted using phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol).
The plasmid DNA was separated in a 0.8% (wt./vol) agarose gel (SeaKem GTG; Medinova, Hellerup, Denmark) in TAE buffer (89 mM Tris, 89 mM acetic acid, 2.5 mM EDTA; pH 8) at 10°C with a current of 135 mA for 4 h. The gels were stained with ethidium bromide (2 μg/ml; Sigma, St. Louis, Mo.) for 15 min, destained in distilled water, and photographed over a 354-nm UV transilluminator. Further, two-dimensional (2-D) gel electrophoresis was conducted according to the method of Hintermann et al. (6), which discriminates between covalently closed circular, open circular, and linear forms of plasmid DNA. Two consecutive steps of agarose gel electrophoresis with a single DNA sample were used. UV irradiation was performed between the steps to introduce single-strand nicks in ethidium bromide-stained DNA, converting covalently closed circular into open circular forms. Thus, differently configured forms of the same plasmid could be identified. Plasmid analyses were repeated at least twice for each strain.
RAPD analysis.
Cells were grown in 1 to 3 ml of heart infusion broth (Difco) overnight at 22°C with aeration. A 200-μl volume of the overnight culture was centrifuged at 14,000 rpm (Marathon Micro A centrifuge; Fisher Scientific, Pittsburgh, Pa.) for 5 min, the supernatant was discarded, and the pellet was suspended in 200 μl of sterile water. At a cell-free station, a master mix containing 2.5 μl of 10× reaction buffer (Promega), 3.5 μl of 25 mM MgCl2 (Promega), 1 to 2 μl of 5 mM deoxynucleoside triphosphate solution (Promega), 1 to 2 μl of a 5 mM primer solution (5′ GGATCTGAAC 3′; Biosynthesis), 0.5 μl of Taq polymerase (Promega), and 8.5 to 9.5 μl of distilled water was made, and 20.0-μl volumes of this mix were placed into sterile 0.5-ml microcentrifuge tubes (USA Scientific, Inc., Ocala, Fla.). All solutions were held on ice. A total of 5.0 μl of each bacterial culture was added to the master mix to give a final reaction volume of 25 μl. Samples were vortexed and overlaid with 20 μl of sterile mineral oil (Sigma) to prevent evaporation. Thermal cycling was performed in a Techne (Princeton, N.J.) model PHC-3 thermal cycler. The cycling profile was as follows: 1 cycle of 94°C for 5 min; 45 cycles of 94°C for 1 min, 36°C for 1 min, 72°C for 2 min; and a final extension cycle of 72°C for 5 min. Fifteen to twenty microliters of PCR product was loaded on a 3% (wt/vol) agarose gel (Fisher Scientific) containing 0.5 μg of ethidium bromide/ml and electrophoresed in 0.5× TBE buffer (prepared from a 10× stock consisting of 0.89 M Tris, 0.89 M boric acid, and 25 mM EDTA) for as long as necessary to yield well-separated bands. The gels were photographed with a Polaroid Quick Shooter model QSP camera (International Biotechnologies, New Haven, Conn.). A 100-bp ladder (BioWhittaker Molecular Applications, Rockland, Maine) was used as a molecular size marker. Each time RAPD-PCR was performed, a clinical strain of V. vulnificus was included as a positive control while heart infusion broth served as a negative control.
Subcutaneously inoculated, iron-dextran-treated mouse model.
Virulence was measured using the s.c.-inoculated, iron-dextran-treated mouse model as previously described (16). Mice were injected intraperitoneally with iron-dextran (Sigma) at 250 μg/g of body weight 2 h before V. vulnificus inoculation. Groups of five mice were initially injected s.c. (lower back) with 103 CFU of bacteria suspended in phosphate-buffered saline containing 0.01% (wt/vol) gelatin. Four parameters were used to determine virulence: CFU per gram of skin lesion, CFU per gram of liver tissue, lesion size and quality score, and body temperature (rectal temperature recorded with a Traceable digital temperature probe [Fisher Scientific]). The most critical criterion for determining virulence was CFU per gram of skin lesion. A strain was labeled as virulent if three or more of five injected mice had 107 CFU/g of lesion or if the average for all of the lesions was 106 CFU/g. The CFU per gram of liver tissue, a decrease in body temperature below 37°C (a sign of severe illness), and the lesion score (0 = no lesion, 1 = discoloration without hemorrhage, 2 = hemorrhagic lesion <2 cm2, and 3 = hemorrhagic lesion >2 cm2) were also noted. If a strain did not meet the virulence criteria at the initial inoculation dose, the dose was increased to 105 CFU/mouse. If the higher dose caused symptoms of virulence, the strain was labeled as attenuated. However, if at a dose of 105 CFU the strain still did not meet the criteria for virulence, it was labeled as avirulent.
Differences in values for virulence measures between oyster and clinical strains were compared using the Student t test or Mann-Whitney U test. Because the 4-point lesion score does not yield a normal distribution, it was not subjected to statistical analysis.
RESULTS
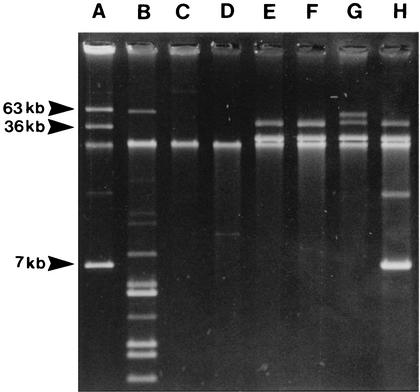
Table 3 lists the virulence markers for each of the V. vulnificus isolates from market oysters and cases of oyster-associated V. vulnificus primary septicemia. Plasmids were detected in 30% of the isolates; the proportions for clinical and oyster isolates were similar (36 and 24%, respectively). The plasmid profiles of the clinical strains CDC 9149-95, CDC 9030-95, CDC 9031-96, and CDC 9075-96 contain a band of approximately 40 kb that is a relaxed form of the 29-kb plasmid; two of these strains also contained a second plasmid (Fig. 1).
TABLE 3.
Virulence markers in V. vulnificus isolates from market oysters and oyster-associated V. vulnificus primary septicemia cases
| Culture identification no. | Plasmid(s) (kb) | 460-bp banda | Mouse s.c. |
|---|---|---|---|
| Oyster | |||
| 99-624 DP-C10 | —b | − | Virulent |
| 99-779 DP-D2 | 4.5 | − | Virulent |
| 99-736 DP-C7 | — | + | Virulent |
| 99-645 DP-C4 | — | + | Virulent |
| 98-624 DP-C9 | — | − | Attenuated |
| 99-581 DP-C7 | — | + | Virulent |
| 99-796 DP-E7 | 23 | ND | Virulent |
| 99-584 DP-B12 | — | − | Avirulent |
| 98-640 DP-E9 | — | − | Virulent |
| 99-743 DP-B6 | — | + | Virulent |
| 98-783 DP-A1 | — | + | Virulent |
| 99-780 DP-E1 | — | + | Virulent |
| 99-625 DP-D8 | — | + | Virulent |
| 99-738 DP-B5 | 112 | + | Virulent |
| 99-537 DP-G7 | — | − | Virulent |
| 99-540 DP-B6 | — | − | Virulent |
| 99-742 DP-A9 | 25 | ND | Virulent |
| 99-578 DP-B1 | — | ND | Virulent |
| 99-623 DP-F5 | — | − | Attenuated |
| 99-520 DP-B8 | — | − | Virulent |
| 99-505 DP-C8 | — | − | Virulent |
| 99-609 DP-A4 | 55 | ND | Virulent |
| 98-641 DP-G8 | — | ND | Virulent |
| 99-622 DP-E4 | 44 | + | Virulent |
| 99-509 DP-A6 | — | − | Virulent |
| Total (%) | 24 | 45 | 88 |
| Clinical | |||
| NSV 5829 (CDC 9149-95) | 29 | + | Virulent |
| NSV 5736 (CDC 9349-95) | 12 | − | Virulent |
| DAL 6-5000 (CDC 9345-95) | — | − | Attenuated |
| ATL 9579 | — | + | Virulent |
| ATL 71503 (CDC 9075-96) | 29, 6.9 | + | Virulent |
| ATL-9824 | — | − | Virulent |
| DAL 79040 (CDC 9070-96) | — | − | Virulent |
| NSV 5830 (CDC 9348-95) | — | + | Virulent |
| ATL 9823 (CDC 9352-94) | — | + | Virulent |
| ATL 71504 (CDC 9076-96) | — | − | Virulent |
| DAL 7-9087 (CDC 9005-97) | — | + | Virulent |
| ATL-9572 | 12 | + | Avirulent |
| ORL 8324 (CDC 9340-95) | 10.5 | − | Virulent |
| ATL-9580 | — | + | Virulent |
| DAL 7-9002 (CDC 9060-96) | — | − | Virulent |
| FLA 9509 (CDC 9003-97) | — | − | Virulent |
| LOS 7343 (CDC 9062-96) | — | ND | Virulent |
| FLA 8869 (CDC 9053-96) | — | − | Attenuated |
| LOS 6966 (CDC 9342-95) | — | + | Virulent |
| ATL 6-1306 (CDC 9031-96) | 50, 29 | + | Virulent |
| ORL 1506 (CDC 9030-95) | 29 | + | Virulent |
| LOS 7318 (CDC 9038-96) | — | − | Virulent |
| DAL 7-9000 (CDC 9067-96) | 41 | ND | Virulent |
| ATL 71491 (CDC 9074-96) | — | − | Virulent |
| ORL 8074 (CDC 9032-95) | 2 | ND | Virulent |
| Total (%) | 36 | 50 | 88 |
−, band absent; +, band present; ND, not determined.
—, no plasmid present.
FIG. 1.
Plasmid profiles of selected V. vulnificus strains. Lanes: A, E. coli 39R861; B, E. coli V517; C, V. cholerae 1075/25; D, V. vulnificus CDC 9344-95; E, V. vulnificus CDC 9149-95; F, V. vulnificus CDC 9030-95; G, V. vulnificus CDC 9031-96; H, V. vulnificus CDC 9075-96. Strains in lanes E to H contain a band of approximately 40 kb that is a relaxed form of the 29-kb plasmid, and lane H also contains a relaxed form of the 6.9-kb plasmid.
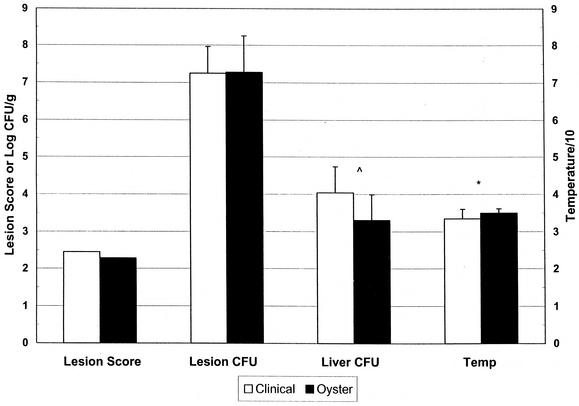
A 460-bp band was found in approximately half of the isolates by RAPD-PCR, with similar proportions in clinical and oyster isolates (50 and 45%, respectively). DNA from 8 (16%) of the 50 strains could not be amplified by this method. Measurement of CFU per gram of skin lesion in the s.c.-inoculated, iron-dextran-treated mouse model indicated that 88% of the isolates were virulent, 8% were attenuated, and only 4% were avirulent; these characteristics were evenly divided among clinical and oyster isolates. The mean log CFU per gram of skin lesion for the clinical isolates was not significantly different from that for oyster isolates (7.24 ± 0.72 and 7.27 ± 0.98, respectively; P = 0.87, Student t test) (Fig. 2), and the mean lesion scores were 2.5 and 2.3, respectively. However, the log CFU per gram of liver tissue for the clinical strains was nearly significantly higher than that for the oyster isolates (4.04 ± 0.70 and 3.30 ± 0.69, respectively; P = 0.06, Student t test). Upon examining the ordered data, we observed that most of the oyster isolates were at the low end of the spectrum, and a Mann-Whitney U test confirmed that despite the insignificant difference in mean CFU per gram of liver tissue, the rank order of clinical isolates was significantly different from that of oyster isolates (P < 0.005). Finally, the mean body temperature at sacrifice of mice infected with clinical isolates was significantly different from that of mice infected with oyster isolates (33.4 ± 2.6 and 34.9 ± 1.2; P = 0.01, Student t test). Lower body temperature is correlated with severe systemic disease. Furthermore, body temperatures of 33°C or less (a correlate for death) were observed with 11 clinical isolates but with only 2 oyster isolates (P = 0.004, chi-squared test).
FIG. 2.
Summary of quantitative data from mouse infection studies. Iron-dextran-treated mice were inoculated s.c. with 103 CFU of V. vulnificus. Data are means ± standard deviations of values for log CFU per gram of skin lesion (Lesion CFU), log CFU per gram of liver tissue (Liver CFU), and rectal temperature (Temp). For all groups, n = 25. *, P = 0.01 (Student t test); ^, P < 0.005 (Mann-Whitney U test).
Taken together, these results indicated that neither plasmid analysis nor RAPD-PCR analysis could distinguish between oyster and clinical isolates of V. vulnificus. Furthermore, nearly all V. vulnificus isolates, regardless of their origin, were virulent according to the s.c. inoculation-iron-dextran method.
DISCUSSION
We undertook this study to determine whether RAPD-PCR genotyping and plasmid analysis could be used to differentiate oyster and clinical isolates of V. vulnificus. Furthermore, we wanted to determine whether oyster and clinical isolates had different virulence potentials as measured in an animal model of disease. Several bacteria, such as Salmonella, Shigella, and Yersinia species, contain plasmids encoding virulence attributes (13, 14). We therefore examined a collection of V. vulnificus strains for the presence of plasmids. Plasmids were found in 30% of the V. vulnificus strains in this collection; their proportions in clinical and oyster isolates were similar. This level is considerably higher than the 12% plasmid carriage for V. vulnificus previously reported by Davidson and Oliver (5) but lower than that determined in a Danish study of both clinical and environmental V. vulnificus strains, in which 11 of 18 strains (60%) contained single plasmids of typical small molecular sizes (4). In our study, a 29-kb plasmid was found in four clinical isolates but in none of the oyster isolates. Apparently, plasmids of different sizes, as seen in traditional one-dimensional electrophoresis, were often shown to be identical in 2-D gel electrophoresis. This result suggests that because they may yield different configurations of the same plasmid (i.e., open circular versus covalently closed circular), existing methods for plasmid extraction from V. vulnificus can still be improved. Southern hybridization using the 29-kb plasmid as a probe may detect additional strains containing few copies of this plasmid. Low copy number could limit detection by the conventional ethidium bromide-based methods that were used to detect the 29-kb plasmid in the present study. However, Southern hybridization using different plasmids from another strain failed to detect additional plasmid-bearing strains among these 50 isolates (data not shown).
The original study by Warner and Oliver (21) determined that a ca. 178-bp amplicon was present in 100% of the clinical isolates tested. Subsequent studies (Y. Yano and J. D. Oliver, data) unpublished found that only 71% of clinical isolates possessed this band. At the same time, however, a band of ca. 460 bp was identified in 86% of these same strains. Thus, in the studies reported here, we focused on the 460-bp band as an indicator of virulence. Approximately half of both the oyster and the clinical cultures produced this band, as determined by RAPD-PCR. Therefore, the presence of this band did not correlate with the source of the strain, and since nearly all of the isolates were virulent when injected s.c. into iron-dextran-treated mice, the band did not correlate with virulence using that model. However, note that all strains employed in the present study were exclusively from primary septicemia cases associated with oyster consumption, whereas the clinical strains examined in the unpublished study were from a variety of sources.
A final goal of the present study was to determine whether the virulence of clinical isolates of V. vulnificus differs from that of oyster isolates, as measured in an animal model. When CFU per gram of skin lesion was measured after s.c. injection of V. vulnificus into iron-dextran-treated mice, 88% of both clinical and oyster isolates were classified as virulent. Most previous animal studies indicated that the majority of V. vulnificus strains are virulent regardless of their source (9, 17, 20); however, there is some evidence that not all strains present in oysters have the ability to cause human disease (8a). In contrast to our use of quantitative microbiology of infected mice, most of the previous studies used 50% lethal dose assays with death as an endpoint. We attempted to devise some combination of virulence criteria (e.g., decrease in body temperature or shorter time to euthanasia due to severity of illness) as a surrogate for death as an endpoint; however, no such marker was found to separate the two groups of V. vulnificus strains. Consistent with the lack of difference in virulence assigned by minimal levels of bacteria in skin lesions, local infection by clinical isolates, measured by determining the mean CFU per gram of skin lesion, was not significantly different from that by V. vulnificus strains from oysters. In contrast, when two criteria for systemic disease, CFU per gram of liver tissue and decreased body temperature (Fig. 2), were employed, clinical strains were capable of causing significantly more severe systemic disease than oyster strains. This result is not entirely unexpected because the clinical isolates were preselected by their ability to cause systemic disease whereas the oyster isolates did not undergo such a selection. Therefore, there may be two populations of virulent V. vulnificus strains in oysters, both of which are capable of causing skin disease but only one having the potential to cause sepsis.
In conclusion, no clear distinctions between V. vulnificus strains isolated from market oysters and those isolated from patients with oyster-associated V. vulnificus primary septicemia were observed with the three approaches used in this study. This collection of encapsulated V. vulnificus isolates should be useful for evaluating additional screening methods for their ability to distinguish between fully virulent and less virulent strains. For reasons of safety, essentially all encapsulated strains should be considered capable of causing human disease until effective screening methods have been identified. Since the incidence of serious V. vulnificus disease is relatively low, even among people in the populations deemed most at risk who consume raw oysters, attention should also be directed toward V. vulnificus levels in oysters and additional, unidentified predisposing conditions of people, with the goal of being able to better predict lethal outcomes of human-oyster-V. vulnificus encounters.
Acknowledgments
We gratefully acknowledge the support of the Interstate Shellfish Sanitation Conference for funding this project.
We acknowledge the Centers for Disease Control and the Food and Drug Administration Southeast Regional Laboratory for providing clinical and oyster strains, respectively. We thank Shih-Shan Lang at the University of Florida for assistance with mouse virulence studies.
REFERENCES
- 1.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchrieser, C., V. V. Gangar, R. L. Murphree, M. L. Tamplin, and C. W. Kaspar. 1995. Multiple Vibrio vulnificus strains in oysters as demonstrated by clamped homogeneous electric field gel electrophoresis. Appl. Environ. Microbiol. 61:1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cook, D. W., P. O'Leary, J. C. Hunsucker, E. M. Sloan, J. C. Bowers, R. J. Blodgett, and A. DePaola. 2002. Vibrio vulnificus and Vibrio parahaemolyticus in U.S. retail shell oysters: a national survey, June 1998 to July 1999. J. Food Prot. 65:79-87. [DOI] [PubMed] [Google Scholar]
- 4.Dalsgaard, A., N. Frimodt-Møller, B. Bruun, L. Høi, and J. L. Larsen. 1996. Clinical manifestations and epidemiology of Vibrio vulnificus infections in Denmark. Eur. J. Clin. Microbiol. Infect. Dis. 15:227-231. [DOI] [PubMed] [Google Scholar]
- 5.Davidson, L. S., and J. D. Oliver. 1986. Plasmid carriage in Vibrio vulnificus and other lactose-fermenting marine vibrios. Appl. Environ. Microbiol. 52:211-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hintermann, G., H.-M. Fischer, R. Crameri, and R. Hütter. 1981. Simple procedure for distinguishing CCC, OC and L forms of plasmid DNA by agarose gel electrophoresis. Plasmid 5:371-373. [DOI] [PubMed] [Google Scholar]
- 7.Hlady, G. 1994. Risk assessment and current data: Florida risk assessment and factors associated with risk, p. 27-33. In W. Watkins and S. McCarthy (ed.), Proceedings of the 1994 Vibrio vulnificus Workshop. U.S. Food and Drug Administration, Washington, D.C.
- 8.Høi, L., A. Dalsgaard, J. L. Larsen, J. M. Warner, and J. D. Oliver. 1997. Comparison of ribotyping and randomly amplified polymorphic DNA polymerase chain reaction (RAPD-PCR) for characterization of Vibrio vulnificus. Appl. Environ. Microbiol. 63:1674-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Jackson, J. K., R. L. Murphree, and M. L. Tamplin. 1997. Evidence that mortality from Vibrio vulnificus infection results from single strains among heterogeneous populations in shellfish. J. clin. Microbiol. 35:2098-2101. [DOI] [PMC free article] [PubMed]
- 9.Kaysner, C. A., C. Abeyta, Jr., M. M. Wekell, A. DePaola, R. F. Stott, and J. M. Leitch. 1987. Virulent strains of Vibrio vulnificus isolated from estuaries of the United States West Coast. Appl. Environ. Microbiol. 53:1349-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linkhous, D. A., and J. D. Oliver. 1999. Pathogenesis of Vibrio vulnificus. FEMS Microbiol. Lett. 174:207-214. [DOI] [PubMed] [Google Scholar]
- 11.Oliver, J. D. 1989. Vibrio vulnificus, p. 569-600. In M. P. Doyle (ed.), Foodborne bacterial pathogens. Marcel Dekker, Inc., New York, N.Y.
- 12.Oliver, J. D., and J. B. Kaper. 2001. Vibrio species, p. 263-300. In M. P. Doyle, L. R. Beuchat, and T. J. Montvile (ed.), Food microbiology: fundamentals and frontiers. ASM Press, Washington, D.C.
- 13.Revell, P. A., and V. L. Miller. 2001. Yersinia virulence: more than a plasmid. FEMS Microbiol. Lett. 205:159-164. [DOI] [PubMed] [Google Scholar]
- 14.Rotger, R., and J. Casadesus. 1999. The virulence plasmids of Salmonella. Int. Microbiol. 2:177-184. [PubMed] [Google Scholar]
- 15.Shapiro, R. L., S. Altekruse, S. Hutwagner, R. Bishop, R. Hammond, S. Wilson, B. Ray, S. Thompson, R. V. Tauxe, P. M. Griffin, and the Vibrio Working Group. 1998. The role of Gulf Coast oysters harvested in warmer months in Vibrio vulnificus infections in the United States, 1988-1996. J. Infect. Dis. 178:752-759. [DOI] [PubMed] [Google Scholar]
- 16.Starks, A. M., T. R. Schoeb, M. L. Tamplin, S. Parveen, T. J. Doyle, P. E. Bomeisl, G. M. Escudero, and P. A. Gulig. 2000. Pathogenesis of infection by clinical and environmental strains of Vibrio vulnificus in iron-dextran-treated mice. Infect. Immun. 68:5785-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stelma, G. N., Jr., A. L. Reyes, J. T. Peeler, C. H. Johnson, and P. L. Spaulding. 1992. Virulence characteristics of clinical and environmental isolates of Vibrio vulnificus. Appl. Environ. Microbiol. 58:2776-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strom, M. S., and R. N. Paranjpye. 2000. Epidemiology and pathogenesis of Vibrio vulnificus. Microbes Infect. 2:177-188. [DOI] [PubMed] [Google Scholar]
- 19.Tamplin, M. L., J. K. Jackson, C. Buchrieser, R. L. Murphree, K. M. Portier, V. Gangar, L. G. Miller, and C. W. Kaspar. 1996. Pulsed-field gel electrophoresis and ribotype profiles of clinical and environmental Vibrio vulnificus isolates. Appl. Environ. Microbiol. 62:3572-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tison, D. L., and M. T. Kelly. 1986. Virulence of Vibrio vulnificus strains from marine environments. Appl. Environ. Microbiol. 51:1004-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warner, J. M., and J. D. Oliver. 1999. Randomly amplified polymorphic DNA analysis of clinical and environmental isolates if Vibrio vulnificus and other Vibrio species. Appl. Environ. Microbiol. 65:1141-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watkins, W., and S. McCarthy (ed.). 1994. Proceedings of the 1994 Vibrio vulnificus Workshop. U.S. Food and Drug Administration, Washington, D.C.
- 23.Zuppardo, A. B., A. DePaola, J. C. Bowers, K. L. Schully, J. A. Gooch, and R. J. Siebeling. 2001. Heterogeneity of environmental, retail, and clinical isolates of Vibrio vulnificus as determined by lipopolysaccharide-specific monoclonal antibodies. J. Food Prot. 64:1172-1177. [DOI] [PubMed] [Google Scholar]