Abstract
The temporal stability and diversity of bacterial species composition as well as the antilisterial potential of two different, complex, and undefined microbial consortia from red-smear soft cheeses were investigated. Samples were collected twice, at 6-month intervals, from each of two food producers, and a total of 400 bacterial isolates were identified by Fourier-transform infrared spectroscopy and 16S ribosomal DNA sequence analysis. Coryneform bacteria represented the majority of the isolates, with certain species being predominant. In addition, Marinolactobacillus psychrotolerans, Halomonas venusta, Halomonas variabilis, Halomonas sp. (106 to 107 CFU per g of smear), and an unknown, gram-positive bacterium (107 to 108 CFU per g of smear) are described for the first time in such a consortium. The species composition of one consortium was quite stable over 6 months, but the other consortium revealed less diversity of coryneform species as well as less stability. While the first consortium had a stable, extraordinarily high antilisterial potential in situ, the antilisterial activity of the second consortium was lower and decreased with time. The cause for the antilisterial activity of the two consortia remained unknown but is not due to the secretion of soluble, inhibitory substances by the individual components of the consortium. Our data indicate that the stability over time and a potential antilisterial activity are individual characteristics of the ripening consortia which can be monitored and used for safe food production without artificial preservatives.
Traditional production of various foodstuffs such as kefir, sourdough, and red-smear cheeses includes the transfer of complex undefined microbial consortia from mature products to freshly prepared matrices. As of now it is not known whether the repeated transfer to fresh substrates, or their indigenous microflora, influences the composition of the respective consortia over time. In the case of red-smear cheeses, the microbial succession during surface ripening starts with the growth of yeasts, which metabolize the lactic acid present and raise the pH on the cheese surface from about 5.0 to about 6.0. When the pH is about 6.0, a salt-tolerant, usually very complex and undefined bacterial consortium begins to develop (8, 13, 29) and eventually covers the entire surface of the cheese. Despite their complex and undefined composition, analysis of bacterial surface-ripening floras of 19 different smeared cheeses yielded no indication that nonculturable bacteria contribute significantly to these floras (11). Bacterial surface floras consist mainly of a large number of coryneform species (3, 4, 14, 33, 37). Identification of coryneform bacteria usually is performed on the basis of physiological, phenotypic, and biochemical characteristics (18, 19, 24, 32, 33). These methods, however, do not always provide satisfactory results (5, 27), and they are both too laborious and too time-consuming to be performed on a routine basis. In contrast, molecular methods such as analysis of the 16S ribosomal DNA (rDNA) sequence yield precise identifications, but the cost per sample is quite high. Alternatively, the corynebacterial core part of bacterial red-smear surface floras can now be identified by Fourier-transform infrared (FT-IR) spectroscopy. This cost-effective technique allows rapid and simple identification of microorganisms (21, 23, 25) and has recently been established for identification of coryneform bacteria (26, 27).
When the traditional “old-young smearing” procedure is used for the production of red-smear cheeses, pathogenic microorganisms such as Listeria monocytogenes may also be transferred from the mature to the fresh cheeses. Due to its ubiquitous nature, its ability to grow at refrigeration temperatures, and its tolerance to low pHs (below pH 5.0) and high (up to 10%) sodium chloride levels (17), it is very difficult to control L. monocytogenes in a cheese environment. As a result, its incidence in red-smear cheeses has not decreased significantly during the past 14 years (30). To increase the safety of these products, several attempts to investigate and use the antilisterial activity of red-smear ripening floras have been carried out. Often, single bacterial strains were isolated from red-smear cheese surface floras and evaluated for their antilisterial activity in vitro (11, 31, 35). Furthermore, in situ evaluation of the antilisterial activity of complex bacterial surface floras and single strains derived from red-smear cheeses has been performed (9, 12, 16).
To date, however, nothing is known about the stability of the composition of such consortia at the species level over time, and no information is available on the stability over time of the antilisterial activity which is associated with some of these complex ripening cultures. The aim of this study was to examine and compare these characteristics of two surface floras of German red-smear soft cheeses.
MATERIALS AND METHODS
Bacterial strains and ripening consortia.
Undefined surface floras (microbial consortia) from two different red-smear soft cheeses produced from pasteurized cow's milk in factories R and K were harvested twice, at 6-month intervals. The consortia of the first and the second samplings were used for the subsequent cheese-ripening experiments (A and B, respectively). The consortia were scraped off the cheese surfaces, and about 10 g of smear was homogenized in 100 ml of sterile NaCl solution (5%). After centrifugation (at 10,400 × g for 25 min), the supernatant was discarded and the cells were resuspended in 20 ml of sterile NaCl solution (5%) plus 4.2 ml of sterile glycerol (87%). The suspensions were then dispensed into microtubes (2 ml/tube) and subsequently stored at −70°C. The preparations were conducted under sterile conditions. A defined consortium isolated by B. Maier and M. Rudolf (our laboratory, unpublished data) from red-smear soft cheese from factory M was used as a control flora. It consists of 12 bacterial strains and 4 yeast strains and was also stored at −70°C. L. monocytogenes WSCL 1364, from Vacherin Mont d' Or cheese (2), was used as an indicator strain for in situ testing of the antilisterial activities of the various ripening consortia. It was cultivated for 24 h at 30°C in brain heart infusion broth (BHI; Merck) and diluted in one-quarter-strength Ringer's solution (Merck) to achieve appropriate cell numbers for brine contamination.
Cheese inoculation and ripening.
All experiments for the evaluation of antilisterial activity were performed independently in duplicate, and duplicate experiments yielded similar results (data not shown). Viable bacterial cell counts of the frozen consortia were determined on PCA3 (plate count agar containing 3% NaCl) after incubation for 72 h at 30°C. Viable yeast cell counts (in the case of flora M) were determined on YGC (yeast-glucose-chloramphenicol agar; Merck), supplemented with 10 μg of bromophenol blue/liter, after incubation for 48 h at 30°C. The undefined consortia (R and K) and the defined consortium (M) were suspended in sterile 5% NaCl solution to a concentration of at least 5 × 107 CFU/ml. This solution was used to inoculate soft cheeses (45% fat content in dry matter), which were obtained from a German dairy right after dry salting. Fresh cheeses were checked for the absence of Listeria (see below). Inoculation was performed according to the work of Eppert et al. (16). Cheeses were incubated under laboratory conditions in glass desiccators as described previously (34) at 16°C from day 1 to day 13 and subsequently at 12°C until day 20. Cheeses were packed in aluminum foil at day 20 and then stored at 10°C for 3 weeks. They were contaminated with L. monocytogenes WSLC 1364 by contaminating the brine at day 1, just before smearing. The contamination levels ranged from 101 to 103 CFU/ml of smear water.
Detection of L. monocytogenes in cheese.
L. monocytogenes was detected according to standard 143A:1995 of the International Dairy Federation (1) by analyzing a 25-g subsample of the cheese surface.
Enumeration of L. monocytogenes in cheese.
When a positive result was obtained, the next Listeria analysis was performed as a direct count without enrichment. The two main surfaces of 5-mm-thick slices of cheese (20 g; 50 cm2) were homogenized in 180 ml of 1.75% (wt/vol) trisodium citrate buffer (C6H5Na3O7 · 2H2O, pH 7.5; Merck). Serial 10-fold dilutions of these suspensions were plated onto Oxford agar and incubated at 37°C for 48 h. Cell counts were calculated per square centimeter of cheese surface.
Evaluation of antilisterial activity.
The 400 isolates (100 per factory [R and K] and sampling [A and B]) tested were maintained as described above. The indicator strain (L. monocytogenes WSCL 1364) and the control strain (Staphylococcus equorum WS 2733), which had been found to produce a bacteriocin with an antagonistic effect against the former (12), were grown at 30°C on tryptose agar (Merck) and plate count agar, respectively, for 24 h and were stored at 4°C as stock cultures for 1 week. To screen for antilisterial activity, coryneform isolates were grown in PC3 broth as described by Valdés-Stauber et al. (35). Gram-negative isolates were propagated for 48 h. Gram-positive and catalase-negative bacteria were grown in M17 (Merck) broth at 30°C for 48 h. The control strain was grown in BHI (Merck) broth at 30°C for 24 h with shaking at 150 rpm (10). The total flora of each consortium was scraped off from countable PCA3 agar plates like those used for isolation of bacteria (see the next section). The 400 isolates, total floras, and the control strain were examined for antilisterial activity according to the work of Valdés-Stauber et al. (35).
Isolation of bacteria from surface floras.
Microtubes of the microbial consortia R and K, from both samplings, were thawed and plated onto PCA3 plates. After incubation at 30°C for 3 days, followed by 4 days at room temperature, 100 colonies per consortium and sampling were selected randomly from countable agar plates and were purified by restreaking onto PCA3 agar. The purified isolates were stored as glycerol stocks at −70°C.
Identification of bacterial isolates.
The purified isolates were subjected to a KOH test (7) and checked for the presence of catalase by using 3% H2O2. Cell morphology was determined by phase-contrast microscopy. Due to their cell morphology, the gram- and catalase-positive isolates were considered putative coryneforms and were identified by FT-IR spectroscopy (27). Additionally, representative strains of the similarity clusters obtained by FT-IR spectroscopy were subjected to 16S rDNA sequence analysis. Strain lysis, 16S rDNA sequence amplification, and purification of the PCR product were performed according to the work of Oberreuter et al. (27). After purification, PCR products were sequenced by SequiServe (Vaterstetten, Germany). Cycle sequencing was performed by using the universal 16S rDNA binding primer 5′f (5′-AGAGTTTGACCTGGCTCA-3′; positions 8 to 26 in the Escherichia coli numbering system) (6).
Gram-positive and catalase-negative isolates were characterized by biochemical tests. These included gas formation from glucose (2%, wt/vol) at 30°C for 48 h in MRS broth (Oxoid) and gas formation from citrate at 30°C for 48 h in M17 broth (Merck) containing 2.5% (wt/vol) diammonium citrate (C6H14N2O7;Merck). Growth was determined at 42°C for 48 h in APT broth (Merck), at 37°C for 48 h in M17 broth containing 6.5% NaCl, and on kanamycin esculin azide agar (Merck) at 37°C for 48 h. Additionally, acid formation from lactose was determined in M17 broth (Merck) at 30°C for 48 h. For identification by FT-IR spectroscopy, cells were incubated at 30°C for 48 h on APT agar under anaerobic conditions and then treated by the method of Oberreuter et al. (27), by using a lactic acid bacteria reference library (H. Seiler, this laboratory, unpublished data). Since the gram-positive and catalase-negative isolates could not be identified by these methods, they were also subjected to 16S rDNA analysis as described above.
The gram-negative isolates were checked for the presence of cytochrome oxidase by using Bactident test strips (Merck) and were subsequently identified by using the BBL Enterotube II or BBL Oxi/Ferm Tube II (Becton Dickinson) biochemical test system. Due to unclear results, the final identification of the gram-negative isolates was performed by 16S rDNA analysis as described above.
RESULTS AND DISCUSSION
Identification of 400 representative bacterial isolates.
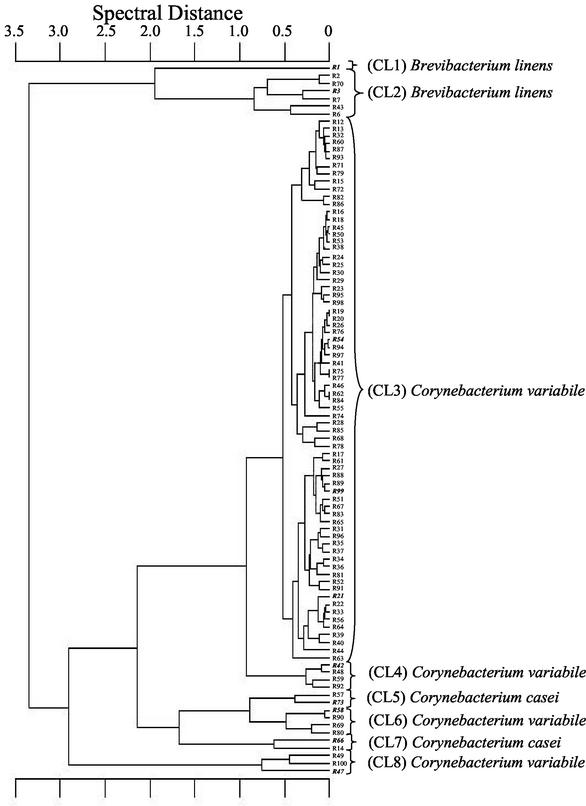
According to the results of the preliminary characterization, the total of 400 isolates was subdivided into three groups: (i) gram-positive, catalase-positive, non-spore-forming, sometimes irregular rods (378 isolates), (ii) gram-positive, catalase-negative, non-spore-forming isolates (12 coccus-shaped and 3 rod-shaped isolates), and (iii) gram-negative isolates (7 isolates) (Fig. 1). All 378 isolates of the first group, representing 94.5% of the total number isolated, were identified as coryneforms at the species level by FT-IR spectroscopy. An example, the FT-IR dendrogram (sampling A) of flora R, is displayed in Fig. 2. In this case, the identification of 94 coryneform isolates resulted in eight major FT-IR clusters. Clusters 1 and 2 are clearly separated from the others and comprise Brevibacterium linens. Clusters 3, 4, 6, and 8 include Corynebacterium variabile. Next to these, clusters 5 and 7 comprise Corynebacterium casei. The positions of these clusters correspond with the results of Brennan et al. (4), who reported a close phylogenetic relationship between the latter two species. All three species were found in more than one cluster. This phenomenon has been described by Kümmerle et al. (23) as an indicator of intraspecific heterogeneity with regard to cellular composition.
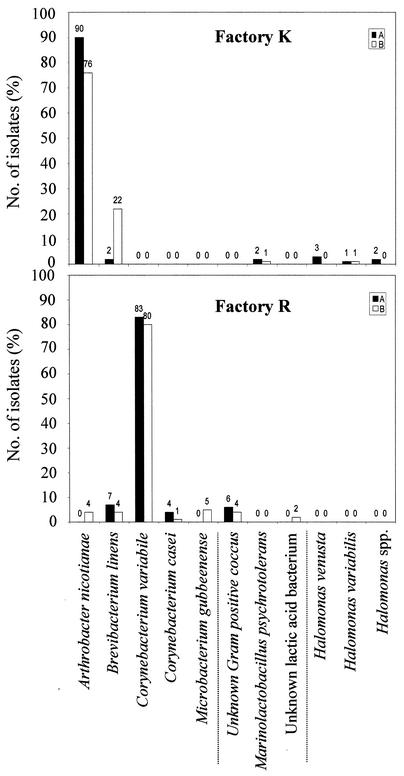
FIG. 1.
Composition of two undefined surface floras derived from German commercial mature red-smear soft cheeses produced in factories R and K. Each flora was derived at the beginning (A) and at the end (B) of a 6-month period from retail red-smear cheeses. The total cell counts of consortium R at samplings A and B were 4.0 × 109 and 3.2 × 109 CFU/g of smear, respectively. The total cell counts of consortium K at samplings A and B were 2.9 × 108 and 3.8 × 108 CFU/g of smear, respectively. The three groups (separated by vertical lines below the graphs) represent coryneform isolates (left), gram-positive and catalase-negative isolates (center), and gram-negative isolates (right).
FIG. 2.
Dendrogram of 94 coryneform bacteria from a complex red-smear surface flora (dairy R; sampling point A in Fig. 1) identified by FT-IR spectroscopy. Bold italics indicate additional 16S rDNA sequence analysis. CL, cluster.
Composition and temporal stability of consortium R.
The composition of the undefined microbial consortium derived from mature red-smear soft cheeses of factory R at the beginning (sampling point A) and at the end (sampling point B) of a 6-month period is given in Fig. 1. At both samplings, the majority of the consortium was formed by coryneform bacteria, representing 94% of the isolates. In both analyses, C. variabile was the dominant species, with almost identical proportions of 83 and 80% of the total isolates, respectively. This species has been isolated from the surface of German red-smear cheeses previously, but in much lower numbers (33, 37). The proportions of all other species ranged between 1 and 7%, with B. linens and C. casei also contributing to the coryneform subflora in the first analysis. While the general structure of the consortium appeared to be stable over 6 months, the diversity at the species level was found to be higher at the end (sampling point B) of this period, with Arthrobacter nicotianae and Microbacterium gubbeenense appearing as additional coryneform species. Although the overall predominance of coryneform bacteria is comparable, the composition as well as the structure of consortium R is completely different from the recent findings for Irish Gubbeen cheese. The latter consortium was dominated by C. casei, which represented about 50% of the isolates, while no C. variabile was isolated (5). Furthermore, those authors reported that M. gubbenense represented a high proportion of the total number of isolates. While the low incidence of microbacteria in consortium R is in contrast to these findings, it is in agreement with other studies where Microbacterium spp. also were found in low numbers only (14, 33, 37). Since no gram-negative bacteria appeared, the 12 gram-positive and catalase-negative cocci were the only noncoryneform isolates from consortium R. They formed gas neither from glucose nor from citrate, and they grew neither at 42°C, nor in the presence of 6.5% NaCl, nor on kanamycin esculin azide agar. Two isolates formed acid from lactose (data not shown). Since none of these 12 isolates could be identified by FT-IR spectroscopy, all were subjected to 16S rDNA analysis. The two lactic acid bacteria revealed 99% sequence similarity to an uncultured bacterium from feedlot manure (28), while the other 10 isolates could not be related to any species described (data not shown).
Composition and temporal stability of consortium K.
With regard to its composition and stability, consortium K differed significantly from consortium R (Fig. 1). The dominant species of consortium R was not isolated at all in consortium K. Instead, in both analyses, consortium K was clearly dominated by A. nicotianae, which had been only a minor constituent of flora R in the second analysis. This species has been isolated from red-smear surface floras previously (14, 33, 37), but none of those authors reported on a comparable predominance. After 6 months, the proportion of A. nicotianae dropped from 90 to 76% and that of B. linens increased significantly (Fig. 1). The latter species represented the only other coryneform bacterium isolated from consortium K and yielded an 11-fold-higher proportion (22%) at the end (sampling point B) of the 6-month period. These numbers mark a significant structural change for consortium K within 6 months. Additionally, small numbers of gram-positive, catalase-negative isolates and some gram-negative isolates were found in both analyses. The three gram-positive, catalase-negative isolates formed acid from lactose but were negative in all other physiological tests and could not be identified by FT-IR spectroscopy (data not shown). By 16S rDNA analysis they were identified as Marinolactobacillus psychrotolerans. This species has never been isolated from red-smear cheese or any other dairy product, but only from marine organisms in Japan (22). Due to its ability to use lactose and its halophilic character, however, it obviously found a niche in the salty smear of a German milk product, representing an interesting case of Beijerinck's law (everything is everywhere, but the environment selects). The gram-negative isolates could not be identified by biochemical tests (data not shown). Three isolates were identified as Halomonas venusta and two as Halomonas variabilis by 16S rDNA analysis. The other two appeared to be members of the genus Halomonas also but could not be identified to the species level (data not shown). To our knowledge, this is the first time that Halomonas spp. have been reported to be present in red-smear cheese-ripening consortia. The presence of these gram-negative bacteria may indicate hygienic problems in factory K. Since these species compensate for the low numbers of coryneform species of consortium K, the overall biodiversity on the species level was comparable in the two consortia, R and K.
In situ antilisterial activities of consortia R and K.
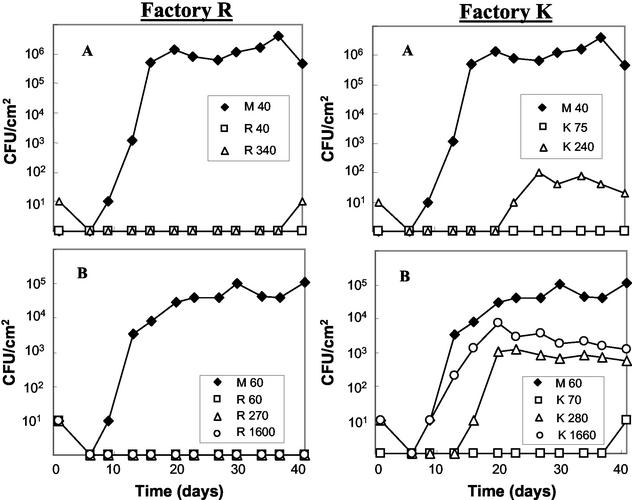
The antilisterial activity of consortium R as determined in situ on soft cheese is displayed in Fig. 3. In both experiments A and B, L. monocytogenes WSLC 1364 grew to high numbers on the control cheeses (M) from initial contamination levels of 40 and 60 CFU/ml of brine, respectively. At the same initial contamination levels, the pathogen was inhibited completely by consortium R. Except for one positive enrichment at day 41 (experiment A), it was also inhibited completely at initial contamination levels of 340 (experiment A) and 270 (experiment B) CFU/ml of brine, respectively. These results indicate a stable antilisterial activity of consortium R over 6 months. In experiment B, the antilisterial activity of consortium R was tested additionally at an initial contamination level of 1,600 CFU/ml of brine. Even then it showed a total inhibition of L. monocytogenes (Fig. 3). The high incidence of Listeria on red-smear cheeses (30) indicates that many of the industrial surface consortia exhibit no significant antilisterial activity or, alternatively, are contaminated by resistant Listeria strains (11). In our study, the contaminating L. monocytogenes strain had been propagated under optimal conditions and the contamination was performed with the first smearing. Hence, the ripening consortium had no possibility to establish itself on the cheese surface in advance. Taking these facts, as well as the high contamination levels, into account, the antilisterial potential of consortium R is extraordinarily high. Its inhibitory effect is also much stronger than that observed by Eppert et al. (16), who did not analyze the composition of the respective ripening cultures.
FIG. 3.
Growth of L. monocytogenes on soft cheese in the presence of the complex, undefined red-smear surface consortia R and K after contamination at day 1. The numbers in the key (e.g., M 40, K 75) refer to the respective consortium and to the initial number of L. monocytogenes cells per milliliter of contaminated brine at day 1. Experimental series A and B were performed with floras harvested from mature retail red-smear soft cheeses from factories R and K, with production dates 6 months apart. Flora M served as a control. It is a defined culture derived from a red-smear soft cheese from factory M.
Data on the in situ antilisterial activity of consortium K, derived at the beginning (sampling point A) and at the end (sampling point B) of the 6-month period, are depicted in Fig. 3. In both cases consortium K inhibited L. monocytogenes at initial contamination levels of 75 (sampling point A) and 70 (sampling point B) CFU/ml of brine. At contamination levels of 240 (sampling point A) and 280 (sampling point B) CFU/ml of brine, however, listerial growth could not be inhibited to the same extent. While comparison with the control consortium M still demonstrates a certain inhibitory effect, Listeria analysis yielded positive results after about 3 and 2 weeks (Fig. 3, experiments A and B, respectively). Our data demonstrate significant differences between consortia R and K, revealing a lower antilisterial potential of the latter, which decreased additionally within 6 months. Interestingly, this change in antilisterial activity was accompanied by a significant change of the species within each consortium (Fig. 1). It is possible that the increasing numbers of B. linens resulted in a loss of antilisterial activity. This observation is of importance for the food industry, since the contribution of a red-smear ripening consortium to food safety may not always be stable over time.
Antilisterial activities of individual isolates of consortium R and consortium K.
The interaction of microorganisms on red-smear cheese surfaces is unknown and, most probably, very complex. It was therefore of considerable interest to test whether the extraordinarily high inhibitory potential of consortium R and the unstable inhibitory potential of consortium K may be due to the secretion of bacteriocins, which has been demonstrated in several studies (12, 15, 16, 20, 34, 36). Therefore, the antagonistic activity against L. monocytogenes was determined in an agar diffusion-screening assay. While the control strain S. equorum WS 2733 yielded clear inhibition zones after 24, 48, and 72 h, none of the 400 isolates tested exhibited antilisterial activity as a single culture at any time point of examination. Since the activities of individual active strains might be insufficient to explain the whole inhibitory potential of undefined red-smear consortia (16), the cumulative antilisterial effect of each consortium was also examined. None of the four consortia exhibited inhibition zones. According to these results, it is unlikely that the antilisterial effects observed in situ (Fig. 3) are due to inhibitory substances of single strains or total consortia secreted into the growth medium. Instead, hitherto unknown factors must be responsible for the inhibition of Listeria in this food ecosystem.
Concluding remarks.
Temporal stability of bacterial composition and antilisterial activity appear to be individual characteristics of undefined industrial cheese-ripening consortia. The continuous carryover of microorganisms between subsequent productions due to the “old-young” smearing procedure does not necessarily lead to a significant shift in species composition within 6 months. A predominant species may represent as much as 90% of the isolates, possibly indicating a certain “backbone function” within the respective consortium, but the functionality of structured ripening consortia is independent of the genus affiliation of the dominating species. Although the antilisterial effect of such cheese-ripening consortia may not constitute a completely stable factor over time, our data clearly indicate a considerable potential of such cultures to be used for safe food production without artificial preservatives.
Acknowledgments
Many thanks to Ronny Scheundel for excellent technical assistance and to Helene Oberreuter, Mareike Wenning, and Stephanie Goerges for sharing their expertise in FT-IR spectroscopy and sequence analysis. We also thank Herbert Seiler for helpful discussions about the identification of coryneforms and lactic acid bacteria.
Danisco GmbH, Niebüll, Germany, and the European Union (grant QLTK1-CT-2001-02228) partially supported this work.
REFERENCES
- 1.Anonymous. 1995. IDF standard 143A:1995. Milk and milk products—detection of Listeria monocytogenes. International Dairy Federation, Brussels, Belgium.
- 2.Bille, J. 1989. Anatomy of a foodborne listeriosis outbreak, p.31-36. In Proceedings of the Food Listeriosis Symposium. Behr's Verlag, Hamburg, Germany.
- 3.Bockelmann, W., U. Krusch, G. Engel, N. Klijn, G. Smit, and K. J. Heller. 1997. The microflora of Tilsit cheese. Part 1. Variability of the smear flora. Nahrung 41:208-212. [Google Scholar]
- 4.Brennan, N. M., R. Brown, M. Goodfellow, A. C. Ward, T. P. Beresford, P. J. Simpson, P. F. Fox, and T. M. Cogan. 2001. Corynebacterium mooreparkense sp. nov. and Corynebacterium casei sp. nov., isolated from the surface of a smear-ripened cheese. Int. J. Syst. Evol. Microbiol. 51:843-852. [DOI] [PubMed] [Google Scholar]
- 5.Brennan, N. M., A. C. Ward, T. P. Beresford, P. F. Fox, M. Goodfellow, and T. M. Cogan. 2002. Biodiversity of the bacterial flora on the surface of a smear cheese. Appl. Environ. Microbiol. 68:820-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck, J. D. 1982. Nonstaining (KOH) method for determination of gram reactions of marine bacteria. Appl. Environ. Microbiol. 44:992-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busse, M. 1989. Die Oberflächenflora von geschmiertem Käse. Milchwissenschaft 99:137-141. [Google Scholar]
- 9.Carminati, D., E. Neviani, G. Ottogalli, and G. Giraffa. 1999. Use of surface-smear bacteria for inhibition of Listeria monocytogenes on the rind of smear cheese. Food Microbiol. 16:29-36. [Google Scholar]
- 10.Carnio, M. C. 1999. Ph.D. thesis. Technical University of Munich, Munich, Germany.
- 11.Carnio, M. C., I. Eppert, and S. Scherer. 1999. Analysis of the bacterial surface ripening flora of German and French smeared cheeses with respect to their anti-listerial potential. Int. J. Food Microbiol. 47:89-97. [DOI] [PubMed] [Google Scholar]
- 12.Carnio, M. C., A. Holtzel, M. Rudolf, T. Henle, G. Jung, and S. Scherer. 2000. The macrocyclic peptide antibiotic micrococcin P1 is secreted by the food-borne bacterium Staphylococcus equorum WS 2733 and inhibits Listeria monocytogenes on soft cheese. Appl. Environ. Microbiol. 66:2378-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Erian, A. F. M. 1972. Bacteriological studies on Limburger cheese. Neth. Milk Dairy J. 26:113-116. [Google Scholar]
- 14.Eliskases-Lechner, F., and W. Ginzinger. 1995. The bacterial flora of surface ripened cheese with special regard to coryneforms. Lait 75:571-584. [Google Scholar]
- 15.Ennahar, S., O. Assobhel, and C. Hasselmann. 1998. Inhibition of Listeria monocytogenes in a smear-surface soft cheese by Lactobacillus plantarum WHE 92, a pediocin AcH producer. J. Food Prot. 61:186-191. [DOI] [PubMed] [Google Scholar]
- 16.Eppert, I., N. Valdés-Stauber, H. Götz, M. Busse, and S. Scherer. 1997. Growth reduction of Listeria spp. caused by undefined industrial red smear cheese cultures and bacteriocin-producing Brevibacterium linens as evaluated in situ on soft cheese. Appl. Environ. Microbiol. 63:4812-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Funke, G., K. Peters, and M. Aravena-Roman. 1998. Evaluation of the RapID CB plus system for identification of coryneform bacteria and Listeria spp. J. Clin. Microbiol. 36:2439-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Funke, G., F. N. Renaud., J. Freney, and P. Riegel. 1997. Multicenter evaluation of the updated and extended API (RAPID) Coryne database 2.0. J. Clin. Microbiol. 35:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giraffa, G., and D. Carminati. 1997. Control of Listeria monocytogenes in the rind of Taleggio, a surface-smear cheese, by a bacteriocin from Enterococcus faecium 7C5. Sci. Alim. 17:383-391. [Google Scholar]
- 21.Helm, D., H. Labischinski, G. Schallehn, and D. Naumann. 1991. Classification and identification of bacteria by Fourier-transform infrared spectroscopy. J. Gen. Microbiol. 137:69-79. [DOI] [PubMed] [Google Scholar]
- 22.Ishikawa, M., K. Nakajima, M. Yanagi, Y. Yamamoto, and K. Yamasato. 2003. Marinolactobacillus psychrotolerans gen., sp. nov., a halophilic and alkaliphilic marine lactic acid bacterium isolated from marine organisms in temperate and subtropical areas of Japan. Int. J. Syst. Evol. Microbiol. 711-720. 53: [DOI] [PubMed]
- 23.Kümmerle, M., S. Scherer, and H. Seiler. 1998. Rapid and reliable identification of food-borne yeasts by Fourier-transform infrared spectroscopy. Appl. Environ. Microbiol. 64:2207-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindenmann, K., A. von Graeventitz, and G. Funke. 1995. Evaluation of the Biolog system for the identification of asporogenous, aerobic gram-positive rods. Med. Microbiol. Lett. 4:287-296. [Google Scholar]
- 25.Naumann, D., D. Helm, and H. Labischinski. 1991. Microbiological characterizations by FT-IR spectroscopy. Nature 351:81-82. [DOI] [PubMed] [Google Scholar]
- 26.Oberreuter, H., J. Charzinski, and S. Scherer. 2002. Intraspecific diversity of Brevibacterium linens, Corynebacterium glutamicum and Rhodococcus erythropolis based on partial 16S rDNA sequence analysis and Fourier-transform infrared (FT-IR) spectroscopy. Microbiology 148:1523-1532. [DOI] [PubMed] [Google Scholar]
- 27.Oberreuter, H., H. Seiler, and S. Scherer. 2002. Identification of coryneform bacteria and related taxa by Fourier-transform infrared (FT-IR) spectroscopy. Int. J. Syst. Evol. Microbiol. 52:91-100. [DOI] [PubMed] [Google Scholar]
- 28.Ouwerkerk, D., and A. V. Klieve. 2001. Bacterial diversity within feedlot manure. Anaerobe 7:59-66. [Google Scholar]
- 29.Reps, A. 1987. Bacterial surface ripened cheeses, p. 151-184. In P. F. Fox (ed.), Cheese: chemistry, physics and microbiology, 1st ed., vol. 2. Elsevier Applied Science, London, United Kingdom.
- 30.Rudolf, M., and S. Scherer. 2001. High incidence of Listeria monocytogenes in European red smear cheese. Int. J. Food Microbiol. 63:91-98. [DOI] [PubMed] [Google Scholar]
- 31.Ryser, E. T., S. Maisnier-Patin, J. J. Gratadoux, and J. Richard. 1994. Isolation and identification of cheese-smear bacteria inhibitory to Listeria spp. Int. J. Food Microbiol. 21:237-246. [DOI] [PubMed] [Google Scholar]
- 32.Seiler, H. 1983. Identification key for coryneform bacteria derived by numerical taxonomic studies. J. Gen. Microbiol. 129:1433-1471. [DOI] [PubMed] [Google Scholar]
- 33.Seiler, H. 1986. Identification of cheese-smear coryneform bacteria. J. Dairy Res. 53:439-449. [Google Scholar]
- 34.Sulzer, G., and M. Busse. 1991. Growth inhibition of Listeria spp. on Camembert cheese by bacteria producing inhibitory substances. Int. J. Food Microbiol. 14:287-296. [DOI] [PubMed] [Google Scholar]
- 35.Valdés-Stauber, N., H. Götz, and M. Busse. 1991. Antagonistic effect of coryneform bacteria from red smear cheese against Listeria species. Int. J. Food Microbiol. 13:119-130. [DOI] [PubMed] [Google Scholar]
- 36.Valdés-Stauber, N., and S. Scherer. 1994. Isolation and characterization of Linocin M18, a bacteriocin produced by Brevibacterium linens. Appl. Environ. Microbiol. 60:3809-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valdés-Stauber, N., S. Scherer, and H. Seiler. 1997. Identification of yeasts and coryneform bacteria from the surface microflora of brick cheeses. Int. J. Food Microbiol. 34:115-129. [DOI] [PubMed] [Google Scholar]