Abstract
We have investigated the effect of overproducing each of the three cold shock proteins (CspL, CspP, and CspC) in the mesophilic lactic acid bacterium Lactobacillus plantarum NC8. CspL overproduction transiently alleviated the reduction in growth rate triggered by exposing exponentially growing cells to cold shock (8°C), suggesting that CspL is involved in cold adaptation. The strain overproducing CspC resumed growth more rapidly when stationary-phase cultures were diluted into fresh medium, indicating a role in the adaptation and recovery of nutritionally deprived cells. Overproduction of CspP led to an enhanced capacity to survive freezing.
It is well established that after a quick downshift in temperature (cold shock), a set of proteins are preferentially expressed, among which a set of small β-barrel proteins referred to as the major cold shock proteins (CSPs) show the highest induction level (14, 20). Members of the CSP family, which includes cold-inducible and non-cold-inducible representatives, seem to play key roles not only in the adaptation to various stresses such as cold temperature, stationary phase, or nutritional deprivation, but also during growth under optimal conditions (15, 16, 26, 32, 38, 40).
Escherichia coli contains nine CSPs (CspA to CspI), of which four (CspA, -B, -G, and -I) are cold shock inducible (31, 38). CspA is a cold shock and nutritional-upshift stress protein (39). CspA, CspC, and CspE are RNA-binding proteins which function as transcriptional antiterminators by preventing the formation of secondary structures in the nascent RNA. Csp-induced transcriptional antitermination is responsible for the increased expression of several genes (4, 27). CspC and CspE, which are constitutively produced at 37°C, likely function as regulators of the expression of some stress response proteins, playing important roles in the global control of carbon flow in cells (26). They also seem to be involved in the acclimation of nutritionally starved cells to fresh medium (3) and chromosome condensation at 37°C (18). CspD is a nutritional-downshift and stationary phase-induced stress response protein which may function as an inhibitor of DNA replication and which plays a regulatory role in chromosomal replication of nutrient-depleted cells (40). CspF and CspH have not yet been characterized. Bacillus subtilis contains three CSPs (CspB, -C, and -D); CspB is essential for cellular growth in a strain lacking CspC and CspD and plays an important role for efficient protein synthesis at optimal and low temperatures (15), while CspB and CspC are major stationary phase-induced proteins (16). CSPs are also found in lactic acid bacteria (22, 23, 34), but their functions are still largely unknown.
Interestingly, it was noted that many bacteria, including lactic acid bacteria, develop an increased ability to survive freezing after cold shock pretreatment (13, 21, 34). In B. subtilis, CspB appeared to be involved in this phenomenon, since cspB knockout reduced cryotolerance (33). In Lactococcus lactis, overexpression of three cold-induced csp genes (cspB, cspD, and cspE) enhanced the survival capacity through successive freeze-thaw cycles, although to a lesser extent than a cold shock pretreatment (35, 36).
Lactobacillus plantarum plays a significant role in a wide range of spontaneous and controlled lactic fermentations in food processing. Nowadays, it is commercially prepared as a starter for the production of dry and semidry sausages, vegetables (sauerkraut, cucumbers, olives, and pickled vegetables), and silage (2, 7, 12, 17, 24). For L. plantarum NC8, we have previously described three cold-inducible csp genes, cspL, cspP, and cspC, encoding highly similar proteins (9, 10). In this paper, we report our investigations on the effects of CspL, CspP, and CspC overproduction on adaptation to cold-shock, stationary-phase, and freezing stresses.
Growth of L. plantarum at various temperatures.
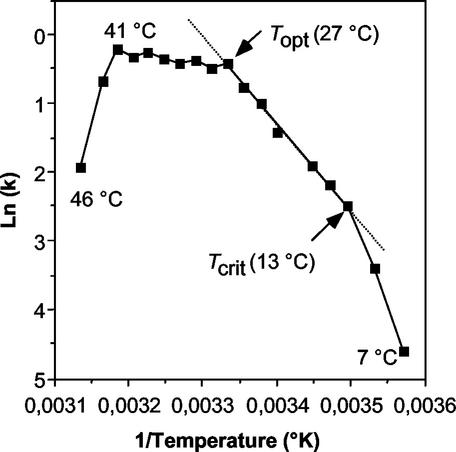
In order to set the optimal and cold-shock temperatures, respectively, an Arrhenius plot of the growth of L. plantarum NC8 (1) was established (Fig. 1) by expressing the log of the growth rate (k) against the reciprocal of the temperature (in kelvins) (30). k (expressed in generations per hour) was determined as the slope of a semilogarithmic plot of the optical density (OD) at 600 nm versus time. The temperatures over which L. plantarum can grow were found to split into the following three ranges: an Arrhenius zone from 13°C (critical temperature, Tcrit) to 27°C (optimal temperature, Topt), within which the activation energy of growth is constant; a cold shock subrange (below 13°C); and a heat shock subrange (above 27°C). According to this Arrhenius plot, a temperature of 27°C corresponds to the optimal temperature (Topt) for L. plantarum. A temperature of 8°C, below the critical temperature (Tcrit), was chosen for the cold-shock experiments.
FIG. 1.
Arrhenius plot of the relationship between growth rate (k) and temperature (kelvins) for L. plantarum NC8. Topt, optimal temperature; Tcrit, critical temperature.
Overproduction of CspL, CspP, and CspC using the nisin controlled expression system.
In order to study the function played by the three CSPs, we overproduced each of them in turn using the nisin controlled expression system, initially developed for protein overproduction in L. lactis (8) and recently adapted for L. plantarum (25). All DNA manipulations were performed according to established procedures (28). The different csp genes were PCR amplified with Dynazyme DNA polymerase (Finnzymes Oy, Espoo, Finland) in a DNA thermal cycler (Perkin-Elmer, Norwalk, Conn.) with the following settings: denaturation at 92°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 1 min for a total of 30 cycles. The pairs of oligonucleotides used were as follows: 5′-CGGAATTCTTTATAGGTGTTAATAACAT-3′ (EcoRI site, underlined) and 5′-GCACTGCAGATTGGTGGCGCTTACTCG-3′ (PstI site, underlined) for cspP, 5′-CCGAATTCAATAAACTATCCCATTTGTAC-3′ (EcoRI site, underlined) and 5′-GCCTTCAAGCAAGTCGCAAT-3′ for cspC, and 5′-ACATGCCATGGAGAATGGTACAGTAAAATGGTTCAA-3′ (NcoI site, underlined) and 5′-TTACAATGCTAACTAATCCCG-3′ for cspL. The cspP PCR fragment was digested with EcoRI-PstI and cloned in the intermediate plasmid pMTL23P (6). An EcoRI-SalI fragment from the resulting plasmid was subsequently cloned between EcoRI and XhoI in pNZ8008 (8). The resulting plasmid, pGIS410, harbored a transcriptional fusion between the nisA promoter and the cspP gene. The cspC PCR fragment was digested with EcoRI-HindIII and subsequently cloned at the respective sites of pNZ8008, leading to pGIS411, which also resulted in a transcriptional fusion with the nisA promoter. Previous Northern blot analysis has revealed that the cspL promoter drives the transcription of two cold-inducible transcripts likely to result from terminator readthrough and/or RNase processing: the shorter one covers the cspL open reading frame (ORF), whereas the longer one extends further downstream into a region containing a putative 77-amino acid (aa) ORF (named orfX) (9). A cspL PCR fragment covering both ORFs was digested with NcoI-HindIII and directly cloned in pNZ8032 (8) to construct an ATG translational fusion (NcoI) with the nisA expression module; the resulting plasmid was named pGIS412. In order to assess a possible role for orfX, plasmid pGIS412 was BamHI digested, filled in, and self-ligated, resulting in an inactivation of orfX (35-aa truncation at the C terminus); this plasmid was named pGIS414. The positioning of an NcoI site at the fusion point mutated the second codon (AAG to GAG) of the cspL ORF, resulting in the replacement of lysine with glutamate. This mutation is unlikely to modify the function of the protein since glutamate is often found at this position in several CSPs. The absence of other mutations in the three fusions was confirmed by DNA sequencing. Plasmids pGIS410, pGIS411, pGIS412, and pGIS414 were next transformed by electroporation (1) into L. plantarum SD1, an NC8 derivative obtained by site-specific chromosomal integration (11) of the regulatory genes nisRK necessary for nisA promoter induction. Overproduction of CSPs was performed by the addition of 25 ng of nisin ml−1 to cultures in the early exponential growth phase (3% inoculum; OD at 600 nm [OD600] = 0.15) as described previously (25). Nisin-induced cells were collected after 5 h at an OD600 of 0.8, and total cellular proteins were extracted from 20-ml cultures by homogenizing cells with glass beads.
Initially, the three genes, cspL, cspP, and cspC, were transcriptionally fused to the nisA promoter, but only the transcriptional fusions with cspP (pGIS410) and cspC (pGIS411) responded to nisin induction in L. plantarum SD1, each yielding an overproduced band of approximately 7 kDa on Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (29; data not shown). As an alternative, the cspL ORF in combination with orfX or the cspL ORF alone were fused at ATG to the nisA expression module as described above, and the resulting plasmids, pGIS412 and pGIS414, allowed overproduction of CspL (data not shown). Both constructions displayed a similar protein profile. This suggests that if orfX is coexpressed with cspL in pGIS412, either its expression level is extremely low or both proteins migrate at the same position (1.5 kDa difference in molecular mass).
CspL overproduction transiently alleviates cold shock impairment of growth.
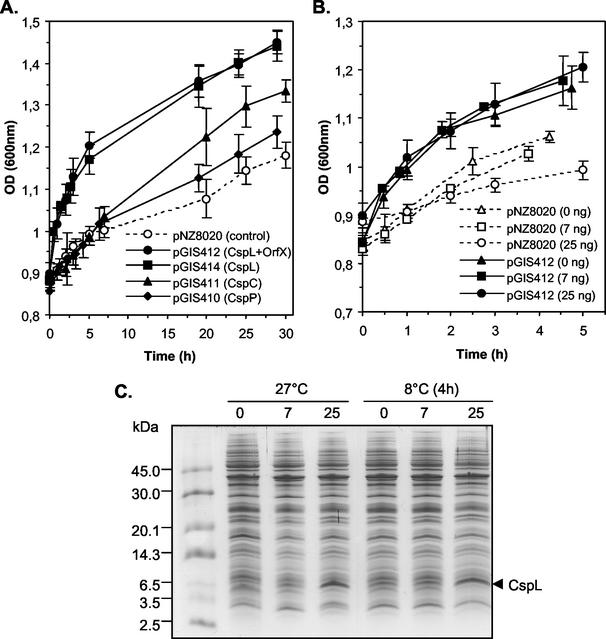
We first monitored the growth in MRS medium (Difco) of mid-exponential-phase cultures (OD600 = 0.8) of CSP-overproducing strains shifted from 27 to 8°C in a precooled water bath (9). The recipient strain SD1 containing the basal cloning vector pNZ8020 was used as a control. Cold shock drastically slowed down the growth of the control and of the overproducing strains (for a comparison, see Fig. 2A and Fig. 3A). Interestingly, the growth of the CspL-overproducing strains (SD1 harboring pGIS412 or pGIS414) was less affected after cold shock than that of the control, while CspP and CspC overproduction had no effect on growth during at least 7 h following cold shock. This observation suggests that preloading with CspL preadapts L. plantarum to cold shock and that OrfX does not contribute to this effect since similar growth curves were obtained for both CspL-overproducing strains (pGIS414 or pGIS412). This preadaptation effect was transient, as the growth rate of the CspL-overproducing strains became similar to that of the control. In contrast, no preadaptation to growth at a low temperature was observed for overproduction of CspC or CspP (Fig. 2A). We noticed that upon prolonged cold shock (over 24 h), the growth rate of the CspC-overproducing strain (SD1 harboring pGIS411) was reproducibly higher than that of the other strains (Fig. 2A). This observation suggests that CspC, although not primarily implicated in cold shock adaptation, may play a positive role during the acclimation phase of growth at cold temperature.
FIG. 2.
Effect of CSP overproduction on growth after cold shock. (A) Cultures of the control strain (dotted line, open circles) and CSP-overproducing strains (solid lines; closed circles, CspL plus OrfX; closed squares, CspL; closed triangles, CspC; closed diamonds, CspP) were inoculated (4%) with overnight precultures and grown at 27°C for 1 h before nisin addition (25 ng ml−1). Exponentially growing cultures (OD600 of 0.85) were transferred from 27 to 8°C and growth was monitored for 30 h. Error bars represent standard deviations from the means (n = 3). (B) Effect of the increase in nisin concentration on growth after cold shock of the CspL-overproducing strain. Cultures of the control strain (pNZ8020; dotted lines and open symbols) and the CspL-overproducing strain (pGIS412; solid lines and closed symbols) were inoculated (4%) with overnight precultures and grown at 27°C for 1 h before nisin addition. The nisin concentrations used for induction were 0 ng ml−1 (triangles), 7 ng ml−1 (squares), and 25 ng ml−1 (circles). Exponentially growing cultures (OD600 of 0.85) were transferred from 27 to 8°C and growth was monitored for 4 to 5 h. The data were obtained from three independent cultures for both the controls and the CspL-overproducing strains. Error bars represent standard deviations from the means (n = 3). (C) Coomassie brilliant blue-stained gel after Tricine-SDS-PAGE showing the dose-dependent nisin induction of CspL synthesis before and after cold shock. Equal amounts of total proteins (13 μg) of cell extracts from the CspL-overproducing strain (pGIS412) grown at 27°C and downshifted for 4 h at 8°C (as indicated on the top of respective lanes) were subjected to electrophoresis. The nisin concentrations used for induction were 0, 7, and 25 ng ml−1 (as indicated on the top of respective lanes). The molecular mass standards (in kilodaltons) (Amersham Pharmacia Biotech) are indicated to the left, and the bands corresponding to CspL are indicated by an arrow.
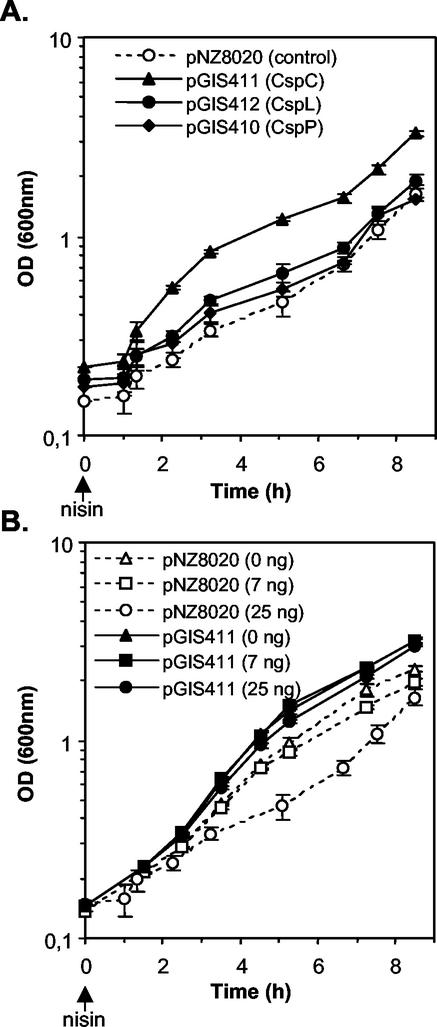
FIG. 3.
Effect of CSP overproduction on L. plantarum growth at optimal temperature. (A) Cultures of the control strain (dotted line, open circles) and the CSP-overproducing strains (solid lines; closed triangles, CspC; closed circles, CspL; closed diamonds, CspP) were grown at 27°C after inoculation with stationary-phase precultures. Nisin induction (25 ng ml−1) was started 1 h after inoculation, and growth was monitored for 8 to 9 h. (B) Growth of control (dotted lines, open symbols) and CspC-overproducing strains (solid lines, closed symbols) upon induction with various nisin concentrations (0 ng ml−1 [triangles], 7 ng ml−1 [squares], and 25 ng ml−1 [circles]). Error bars represent the standard deviations from the means (n = 3).
In order to further characterize the effect of CspL overproduction upon cold shock adaptation, a nisin dose-response experiment was performed (Fig. 2B and C). As expected, it was found that CspL production visualized on a Tricine-SDS-PAGE gel was dose dependent (Fig. 2C). The amount of CspL present in the overproducing cells after a cold shock of 4 h at 8°C was similar to those observed before the temperature downshift (27°C) for 7 and 25 ng of nisin ml−1. The noninduced culture contained more CspL after 4 h at 8°C than before the downshift, likely resulting from the chromosomal cspL cold shock induction. Higher nisin concentrations could not be used due to strong growth inhibition (data not shown). At low nisin doses (0 to 25 ng ml−1), all cultures of the CspL-overproducing strain were preadapted to cold shock in comparison to their respective controls grown under the same conditions (Fig. 2B). This preadaptation in the absence of nisin shows that the nisA promoter displays a basal level of activity, as previously shown in L. plantarum (25), and that a low amount of CspL is sufficient for this preadaptation effect.
We previously demonstrated that cspL mRNA was the most cold shock induced of the three csp transcripts (9). This suggests that CspL could be a key protein needed for cellular adaptation to lower temperatures. The major cold-inducible protein CspA of E. coli is an RNA chaperone playing a crucial role in efficient translation of mRNAs at low temperatures (19). It is also a transcriptional antiterminator playing an important role in cold shock adaptation by reprogramming gene expression to induce several other cold-induced proteins (4). We speculate that CspL plays a similar role and that its overproduction directly or indirectly alleviates the negative effects of cold shock on protein synthesis.
CspC overproduction improves growth resumption at 27°C.
Stationary-phase cultures (OD600 = 6) grown without nisin were used to inoculate fresh MRS medium prewarmed at 27°C, and nisin was added to these cultures 1 h later. The growth curves obtained clearly indicated that the CspC-overproducing strain (SD1 harboring pGIS411) resumed growth more rapidly than the control while the growth of the two other overproducing strains was not significantly improved (Fig. 3A). Analysis of the nisin dose response on growth at 27°C showed that the CspC-overproducing strain induced with a nisin concentration range of 0 to 25 ng ml−1 displayed a higher growth rate and resumed growth more rapidly than the respective controls (Fig. 3B). Increasing nisin concentration resulted in a progressive growth inhibition of the control strain without affecting the growth rate of the CspC-overproducing strain. This result suggests that increasing the CspC amount counteracts the growth inhibition resulting from nisin addition to the growth medium.
During the lag phase following inoculation of stationary-phase bacteria into fresh medium, many physiological functions must be restored, and an adaptation process must therefore take place. Our results indicate that CspC could play an important role in this adaptation. We previously showed that L. plantarum cells contain a substantial amount of the cspC transcript during early exponential growth at 27°C and that cspC mRNA abundance drastically declines in stationary phase (9). The presence of CspC in exponentially growing cells at 27°C was recently established using two-dimensional gel electrophoresis followed by microsequencing (data not shown). In E. coli, the cspA mRNA and CspA protein amounts are also very high during early exponential growth at 37°C. Furthermore, it has been shown that a double cspA cspE mutant lagged longer than the single cspE mutant after dilution into a fresh medium (3).
CspP overproduction enhances cryotolerance.
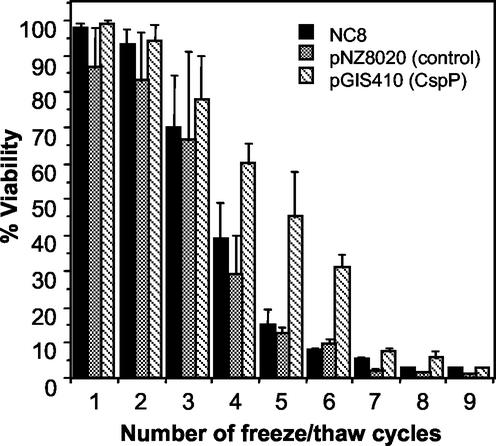
The potential role of CSPs in cryotolerance was investigated by performing freeze-thaw experiments with the three CSP-overproducing strains and their respective controls (NC8 and SD1 harboring pNZ8020). The recombinant L. plantarum SD1 strains were subjected to the nisin induction procedure and frozen at −80°C in their growth medium without cryoprotective agents. For each sample, CFU were measured before freezing by plating serial dilutions in the same medium, and the colonies were counted after incubation at 27°C for 48 h. Every 24 h (referred to here as a cycle), the samples were thawed at room temperature to allow CFU counting as described above and were immediately refrozen. The freezing experiments were performed in triplicate with samples from individual cultures. An increased cryotolerance was observed for the CspP-overproducing strain (SD1 harboring pGIS410) after four to six challenges (Fig. 4), while the CspL- and CspC-overproducing strains did not differ from the control (data not shown). The implication of CSPs in cryotolerance has already been reported for other bacteria. For B. subtilis, knockout of cspB led to an increased sensitivity to freezing stress (33). In L. lactis, overproduction of CspB, CspD, and CspE also enhances cryotolerance (5- to 10-fold), although to a lesser extent than cold shock preadaptation (35). How could CspP play a role in cryoprotection? A direct cryoprotective role for CspP as an antifreeze protein cannot be rule out. Yet CSPs are thought to be general RNA-binding proteins involved in the maintenance of active growth conditions by favoring transcription, translation, and/or ribosome assembly. In this way, they may indirectly stimulate the production of critical factors, ensuring cryotolerance through the maintenance of membrane, DNA, RNA, or protein integrity upon freezing. A direct cryoprotective effect may also be invoked. Indeed, CspE of E. coli is quite abundant at optimal temperature and seems to be involved in chromosomal condensation (18), a function which could be important to help DNA face off freezing. In addition, as RNA-binding proteins, CSPs may protect some mRNAs from degradation by direct binding (26).
FIG. 4.
Cryoprotective effect of CspP overproduction in L. plantarum. The CFU ratios before and after freezing (% viability) are expressed as a function of the number of freeze-thaw cycles. Error bars represent the standard deviations from the means (n = 3).
Concluding remarks.
Our studies support the claim that different members of the CSP family perform specific functions in adapting L. plantarum to specific environmental stresses. In B. subtilis, CspB is important for cellular growth at an optimal temperature and for adaptation to cold shock and freezing and survival during stationary phase (15, 16, 33). Its absence in knockout mutants is compensated for by CspC at low temperature and during stationary phase and by CspD at 37°C (15, 16). In E. coli, CspA is important for cold shock adaptation and growth resumption after dilution at optimal temperature. Its absence in knockout mutants is compensated for at low temperature by CspB, CspG, and CspI and after the dilution effect by CspE. No growth defect was observed until four csp genes (cspABEG) were deleted (37). As for CspD, it is important during nutritional deprivation (3, 5, 31, 38, 40). In contrast to these functional redundancies, we show that overproduction of each CSP causes distinct phenotypic effects in L. plantarum. It is attractive to speculate that the different CSP proteins may antiterminate transcription of different sets of genes, thus allowing the cell to adequately respond to different environmental challenges. However, we cannot exclude the idea that the overproduction of a single CSP could modulate a regulation network in a complex manner involving more than one CSP such as that observed through the overproduction of CSPs in L. lactis (35).
Although the new properties of the recombinant strains have not been validated in industrial processes, an increased cryotolerance could be relevant for improving starter production, and a faster growth rate during the first hours following cold shock or inoculation would allow for the development of cultures showing improved performance in fermentations carried out at cold or optimal temperature. The observations reported in this study could be the starting point for the construction of food-grade recombinant starter cultures overproducing CSPs.
Acknowledgments
We thank M. Kleerebezem for providing plasmids pNZ8020, pNZ8008, and pNZ8032.
S.D. holds a fellowship of the “Fonds pour la Formation à la Recherche dans l'Industrie et dans l'Agriculture” (FRIA). B.H. is a research associate at the FNRS.
REFERENCES
- 1.Aukrust, T., and H. Blom. 1992. Transformation of Lactobacillus strains used in meat and vegetable fermentations. Food Res. Int. 25:253-261. [Google Scholar]
- 2.Bacus, J. N., and W. L. Brown. 1985. The lactobacilli: meat products, p. 57-72. In S. E. Gilliland (ed.), Bacterial starter cultures for foods. CRC Press Inc., Boca Raton, Fla.
- 3.Bae, W., S. Phadtare, K. Severinov, and M. Inouye. 1999. Characterization of Escherichia coli cspE, whose product negatively regulates transcription of cspA, the gene for the major cold shock protein. Mol. Microbiol. 31:1429-1441. [DOI] [PubMed] [Google Scholar]
- 4.Bae, W., B. Xia, M. Inouye, and K. Severinov. 2000. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. USA 97:7784-7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandi, A., R. Spurio, C. O. Gualerzi, and C. Pon. 1999. Massive presence of the Escherichia coli “major cold-shock protein” CspA under non-stress conditions. EMBO J. 18:1653-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers, S. P., S. E. Prior, D. A. Barstow, and N. P. Minton. 1988. The pMTL nic-cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene 68:139-149. [DOI] [PubMed] [Google Scholar]
- 7.Daeschel, M. A., R. E. Anderson, and H. P. Fleming. 1987. Microbial ecology of fermenting plant materials. FEMS Microbiol. Rev. 46:357-367. [Google Scholar]
- 8.de Ruyter, P. G. G. A., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derzelle, S., B. Hallet, K. P. Francis, T. Ferain, J. Delcour, and P. Hols. 2000. Changes in cspL, cspP, and cspC mRNA abundance as a function of cold shock and growth phase in Lactobacillus plantarum. J. Bacteriol. 182:5105-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derzelle, S., B. Hallet, T. Ferain, J. Delcour, and P. Hols. 2002. Cold shock induction of the cspL gene of Lactobacillus plantarum involves transcriptional regulation. J. Bacteriol. 184:5518-5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupont, L., B. Boizet-Bonhoure, M. Coddeville, F. Auvray, and P. Ritzenthaler. 1995. Characterization of genetic elements required for site-specific integration of Lactobacillus delbrueckii subsp. bulgaricus bacteriophage mv4 and construction of an integration-proficient vector for Lactobacillus plantarum. J. Bacteriol. 177:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleming, H. P., R. F. McFeeters, and M. A. Daeschel. 1985. The lactobacilli, pediococci, and leuconostocs: vegetable products, p. 97-118. In S. E. Gilliland (ed.), Bacterial starter cultures for foods. CRC Press Inc., Boca Raton, Fla.
- 13.Goldstein, J., N. S. Pollitt, and M. Inouye. 1990. Major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 87:283-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graumann, P., K. Schröder, R. Schmid, and M. A. Marahiel. 1996. Cold shock stress-induces proteins in Bacillus subtilis. J. Bacteriol. 178:4611-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graumann, P., T. M. Wendrich, M. H. W. Weber, K. Schröder, and M. A. Marahiel. 1997. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperature. Mol. Microbiol. 25:741-756. [DOI] [PubMed] [Google Scholar]
- 16.Graumann, P., and M. A. Marahiel. 1999. Cold shock proteins CspB and CspC are major stationary-phase-induced proteins in B. subtilis. Arch. Microbiol. 171:135-138. [DOI] [PubMed] [Google Scholar]
- 17.Hammes, W. P., A. Bantleon, and S. Min. 1990. Lactic acid bacteria in meat fermentation. FEMS Microbiol. Rev. 87:165-174. [Google Scholar]
- 18.Hu, K. H., E. Liu, K. Dean, M. Gingras, W. DeGraff, and N. J. Trun. 1996. Overproduction of three genes leads to camphor resistance and chromosome condensation in Escherichia coli. Genetics 143:1521-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang, W., Y. Hou, and M. Inouye. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272:196-202. [DOI] [PubMed] [Google Scholar]
- 20.Jones, P. G., R. A. VanBogelen, and F. C. Neidhardt. 1987. Induction of proteins in response to low temperature in Escherichia coli. J. Bacteriol. 169:2092-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, W. S., and N. W. Dunn. 1997. Identification of a cold shock gene in lactic acid bacteria and the effect of cold shock on cryotolerance. Curr. Microbiol. 35:59-63. [DOI] [PubMed] [Google Scholar]
- 22.Kim, W. S., N. Khunajakr, J. Ren, and N. W. Dunn. 1998. Conservation of the major cold shock protein in lactic acid bacteria. Curr. Microbiol. 37:333-336. [DOI] [PubMed] [Google Scholar]
- 23.Mayo, B., S. Derzelle, M. Fernandez, C. Leonard, T. Ferain, P. Hols, J. E. Suarez, and J. Delcour. 1997. Cloning and characterization of cspL and cspP, two cold-inducible genes from Lactobacillus plantarum. J. Bacteriol. 179:3039-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mäyrä-Mäkinen, A., and M. Bigret. 1993. Industrial use and production of lactic acid bacteria, p. 65-96. In S. Salminen and A. von Wright (ed.), Lactic acid bacteria: classification and physiology. Marcel Dekker, Inc., New York, N.Y.
- 25.Pavan, S., P. Hols, J. Delcour, M. C. Geoffroy, C. Grangette, M. Kleerebezem, and A. Mercenier. 2000. Adaptation of the nisin-controlled expression system in Lactobacillus plantarum: a tool to study in vivo biological effects. Appl. Environ. Microbiol. 66:4427-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phadtare, S., and M. Inouye. 2001. Role of CspC and CspE in regulation of expression of RpoS and UspA, the stress response proteins in Escherichia coli. J. Bacteriol. 183:1205-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phadtare, S., S. Tyagi, M. Inouye, and K. Severinov. 2002. Three amino acids in Escherichia coli CspE surface-exposed aromatic patch are critical for nucleic acid melting activity leading to transcription antitermination and cold acclimation of cells. J. Biol. Chem. 277:46706-46711. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 30.Thammavongs, B., D. Corroler, J.-M. Panoff, Y. Auffray, and P. Boutibonnes. 1996. Physiological response of Enterococcus faecalis JH2-2 to cold shock: growth at low temperatures and freezing/thawing challenge. Lett. Appl. Microbiol. 23:398-402. [DOI] [PubMed] [Google Scholar]
- 31.Wang, N., K. Yamanaka, and M. Inouye. 1999. CspI, the ninth member of the CspA family of Escherichia coli is induced upon cold shock. J. Bacteriol. 181:1603-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber, M. H. W., and M. A. Marahiel. 2002. Coping with the cold. Phil. Trans. R. Soc. Lond. B 357:895-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willimsky, G., H. Bang, G. Fisher, and M. A. Marahiel. 1992. Characterization of cspB, a Bacillus subtilis inducible cold shock gene affecting cell viability at low temperatures. J. Bacteriol. 174:6326-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wouters, J. A., B. Jeynov, F. M. Rombouts, W. M. de Vos, O. P. Kuipers, and T. Abee. 1999. Analysis of the role of 7 kDa cold-shock proteins of Lactococcus lactis MG1363 in cryoprotection. Microbiology 145:3185-3194. [DOI] [PubMed] [Google Scholar]
- 35.Wouters, J. A., M. Mailhes, F. M. Rombouts, W. M. de Vos, O. P. Kuipers, and T. Abee. 2000. Physiological and regulatory effects of controlled overproduction of five cold shock proteins of Lactococcus lactis MG1363. Appl. Environ. Microbiol. 66:3756-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wouters, J. A., H. Frenkiel, W. M. de Vos, O. P. Kuipers, and T. Abee. 2001. Cold shock proteins of Lactococcus lactis MG1363 are involved in cryoprotection and in the production of cold-induced proteins. Appl. Environ. Microbiol. 67:5171-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia, B., H. Ke, and M. Inouye. 2001. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol. Microbiol. 40:179-188. [DOI] [PubMed] [Google Scholar]
- 38.Yamanaka, K., L. Fang, and M. Inouye. 1998. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol. Microbiol. 27:247-255. [DOI] [PubMed] [Google Scholar]
- 39.Yamanaka, K., and M. Inouye. 2001. Induction of CspA, an E. coli major cold-shock protein, upon nutritional upshift at 37°C. Genes Cells 6:279-290. [DOI] [PubMed] [Google Scholar]
- 40.Yamanaka, K., W. Zheng, E. Crooke, Y. H. Wang, and M. Inouye. 2001. CspD, a novel DNA replication inhibitor induced during the stationary phase in Escherichia coli. Mol. Microbiol. 39:1572-1584. [DOI] [PubMed] [Google Scholar]