Abstract
Lactococcus raffinolactis, unlike most lactococci, is able to ferment α-galactosides, such as melibiose and raffinose. More than 12 kb of chromosomal DNA from L. raffinolactis ATCC 43920 was sequenced, including the α-galactosidase gene and genes involved in the Leloir pathway of galactose metabolism. These genes are organized into an operon containing aga (α-galactosidase), galK (galactokinase), and galT (galactose 1-phosphate uridylyltransferase). Northern blotting experiments revealed that this operon was induced by galactosides, such as lactose, melibiose, raffinose, and, to a lesser extent, galactose. Similarly, α-galactosidase activity was higher in lactose-, melibiose-, and raffinose-grown cells than in galactose-grown cells. No α-galactosidase activity was detected in glucose-grown cells. The expression of the aga-galKT operon was modulated by a regulator encoded by the upstream gene galR. The product of galR belongs to the LacI/GalR family of transcriptional regulators. In L. lactis, L. raffinolactis GalR acted as a repressor of aga and lowered the enzyme activity by more than 20-fold. We suggest that the expression of the aga operon in lactococci is negatively controlled by GalR and induced by a metabolite derived from the metabolism of galactosides.
The gram-positive lactic acid bacteria (LAB) comprise 11 bacterial genera, including Lactococcus (28). Several strains of Lactococcus lactis are commonly used in the production of fermented dairy products to convert the milk sugar lactose into lactic acid (17). Lactococcus raffinolactis, formerly known as Streptococcus raffinolactis, is a LAB naturally found in raw milk, but this lactococcal species is not currently used by the dairy industry, mainly because of its lack of caseinolytic activity (15, 17, 26). L. raffinolactis cells are ovoid, and they are found in pairs or short chains (17). This species does not grow at 40°C, at pH 9.2, or in the presence of 4% NaCl and cannot hydrolyze arginine. To our knowledge, no genetic studies are available on this LAB.
L. raffinolactis has also the peculiar property of being able to ferment α-galactosides, such as melibiose and raffinose. This characteristic is attributed mainly to the activity of an α-galactosidase (Aga). Following the hydrolysis of α-galactosides by Aga, the α-galactose subunit is released and can be degraded through two distinct metabolic pathways: the tagatose 6-phosphate pathway and the Leloir pathway. The substrate for the tagatose 6-phosphate pathway is the galactose 6-phosphate, resulting from the transport and phosphorylation of melibiose by the sugar phosphotransferase transport system (PTS). Galactose 6-phosphate is metabolized to triose-phosphates by three enzymes: galactose 6-phosphate isomerase (LacA and LacB), tagatose 6-phosphate kinase (LacC), and tagatose 1,6-bisphosphate aldolase (LacD). On the other hand, intracellular nonphosphorylated galactose molecules are degraded by the Leloir pathway. α-Galactose is first transformed into galactose 1-phosphate by a galactokinase (GalK). Then, two additional enzymes, namely, galactose 1-phosphate uridylyltransferase (GalT) and UDP-galactose 4-epimerase (GalE), are responsible for converting the galactose 1-phosphate to glucose 1-phosphate, a glycolysis precursor. Both pathways are found in L. lactis (2, 29, 30), and their genetic determinants usually form an operon (9, 13, 14, 32).
A few α-galactosidase genes have also been cloned and sequenced in LAB. In Streptococcus mutans, the α-galactosidase gene is associated with the multiple-sugar metabolism operon (24). This Aga is essential for growth in the presence of melibiose and raffinose. The expression of S. mutans α-galactosidase is activated by a transcriptional regulator of the AraC/XylS family. In Streptococcus pneumoniae, Aga is essential for raffinose utilization and its activity is stimulated by raffinose and catabolite repressed by sucrose but not glucose (23). The expression of Aga is also stimulated by two gene products, one of which belongs to the AraC/XylS family. In Carnobacterium piscicola, the α-galactosidase determinant is grouped with two β-galactosidase genes and both enzymatic activities are repressed in the presence of glucose or lactose during growth (6). Similarly, the α-galactosidase gene from Lactobacillus plantarum (melA) is clustered with lacA and lacM, encoding a heterodimeric β-galactosidase (21, 27). Transcription of melA is independent of surrounding genes and is induced by melibiose and partially repressed by glucose (27).
In this work, we present the characterization and transcriptional analysis of a 12-kb chromosomal segment of L. raffinolactis ATCC 43920 (also known as NCDO617) containing the α-galactosidase gene and its transcriptional regulator as well as genes involved in the Leloir pathway of galactose degradation. This study provides the first genetic analysis of L. raffinolactis through its α-galactosides metabolism.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown at 37°C in Luria broth (LB), and lactococci were grown at 30°C in M17 medium (0.5% casein peptone, 0.5% soy peptone, 0.5% meat peptone, 0.25% yeast extract, 0.05% ascorbic acid, 0.025% magnesium sulfate, 1.9% sodium β-glycerophosphate) (Quélab) supplemented with the appropriate sugar. Carbohydrate fermentation was tested in bromocresol purple medium (2% tryptone, 0.5% yeast extract, 0.4% NaCl, 0.15% Na-acetate, 40 mg of purple bromocresol/liter). Sugars were filter sterilized and added to autoclaved media at a final concentration of 0.5%. When required, antibiotics (Sigma-Aldrich) were added as follows: for E. coli, 50 μg of ampicillin per ml, and for L. lactis, 5 μg of chloramphenicol per ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant characteristic(s)a | Sourceb or reference(s) |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | supE44 Δlac U169 (f80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Invitrogen Life Technologies |
| MC1061 | F−araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL (Strr) hsdR2 (rK− mK+) mcrA mcrB1 | ATCC |
| Lactococcus lactis | ||
| MG1363 | Laboratory strain, plasmid free, Lac− | 12 |
| SMQ-754 | Industrial strain, Lac+ | This study |
| KF292 | Isolated from soya sprouts, Raf+ | 19, 20 |
| Lactococcus raffinolactis | ||
| ATCC 43920 | Plasmid free | ATCC |
| Streptococcus thermophilus | ||
| SMQ-301 | Industrial strain, Lac+ | 31, 32 |
| Plasmids | ||
| pBS | Cloning vector for DNA sequencing, Apr | Stratagene |
| pNZ123 | Shuttle cloning vector, Cmr | 8 |
| pRAF102 | pBS plus 5-kb ClaI/SpeI fragment from L. raffinolactis ATCC 43920 encoding galT, Apr | This study |
| pRAF110 | pBS plus 8-kb KpnI/NheI fragment from L. raffinolactis ATCC 43920 encoding aga, Apr | This study |
| pRAF300 | pNZ123 plus 4-kb EcoRI/HindIII fragment from L. raffinolactis ATCC 43920 encoding aga, Cmr | 5 |
| pRAF301 | pNZ123 plus 2.5-kb aga amplicon from L. raffinolactis ATCC 43920, Cmr | 5 |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Strr, streptomycin resistance; Lac, lactose fermentation; Raf, raffinose fermentation.
ATCC, American Type Culture Collection, Manassas, Va.
DNA techniques.
Routine DNA manipulations were carried out according to standard procedures (25). Restriction enzymes, alkaline phosphatase, RNase-free DNase, RNase inhibitor (Roche Diagnostics), and T4 DNA ligase (Invitrogen Life Technologies) were used according to the suppliers' instructions. All primers used in this study were obtained from Invitrogen Life Technologies and are listed in Table 2. Transformations of E. coli (25) and L. lactis (16) were performed as described elsewhere. Plasmid DNA from E. coli and L. lactis was isolated as previously described (5, 11). Total lactococcal DNA was obtained as described by Boucher et al. (5). When needed, large amounts of E. coli plasmid DNA were isolated with a Qiagen Plasmid Maxi kit.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| CB3 | GAATTCGAATTCCCCCCCCCCCCCCCCCCC |
| raf5 | AGTGCAGCGCACCAAGGTGA |
| raf9 | CATTGAAGGCACGATTCCCA |
| raf11 | TTTAGCCACAGCACAAAGGA |
| raf12bam | GGATCCGGATCCATGACACTAATCACATTTGA |
| raf13 | CCATCACCGAAGAGGGCTGT |
| raf32 | TGCTGTGATCAAGACGCATT |
| raf33 | AACGCACGATAAATCCCGAT |
| raf34 | CGTTGAGTTCAAATTGCCAG |
| raf48 | CCCTTAGAAGAGTTGCTTGG |
| raf57 | CCTCCGGTCTAGCGTGCGGT |
| raf67 | CGGTATTGTCCACGCGCCAA |
| raf73 | CGCCGTGACCATAACTTGGA |
Sequencing of the aga locus from L. raffinolactis ATCC 43920.
The 4-kb EcoRI/HindIII DNA fragment from L. raffinolactis containing the aga gene (5) was used as a probe in Southern hybridizations to target a larger chromosomal region. The fragment was labeled using a DIG High-Prime DNA labeling kit (Roche Diagnostics). Prehybridization, hybridization, and posthybridization washes and detection by chemiluminescence were performed as suggested by the manufacturer (Roche Diagnostics). An 8-kb KpnI/NheI fragment was isolated as previously described (5) and cloned into pBS to construct pRAF110, and the DNA sequences on both strands were determined by primer walking. Primer raf34, complementary to the galT gene, was then used to target additional chromosomal restriction fragments covering the area downstream of this gene on Southern hybridizations. A 5-kb ClaI/SpeI fragment was cloned into pBS to construct pRAF102, and the cloned DNA was sequenced on both strands by primer walking. Finally, a 3-kb KpnI fragment was used as the substrate for inverse PCR (4), and the resulting amplicon was also sequenced. DNA sequencing was carried out by the DNA sequencing service at the Université Laval by means of an ABI Prism 3100 apparatus. Sequence analyses were performed using the Wisconsin Package software (version 10.3) of the Genetics Computer Group (7).
Transcriptional analysis of the aga locus.
Total RNA was isolated from L. raffinolactis by means of the same procedure used with L. lactis (5). Approximately 5 μg of material was separated on agarose-formaldehyde electrophoresis gel as described by Sambrook and Russell (25). Nucleic acids were transferred to a positively charged nylon membrane (Roche Diagnostics) and fixed by UV exposure. PCR amplicons were labeled with 32P by using High Prime (Roche Diagnostics) and used as probes. Prehybridization (3 h) and hybridization (overnight) were performed in DIG Easy Hyb (Roche Diagnostics) at 50°C, and two posthybridization washes were made at the same temperature in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 sodium citrate)-0.1% sodium dodecyl sulfate prior to film exposure (Kodak X-Omat AR). Stripping of the membrane was performed by immersion in boiling 0.1% sodium dodecyl sulfate (25). The membrane was successively hybridized with the different probes in the following order: B, D, A, and C (Fig. 1). Probes A, B, C, and D were obtained by PCR amplification of L. raffinolactis DNA with primers raf32 and raf33, primers raf5 and raf13, primers raf9 and raf11, and primers raf48 and raf57, respectively.
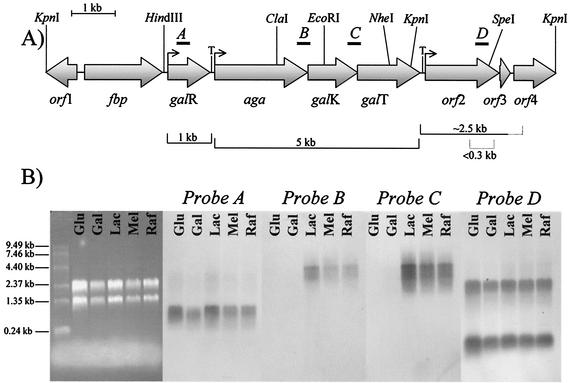
FIG. 1.
Characterization of the aga locus of L. raffinolactis ATCC 43920. (A) Over 12 kb of genomic DNA of L. raffinolactis was analyzed. Nine ORFs were identified, and putative functions were assigned to corresponding proteins according to conserved motifs and similarity with proteins of known functions (Table 3). Brackets located under the figure identify transcripts, with dotted lines indicating the putative limits of transcription. Positions of restriction sites used for cloning or sequencing are indicated above the figure. Bent arrow, promoter; T, terminator. (B) Northern analysis of the aga locus. Total RNA was isolated from L. raffinolactis cultivated in the presence of glucose (Glu), galactose (Gal), lactose (Lac), melibiose (Mel), or raffinose (Raf). The agarose gel (left side) is presented to indicate the relative amount of RNA loaded per lane. mRNAs were detected using 32P-labeled probes targeting galR (probe A), aga-galK (probe B), galK-galT (probe C), and orf2 (probe D).
Localization of the transcriptional initiation site of aga.
The transcriptional initiation site of aga was established through 5′ rapid amplification of cDNA ends (RACE) essentially as described by Sambrook and Russell (25). RNA was isolated from L. raffinolactis cells grown in raffinose. DNase treatments and cDNA synthesis were performed using 20 μg of total RNA and the aga-specific primer raf67 (5). Free nucleotides and primers were removed by precipitating the cDNA twice in 2.5 M ammonium acetate with 3 volumes of 95% ethyl alcohol. cDNA was then dissolved in double-distilled water to a final volume of 20 μl. cDNA (13 μl) was used for poly(dG) tailing with terminal deoxyribonucleotidyl transferase, as recommended by the manufacturer (Amersham Pharmacia Biotech). Tailed DNA was diluted to 1 ml with 10 mM Tris-HCl, pH 8.5, and 5 μl was used for PCR amplification with a poly(dC) primer (CB3) and the aga-specific primer raf73. The PCR product was purified with silica as previously described (10) and sequenced using primer raf73. The transcriptional initiation site of aga was also determined by primer extension analysis essentially as described by Sambrook and Russell (25).
α-Galactosidase assay.
α-Galactosidase activity was evaluated as described by Boucher et al. (5). Briefly, 10-ml cell cultures (optical density at 600 nm, 0.5) were washed and lysed with glass beads. The cell lysate was cleared by centrifugation, and the protein content was determined with the Bio-Rad DC protein assay reagent. The α-galactosidase activity was assayed at 30°C and at pH 7.0 with p-nitrophenyl-α-d-galactopyranoside (Sigma) as the substrate (22).
β-Galactosidase and phospho-β-galactosidase assays.
Both the β-galactosidase and phospho-β-galactosidase enzymatic activities were detected as described for the α-galactosidase assay but with cells grown in LM17 medium (optical density at 600 nm, 0.5). The enzyme assays were performed at 30°C and at pH 7.0 with o-nitrophenyl-β-d-galactopyranoside (Sigma-Aldrich) as the substrate for β-galactosidase and o-nitrophenyl-β-d-galactopyranoside 6-phosphate (Sigma-Aldrich) as the substrate for phospho-β-galactosidase. The presence of the enzyme activity was confirmed by the appearance of a yellow color after the cells were incubated overnight. The industrial strain Streptococcus thermophilus SMQ-301 (31) was used as a positive control for β-galactosidase, and L. lactis subsp. cremoris SMQ-754 was used as a positive control for phospho-β-galactosidase.
Nucleotide sequence accession number.
The GenBank accession number assigned to the nucleotide sequence of the aga locus of L. raffinolactis ATCC 43920 is AY164273.
RESULTS AND DISCUSSION
Confirmation of L. raffinolactis ATCC 43920.
The L. raffinolactis strain used in this study grew well in M17 medium and fermented the two α-galactosides raffinose and melibiose, as well as lactose (β-galactoside), galactose, and glucose. Sequencing of the 16S rRNA gene was performed on L. raffinolactis ATCC 43920, and the data matched the previously published 16S rRNA sequence for L. raffinolactis NCDO617 (GenBank accession no. X54261).
Sequence analysis of the aga locus from L. raffinolactis ATCC 43920.
A 4-kb DNA region involved in α-galactoside fermentation was previously cloned and sequenced from L. raffinolactis ATCC 43920 and used as a dominant selectable marker in a food-grade cloning vector for the genetic modification of L. lactis (5). In our study, this region was further characterized, and an additional 8 kb of DNA was sequenced (Fig. 1A) This DNA segment possessed a G+C content of 39.7%, which is in agreement with the previous estimated values of 40.3 to 41.5% (17). The analysis of the 12-kb DNA fragment revealed nine open reading frames (ORFs) (Fig. 1A and Table 3). Putative functions were attributed to products of eight ORFs based on motifs in the peptide chains, and/or extensive amino acid similarity with proteins of known function (see Table 3 for details).
TABLE 3.
Characterization of ORFs identified on the L. raffinolactis chromosomea
| Product of ORF | Length (aa) | pI | Mol. mass (kDa) | Potential RBS/N terminusb | Proposed function | Characteristic(s) |
|---|---|---|---|---|---|---|
| Orf1 | >241 (incomplete) | tattcgattatttGGAGGttt tatg/MTIGKDDFIR | Unknown | |||
| Fbp | 640 | 6.00 | 73.6 | cccatcAGGAAAGaagacaag catg/MQENKYLSLL | Fructose-1,6- bisphosphatase | Up to 51% aa identity over the entire protein sequence with orthologues found in various gram-positive organisms, including L. lactis subsp. lactis (GB no. AE006263) |
| GalR | 345 | 5.86 | 38.4 | gcaagtAAAGGAAttgtaag ctatg/MASIREIAKL | Transcriptional regulator | LacI/GalR family; up to 34% aa identity over the entire protein sequence with orthologues found in various gram-positive organisms, such as S. thermophilus (GB no. AAM28581.1) and S. mutans (GB no. JC5310) |
| Aga | 735 | 5.02 | 83.3 | aaatAAAGAAAGCGGG tcactcatg/MTLITFDENN | α-Galactosidase | Up to 54% aa identity over the entire protein sequence with orthologues found in various gram-positive organisms, such as Geobacillus stearothermophilus(GB no. AF130985, AY013287, AY013286) |
| GalK | 392 | 4.98 | 42.7 | aaatttACAGccctcttcggt gatg/MSTKEMKQEV | Galactokinase | Up to 66% aa identity over the entire protein sequence with orthologues found in various gram-positive organisms, such as S. thermophilus (GB no. AF152357) and L. lactis (GB no. AJ011653, AE006428) |
| GalT | 492 | 5.24 | 55.0 | ttgcAGATGGAGctagaaaa ttatg/MNISQAVIDF | Galactose 1-phosphate-uridylyltransferase | Up to 54% aa identity over the entire protein sequence with orthologues found in various gram-positive organisms, such as S. pneumoniae (GB no. AE007475) and L. lactis (GB no. AF082008, AE006428) |
| Orf2 | 608 | 5.33 | 70.0 | aagtgtGAGGAGGAGAta aaatatg/MIRQAINLTE | Transcriptional regulator | Contains a PTS regulatory domain and a PTS EIIA domain from the fructose subfamily |
| Orf3 | 94 | 4.40 | 9.9 | tcaagaAGGTGAGGaataaa atatg/MKLAAVCGSG | PTS EIIB component | Contains a putative phosphorylation site and a cysteine residue at position 7 |
| Orf4 | >297 (incomplete) | ctagattAGGAGAAtaatca acatg/MKDVLDVLID | PTS EIIC component | Contains putative transmembrane helices |
aa, amino acids; RBS, ribosome binding site; GB no., GenBank accession number.
Bold type indicates start codons; uppercase letters indicate A+G-rich stretches.
Sequence analysis also revealed the presence of three putative promoters upstream from the genes aga (TTGACA-N17-TATAAT) (5), orf2 (TTGAAA-N17-TAAAGT), and galR (TTGTTT-N20-TAAAAT). Two regions containing inverted repeats, able to form a stem-loop structure and to act as intrinsic transcriptional terminators, were found downstream from galR (ΔG25°C = −17.6 kcal/mol) and galT (ΔG25°C = −13.2 kcal/mol). Two catabolite-responsive element sequences were also identified in the 12-kb fragment of L. raffinolactis. Catabolite-responsive element sequences were found in the −35 region of the promoters upstream from aga and orf2, indicating that these genes might be subjected to catabolite repression (18). Finally, a potential binding site for GalR was identified in the −10 box region of the aga promoter (Fig. 2). The N-terminal helix-turn-helix motif of GalR and its putative binding site are similar to cognate regions characterized for S. mutans (1) and S. thermophilus (34).
FIG. 2.
Identification of a potential GalR binding site upstream of aga. (A) Alignment of the putative GalR binding site from L. raffinolactis and S. thermophilus (34) with the DNA sequence protected by GalR from DNase I digestion in S. mutans (1). The −10 and −35 boxes are indicated in bold. For L. raffinolactis, only the −10 box is provided. The transcription start nucleotide is boxed in black, and the ATG start codon is underlined. (B) Alignment of the N termini of S. thermophilus (S. t.), S. mutans (S. m.), and L. raffinolactis (L. r.) GalR proteins. The helix-turn-helix motif is in bold. |, identical positions; +, conservative substitutions.
These results suggested that (i) aga, galK, and galT formed a transcriptional unit expressed from a promoter located upstream from aga and ending at the terminator identified downstream from galT; (ii) galR was transcribed independently from the aga-galKT operon and its transcription ended in the galR-aga intergenic region; and (iii) orf2, orf3, and orf4 may also be expressed independently from at least one promoter located upstream from orf2.
mRNA analysis of the 12-kb DNA fragment from L. raffinolactis ATCC 43920.
To determine the number and size of transcripts in this chromosomal region, Northern blot analyses were performed on L. raffinolactis cells grown on various sugars (Fig. 1B). The gene galR was expressed, under every condition tested, as a monocistronic mRNA 1 kb in size. The transcript size was in agreement with the gene size and suggested transcription from a promoter located upstream from galR and termination at the stem-loop structure identified in the galR-aga intergenic region.
The aga-galKT mRNA was 5 kb in size, which corresponded to a transcriptional unit that started at the proposed promoter upstream from aga and ended at the terminator downstream from galT. The transcription of this operon was stimulated by galactosides such as lactose, melibiose, and raffinose (Fig. 1B). Overexposure of the Northern membrane revealed a weak aga-galKT transcript of the same size in cells grown on galactose, while no signal was detected in glucose-grown cells (data not shown).
Finally, the three genes downstream from galT were expressed independently from the aga operon in cells grown on all the sugars tested (Fig. 1B).
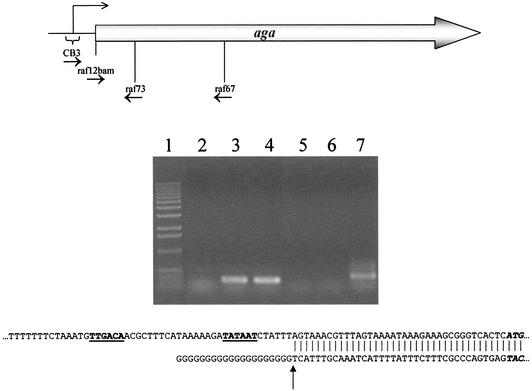
Identification of the transcription start site of the aga operon.
The RACE-PCR technique was used to identify the transcription initiation site of the aga-galKT operon (Fig. 3). The transcription was initiated at the A position located 7 bases downstream from the last T position of the −10 box identified on the DNA coding strand. An identical transcription initiation site was confirmed by primer extension analysis (data not shown).
FIG. 3.
Localization of the transcription initiation site of aga by RACE-PCR. Total RNA was isolated from L. raffinolactis grown in M17 medium plus 0.5% raffinose and used to synthesize cDNA with primer raf67. cDNA was tailed with dGTP using terminal transferase and PCR amplified with raf73 and a poly(C) primer (CB3) or primer raf12bam as a control. Lane 1, 1-kb DNA ladder (Invitrogen Life Technologies); lanes 2 to 4, controls with raf73 and raf12bam; lane 2, negative control (no DNA); lane 3, positive control (L. raffinolactis DNA); lane 4, positive control [poly(G)-cDNA]; lanes 5 to 7, amplification with raf73 and CB3; lane 5, negative control (no DNA); lane 6, CB3 specificity control (L. raffinolactis DNA); lane 7, poly(G)-cDNA. The amplicon obtained in lane 7 was sequenced with primer raf73, and its sequence was aligned with that of L. raffinolactis genomic DNA. The transcriptional initiation site is indicated by an arrow.
α-Galactosidase activity in L. raffinolactis ATCC 43920.
The α-galactosidase activity was measured in cell lysates of L. raffinolactis grown in the presence of various sugars (Table 4). Aga expression was strongly stimulated by the galactosides lactose, melibiose, and raffinose but poorly stimulated by galactose (Table 4), which induced fivefold less Aga activity than did melibiose. Virtually no Aga activity was detected in L. raffinolactis cells cultivated in the presence of glucose (Table 4). The finding that lactose induced aga in L. raffinolactis contrasted with the observation that lactose did not induce aga in the industrial lactose-positive strain L. lactis SMQ-741 (5). This difference most likely resulted from the fact that these two lactococcal species do not metabolize lactose the same way. L. lactis transports lactose by the PTS, and the galactose 6-phosphate produced following hydrolysis by phospho-β-galactosidase is metabolized via the tagatose 6-phosphate pathway (2, 29). Both β-galactosidase and phospho-β-galactosidase activities were detected in lactose-grown L. raffinolactis cells (data not shown), suggesting that the galactose moiety of lactose could be metabolized by the Leloir and the tagatose 6-phosphate pathways. Thus, the metabolism of lactose by L. raffinolactis generated intracellular galactose 6-phosphate as well as free galactose and galactose derivatives of the Leloir pathway. These results suggested that galactose or an intermediate of the Leloir pathway, but not galactose-6 phosphate, is involved in aga induction in lactococci. However, since extracellular galactose barely induced the aga operon in L. raffinolactis (Table 4), we suggest that aga induction in lactococci is optimally driven by a metabolite derived from galactoside metabolism.
TABLE 4.
Effect of the fermentation substrate on α-galactosidase activity in L. raffinolactis ATCC 43920
| Sugar | Activitya |
|---|---|
| Glucose | 16 ± 18 |
| Galactose | 191 ± 12 |
| Lactose | 643 ± 34 |
| Melibiose | 930 ± 93 |
| Raffinose | 513 ± 44 |
Values are the means ± standard deviations of 12 measurements obtained with two cell extract quantities in two independent experiments. Activity is expressed in nanomoles of p-nitrophenol formed per milligram of protein per minute.
α-Galactosidase activity in L. lactis MG1363 in the presence or absence of GalR.
A potential GalR binding site was found in the promoter region of the aga operon. To determine whether GalR acted as a transcriptional regulator of the aga operon, gene inactivation and allelic replacement were attempted in L. raffinolactis. Despite several attempts using various strategies, we failed to obtain a galR mutant derivative in this species. Thus, the function of L. raffinolactis GalR was assessed in L. lactis subsp. cremoris MG1363, which does not display any α-galactosidase activity.
The aga gene was cloned into the high-copy-number plasmid vector pNZ123, alone (pRAF301) or in combination with galR (pRAF300). aga was cloned on a high-copy-number vector to minimize the potential effects of endogenous L. lactis regulators. The recombinant plasmids were separately introduced into L. lactis MG1363, and the α-galactosidase activity was evaluated after growth in the presence of glucose, galactose, and melibiose (Table 5). It is noteworthy that this L. lactis strain cannot utilize lactose. Interestingly, the α-galactosidase activity was at least 20-fold higher in cells containing pRAF301 (aga) than in those with pRAF300 (aga and galR). Therefore, aga was expressed from its own promoter without the need for GalR-mediated activation, and GalR clearly repressed Aga activity in L. lactis. These results suggest that GalR acted as a repressor in L. raffinolactis.
TABLE 5.
Regulation of α-galactosidase activity by GalR in L. lactis subsp. cremoris MG1363
| Recombinant plasmid | Sugar | Activitya |
|---|---|---|
| pRAF300 (aga plus galR) | Glucose | 27 ± 27 |
| Galactose | 372 ± 107 | |
| Melibiose | 314 ± 21 | |
| pRAF301 (aga) | Glucose | 1807 ± 469 |
| Galactose | 9998 ± 1713 | |
| Melibiose | 6719 ± 816 |
Values are the means ± standard deviations of 12 measurements obtained with two cell extract quantities in two independent experiments. Activity is expressed in nanomoles of p-nitrophenol formed per milligram of protein per minute.
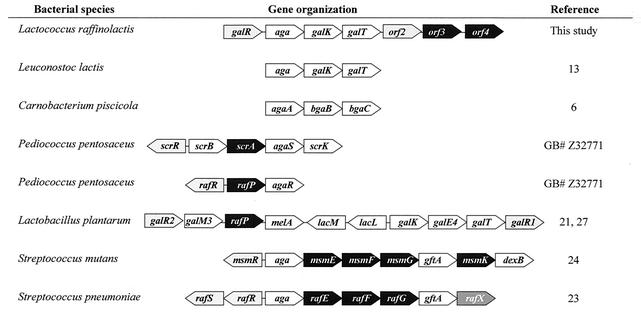
Organization of the α-galactosidase genes in LAB.
The genes coding for the Leloir pathway enzymes generally form operons (14, 32). In L. lactis subsp. cremoris MG1363, the order of the five genes required for galactose utilization is galPMKTE, where galP (formerly galA) encodes a galactose transporter (5, 13, 14). The gal genes of L. lactis subsp. lactis NCDO2054 and IL1403 are similarly organized but include the insertion of two genes (lacA and lacZ) between galT and galE (3, 33). In L. raffinolactis ATCC 43920, the gal genes are not grouped around galM, and galE genes could not be found on the 12-kb segment of the DNA sequenced.
Conversely, α-galactosidase determinants are heterogeneous in terms of their genetic positioning. In L. raffinolactis, the α-galactosidase gene is clustered with galK and galT to form an operon. The α-galactosidase gene organization is known for six other LAB genera (Fig. 4). In a general manner, α-galactosidase genes were found with genes coding for a diversity of enzymes and transporters involved in the metabolism of various sugars. In Lactobacillus plantarum, the melA gene encoding α-galactosidase was also located near genes involved in the Leloir pathway of galactose metabolism, namely, galM3, galK, galE4, and galT (21, 27). Surprisingly, the organization found in Leuconostoc lactis appeared to be similar to that found in L. raffinolactis (13). Unfortunately, the DNA sequence required to support investigations concerning possible horizontal gene transfer between the two bacteria was not available.
FIG. 4.
Genetic organization of α-galactosidase genes in LAB. The genes are indicated by polygons of the same size regardless of their length. White, carbohydrate degradation enzymes; black, sugar transporters; gray, transcriptional regulators. The following genes code for the indicated proteins: aga, melA α-galactosidase; galK, galactokinase; galT, galactose 1-phosphate uridylyltransferase; galM, mutarotase; bgaB, bgaC, lacL, and lacM β-galactosidase; scrB, sucrose phosphorylase; scrK, fructokinase; gftA, sucrose 6-phosphate hydrolase; dexB, dextran glucosidase; galR, scrR, rafR, rafS, msmR, and orf2 transcriptional regulators; orf3 and orf4, putative PTS EIIB and EIIC domains; scrA, PTS EIISuc; rafP (also named lacS2 in Lactobacillus plantarum WCFS1), galactoside-pentose-hexuronide transporter; msmEFGK and rafEFG, ABC transporters; rafX, unknown. GB#, GenBank accession number.
α-Galactoside metabolism is also found in some L. lactis subsp. lactis strains isolated from fruits and vegetables where raffinose is readily available (19, 20). In the α-galactosides-fermenting strain KF292 of L. lactis, we have found a DNA region encompassing the galR and aga genes that is over 90% identical to a DNA segment of L. raffinolactis ATCC 43920 (data not shown). Most L. lactis strains used by the dairy industry are melibiose and raffinose negative, and they were isolated from raw milk or fermented milk products. On the other hand, L. lactis strains found on plant material are able to utilize these sugars. Therefore, the presence or absence of an α-galactosides metabolism in lactococcal strains is likely related to their distinct ecological adaptation.
Conclusion.
Over the years, research on LAB genetics has focused mainly on the industrial applications of these organisms. Consequently, certain lactococcal species, such as L. raffinolactis, remain uncharacterized at the genetic level. This study provides the first genetic analysis of L. raffinolactis. In this species, α-galactoside metabolism was driven by an operon that contains the three genes aga, galK, and galT. The gene aga encoded an α-galactosidase, while galK and galT most likely encoded, respectively, a galactokinase and a galactose 1-phosphate-uridylyltransferase, two enzymes of the Leloir pathway. The expression of this operon is controlled by a gene located upstream, galR. The product of galR belonged to the LacI/GalR family of transcriptional regulators and acted as a repressor of the gene cluster.
Acknowledgments
We thank the Natural Sciences and Engineering Research Council of Canada (Research Partnerships Program—Research Network on Lactic Acid Bacteria), Agriculture and Agri-Food Canada, Novalait, Inc., Dairy Farmers of Canada, and Institut Rosell-Lallemand, Inc., for their financial support. I.B. is a recipient of a Fonds FCAR graduate scholarship.
We are grateful to William Kelly for providing the α-galactoside-fermenting strain L. lactis KF292.
REFERENCES
- 1.Ajdic, D., and J. J. Ferretti. 1998. Transcriptional regulation of the Streptococcus mutans gal operon by the GalR repressor. J. Bacteriol. 180:5727-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bissette, D. L., and R. L. Anderson. 1974. Lactose and d-galactose metabolism in group N streptococci: presence of enzymes for both the d-galactose 1-phosphate and d-tagatose 6-phosphate pathways. J. Bacteriol. 117:318-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouchard, J. D., and S. Moineau. 2000. Homologous recombination between a lactococcal bacteriophage and the chromosome of its host strain. Virology 270:65-75. [DOI] [PubMed] [Google Scholar]
- 5.Boucher, I., M. Parrot, H. Gaudreau, C. P. Champagne, C. Vadeboncoeur, and S. Moineau. 2002. Novel food-grade plasmid vector based on melibiose fermentation for the genetic engineering of Lactococcus lactis. Appl. Environ. Microbiol. 68:6152-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coombs, J., and J. E. Brenchley. 2001. Characterization of two new glycosyl hydrolases from the lactic acid bacterium Carnobacterium piscicola strain BA. Appl. Environ. Microbiol. 67:5094-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vos, W. M. 1987. Gene cloning and expression in lactic streptococci. FEMS Microbiol. Rev. 46:281-295. [Google Scholar]
- 9.de Vos, W. M., and E. E. Vaughan. 1994. Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol. Rev. 15:217-237. [DOI] [PubMed] [Google Scholar]
- 10.Duplessis, M., and S. Moineau. 2001. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 41:325-336. [DOI] [PubMed] [Google Scholar]
- 11.Émond, É., R. Lavallée, G. Drolet, S. Moineau, and G. LaPointe. 2001. Molecular characterization of a theta replication plasmid and its use for development of a two-component food-grade cloning system for Lactococcus lactis. Appl. Environ. Microbiol. 67:1700-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grossiord, B. P., E. J. Luesink, E. E. Vaughan, A. Arnaud, and W. M. de Vos. 2003. Characterization, expression, and mutation of the Lactococcus lactis galPMKTE genes, involved in galactose utilization via the Leloir pathway. J. Bacteriol. 185:870-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossiord, B., E. E. Vaughan, E. Luesink, and W. M. de Vos. 1998. Genetics of galactose utilization via the Leloir pathway in lactic acid bacteria. Lait 78:77-84. [Google Scholar]
- 15.Holler, B. J., and J. L. Steele. 1995. Characterization of lactococci other than Lactococcus lactis for possible use as starter cultures. Int. Dairy J. 5:275-289. [Google Scholar]
- 16.Holo, H., and I. F. Nes. 1989. High frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holt, J. G., N. R. Krieg, P. H. A. Sneath, J. T. Staley, and S. T. Williams. 1994. Bergey's manual of determinative bacteriology, 9th ed. The Williams & Wilkins Co., Baltimore, Md.
- 18.Hueck, C. J., W. Hillen, and M. H. Saier, Jr. 1994. Analysis of a cis-active sequence mediating catabolite repression in Gram-positive bacteria. Res. Microbiol. 145:503-518. [DOI] [PubMed] [Google Scholar]
- 19.Kelly, W. J., G. P. Davey, and L. J. H. Ward. 1998. Characterization of lactococci isolated from minimally processed fresh fruit and vegetables. Int. J. Food Microbiol. 45:85-92. [DOI] [PubMed] [Google Scholar]
- 20.Kelly, W. J., G. P. Davey, and L. J. H. Ward. 1998. Conjugative transfer of raffinose metabolism in Lactococcus lactis. FEMS Microbiol. Lett. 167:145-149. [Google Scholar]
- 21.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. De Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCleary, B. V. 1988. Alpha-galactosidase from luciferine and guar seed. Methods Enzymol. 160:627-632. [Google Scholar]
- 23.Rosenow, C., M. Maniar, and J. Trias. 1999. Regulation of the α-galactosidase activity in Streptococcus pneumoniae: characterization of the raffinose utilization system. Genome Res. 9:1189-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russell, R. R. B., J. Aduse-Opoku, I. C. Sutcliffe, L. Tao, and J. J. Ferretti. 1992. A binding protein-dependent transport system in Streptococcus mutans responsible for multiple sugar metabolism. J. Biol. Chem. 267:4631-4637. [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Schleifer, K. H., J. Kraus, C. Dvorak, R. Kilpper-Balz, M. D. Collins, and W. Fischer. 1985. Transfer of Streptococcus lactis and related streptococci into the genus Lactococcus gen. nov. Syst. Appl. Microbiol. 6:183-195. [Google Scholar]
- 27.Silvestroni, A., C. Connes, F. Sesma, G. Savoy de Giori, and J.-C. Piard. 2002. Characterization of the melA locus for α-galactosidase in Lactobacillus plantarum. Appl. Environ. Microbiol. 68:5464-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stiles, M. E., and W. H. Holzapfel. 1997. Lactic acid bacteria of foods and their current taxonomy. Int. J. Food Microbiol. 36:1-29. [DOI] [PubMed] [Google Scholar]
- 29.Thompson, J. 1979. Lactose metabolism in Streptococcus lactis: phosphorylation of galactose and glucose moieties in vivo. J. Bacteriol. 140:774-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson, J. 1980. Galactose transport systems in Streptococcus lactis. J. Bacteriol. 144:683-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tremblay, D. M., and S. Moineau. 1999. Complete genomic sequence of the lytic bacteriophage DT1 of Streptococcus thermophilus. Virology 255:63-76. [DOI] [PubMed] [Google Scholar]
- 32.Vaillancourt, K., S. Moineau, M. Frenette, C. Lessard, and C. Vadeboncoeur. 2002. Galactose and lactose genes from the galactose-positive bacterium Streptococcus salivarius and the phylogenetically related galactose-negative bacterium Streptococcus thermophilus: organization, sequence, transcription, and activity of the gal gene products. J. Bacteriol. 184:785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaughan, E. E., R. D. Pridmore, and B. Mollet. 1998. Transcriptional regulation and evolution of lactose genes in the galactose-lactose operon of Lactococcus lactis NCDO2054. J. Bacteriol. 180:4893-4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaughan, E. E., P. T. C. van den Bogaard, P. Catzeddu, O. P. Kuipers, and W. M. de Vos. 2001. Activation of silent gal genes in the lac-gal regulon of Streptococcus thermophilus. J. Bacteriol. 183:1184-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]