The gram-negative bacterial species Salmonella enterica and Escherichia coli are members of the family Enterobacteriaceae that spend a good part of their lives as residents of animal hosts. S. enterica is the etiologic agent of gastroenteritis and typhoid fever in humans (88), whereas E. coli is most commonly known as a commensal of the lower intestine of mammals, although pathogenic variants also exist (77). The animal host is believed to be the primary habitat of these two enteric species (86), which are genetically endowed to do well in this environment. For example, Salmonella has genes that mediate invasion of and survival within host cells, including genes that promote resistance to different microbicidal host products (62, 88). Likewise, the E. coli genome encodes proteins that mediate resistance to acid pH as well as growth on lactose, which is critical for a commensal of mammals (11, 57).
In this review, we discuss whether Salmonella and E. coli live in stable, dividing populations in nonhost environments and whether such environments constitute dead ends for these species (e.g., as a consequence of residing in the vertebrate lower intestine, whose contents are regularly excreted [86, 102]). In addition, we examine the role that genes specific to Salmonella and E. coli play in the different abilities of these species to proliferate outside animal hosts.
DO SALMONELLA AND E. COLI EXIST IN A VBNC STATE?
The host environment provides Salmonella and E. coli with a warm constant temperature, as well as high concentrations of free amino acids and sugars, which are conducive to bacterial growth (86). Once excreted from an animal host, Salmonella and E. coli find themselves battling for survival, facing limited nutrient availability, osmotic stress, large variations in temperature and pH, and predation (61, 83, 86). It has been proposed that bacteria can survive such stressful conditions by entering a viable but nonculturable (VBNC) state.
The VBNC hypothesis describes an apparent dormant state in which bacterial cells are metabolically active but cannot be cultured by known laboratory methods (78). It has been suggested that bacteria enter this state in response to harsh environmental conditions, such as a temperature change, high salinity, or nutrient deprivation (78, 82, 93). The controversy surrounding the VBNC concept involves whether bacterial cells can be resuscitated out of the dormant state and can start dividing in the external environment, which in the case of human enteric bacteria may constitute a public health concern.
Nutrient addition and temperature upshift are reportedly effective methods for rescuing bacterial populations from the VBNC state (78, 82, 93), but they may simply support the growth of a small number of culturable cells that survive the stress conditions and are not detected because of limitations of laboratory cultivation methods (13, 78). Thus, the apparent resuscitation of nonculturable cells may result from dispersion of clumps of culturable cells or from the response of undetected culturable cells to the new growth conditions, either of which would give the illusion of recovery (12). If one accepts that viability means “the ability of a single cell to attain a population discernible by the observer” (15), then the use of staining techniques to determine cell viability may not provide an accurate measurement of whether cells are alive (12); if this is true, few studies have actually detected viable cells emerging from the VBNC state.
Raw sewage is often discharged into low-temperature marine and aquatic environments, raising the question of whether human enteric pathogens (i.e., Salmonella) and indicator organisms (i.e., E. coli) are able to adapt to and persist in these extreme environments. Exposure of Salmonella and E. coli to polar marine conditions in Antarctica for 54 to 56 days altered their physiology to a state characteristic of VBNC cells. Although no viable colonies were observed after 54 days, the level of bacterial respiration was 90% of the initial levels after nutrient addition at day 46 (93). A Salmonella environmental isolate from the Potomac River in Washington, D.C., was found to be in the VBNC state following dilution into a laboratory microcosm containing sterilized Potomac River water. While the metabolic activity remained constant, the bacterial count decreased to zero after 4 days and resuscitation by nutrient addition to the river water on day 21 was unsuccessful (82), indicating that the cells may not have been viable.
Proponents of the VBNC hypothesis have suggested that bacterial species, particularly pathogens, pose a health risk when they are present in a dormant state in the environment because of the likelihood of resuscitation upon access to an animal host (78, 82, 93). However, evidence against recovery of VBNC cells brings into question the premise that dormant pathogens are capable of causing disease. For example, seawater and UV-C radiation induce in Salmonella the VBNC state, which is characterized by a lack of culturability, 95% respiratory activity, and unrecoverable virulence in mice (16). Likewise, VBNC S. enterica serovar Oranienburg detected in dried processed squid was avirulent in BALB/c mice, and virulence was detected only in mice treated with morphine (4), which suppresses the mouse immune system. This raises the possibility that the virulence phenotype observed in morphine-treated mice is not due to resuscitation from the vegetative state but rather is the result of a bacterial product, such as lipopolysaccharide, which is present in VBNC organisms.
Enteropathogenic E. coli exposed to seawater and sunlight for 26 h (i.e., unculturable, yet metabolically active) was positive for enterotoxin in a rabbit intestinal loop assay (76). Nonetheless, the presence of enterotoxin does not provide strong evidence for the VBNC hypothesis, given that a toxin may remain stable in otherwise dead bacterial cells. Moreover, if VBNC pathogens do pose a health risk, one would expect to find a greater occurrence of outbreaks of waterborne bacterial diseases in developed nations (13). Recovery from a VBNC state occurs rarely, if at all, and long-term contamination appears to result from regular deposition of bacteria from human and animal sources, which may be the case in developing countries.
E. COLI HAS A LOW RATE OF SURVIVAL OUTSIDE ANIMAL HOSTS
E. coli resides in the lower intestine of warm-blooded animals (92), an environment that provides a vast supply of nutrients for bacterial growth (22, 71, 86, 87). The doubling time of E. coli in this primary habitat has been estimated to be 2 days (86). It has been suggested that one-half of the total E. coli population resides in the primary habitat of the host and one-half is in the external environment (i.e., the secondary habitat) (86). According to this notion, E. coli grows and divides in its primary habitat, but it has a net negative rate of growth in the secondary habitat, with half-lives of approximately 1 day in water (29), 1.5 days in sediment (35), and 3 days in soil (101). These estimates imply that E. coli does not live in nonhost environments but that continuous bulk transfer from human and animal sources maintains a stable population outside animal hosts (86).
The genetic diversity of the E. coli population in individual hosts has been attributed to turnover of both resident strains (i.e., strains that persist for months to years) and transient strains (i.e., strains that persist for days to weeks) as a result of ingestion of contaminated food or water (19, 91). Hosts typically acquire E. coli during birth (9) and subsequently maintain one or two resident strains and three or four transient strains, all of which appear to change over time (2, 18, 19, 55, 89, 90). However, the mechanism of turnover of the host intestinal flora in the absence of antibiotic treatment remains unclear, as experimental implantation of new strains into healthy subjects is difficult (33, 34, 89, 90). Furthermore, the vast majority of ingested strains fail to colonize, as reflected by the relatively constant intestinal composition (90). Indeed, the cycle time of E. coli through a host has been estimated to range from 26 h to 66 years (85).
Survival of E. coli in the secondary habitat requires the ability to overcome low nutrient availability and wide temperature fluctuations. E. coli populations decline rapidly in freshwater microcosms (47, 59); however, addition of readily usable carbon sources, as well as removal of competitor microflora, allows growth, suggesting that E. coli is unable to acquire and compete for nutrients under starvation conditions (59).
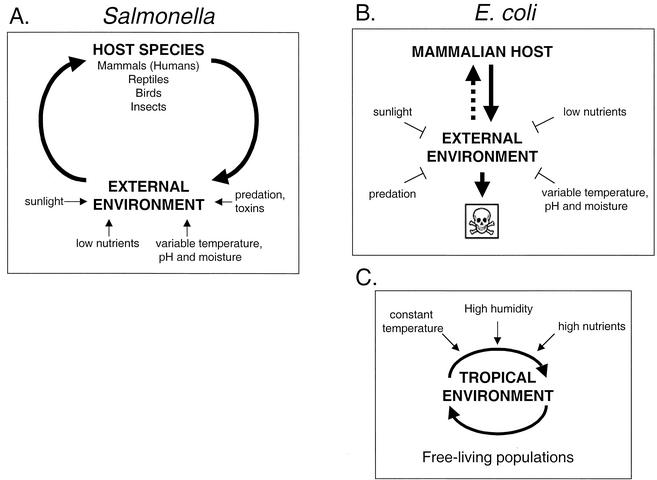
The E. coli populations found in the secondary habitat are apparently maintained by constant arrival of microorganisms from the primary habitat (86), as a lack of nutrients and harsh ecological conditions prevent E. coli from sustaining a dividing population outside the animal host. This model implies that E. coli does not live in nonhost environments and that its presence in such locations results from excretion of waste by animal hosts. This is the logic behind the use of E. coli as the indicator organism for environmental fecal contamination (i.e., as an indicator species, E. coli is assumed not to be a permanent resident of soil and water) (49). Cumulatively, the results of experiments suggest that there is a low probability that E. coli will colonize a new host and that this is largely due to its relatively low survival rate in nonhost environments (Fig. 1B).
FIG. 1.
Life cycles of Salmonella and E. coli. (A) Salmonella actively cycles through host and nonhost environments. (B) E. coli has a low rate of survival in nonhost environments and a low probability of colonizing a new host. (C) Free-living E. coli populations exist in tropical environments in the absence of human or animal contamination.
However, are there nonhost environments that mimic the physical and chemical milieu of the colon? If so, do E. coli populations live in such ecological niches as members of the natural flora?
E. COLI LIVES IN TROPICAL FRESHWATER AND SOIL
As in the mammalian host environment, nutrients in tropical ecosystems are maintained at high concentrations, and together with constant warm air, soil, and water temperatures, this provides an ideal habitat for survival, growth, and proliferation of E. coli (Fig. 1C). Indeed, high concentrations of E. coli have been found in numerous tropical locations in the absence of known fecal sources (49). For example, the waters of Puerto Rico are highly contaminated with human waste, yet at sampling sites upstream of known sewage drainage points there are large numbers of fecal coliforms, similar to the numbers at sites downstream (49). Likewise, the number of viable E. coli cells in the upper one-third of the Mameyes River in Puerto Rico (in a cloud rain forest) shows a strong positive correlation with water temperature and nutrient concentrations (17). The total bacterial count found at this unspoiled tropical location exceeds by twofold the total count reported for the polluted, temperate Anacostia River in Washington, D.C. High respiration rates and extended survival suggest that tropical waters harbor natural populations of E. coli (17). However, studies performed with a pure culture of an E. coli strain that was not isolated from the Mameyes River showed that the size of the population decreased by 90% after 12 days, raising the possibility that a natural isolate in the region may be genetically adapted for increased survival in this environment (49). In spite of this, the survival time and the percentage of physiologically active cells (55 to 90%) suggest that E. coli is able to grow in a tropical freshwater environment (49, 60).
Water sources in uninhabited rain forests provide elevated concentrations of nutrients for thriving microbial communities in the absence of human pollution (81). Epiphyte species living in trees 15 m above the ground, such as bromeliads, form large cups with their leaves, which collect rainwater and runoff, thus serving as microcosms that are rich in nutrients. For example, bromeliad water, which can contain up to 50 times the typical concentration of nitrites and nitrates, was shown to contain an average of 1.5 × 106 coliforms per 100 ml, 51.6% of which were undergoing respiration and 49.1% of which were engaged in protein synthesis. E. coli comprises 72% of the total bacterial species living in bromeliad water microcosms. The presence of active E. coli populations in bromeliad water at different forest elevations throughout the year suggests that these populations did not originate from the small population of Puerto Rican birds or tree-climbing mammals (81).
Natural E. coli populations are not limited to tropical freshwater. The soil of the North Fork of the New River in Ft. Lauderdale, Fla., is a major source of E. coli (97). This subtropical riverbank environment is characterized by warm and humid conditions with cyclic periods of wet and dry weather, which are conducive to E. coli growth (26, 97). While the presence of E. coli in this soil correlates with storm sewer flooding due to heavy rainfall, high bacterial levels have also been detected 2 days following flooding, implying that E. coli is capable of multiplying in bank soil, perhaps as a member of the natural flora (97). In summary, the studies described above support the notion that in the nutrient-rich tropics, E. coli is a member of the natural microflora (81).
SALMONELLA PASSAGE BETWEEN HOST SPECIES
Salmonella has been isolated from a large number of animal species (88). The majority of the 1.3 billion annual cases of Salmonella-caused human gastroenteritis result from ingestion of contaminated food products, such as undercooked beef, pork, chicken, seafood, and eggs (72). Salmonella infections are also contracted following consumption of fresh fruits or vegetables that have been contaminated by infected fertilizer (100). Furthermore, as several reptile species are known Salmonella reservoirs (53, 54), reptilian pets have served as a source of Salmonella for infected owners (84). This indicates that Salmonella species lack special host adaptations and are capable of colonizing a wide variety of macroorganisms (32). The ubiquitous nature of Salmonella may facilitate a cyclic lifestyle consisting of passage through a host into the environment and back into a new host (104). Thus, unlike the commensal E. coli, which does not appear to survive in nonhost environments for extended periods of time in climatic regions other than the tropics, long-term survival of Salmonella in the secondary habitat ensures its passage to the next host (Fig. 1A).
The health and economic consequences associated with Salmonella contamination of food products have led to numerous studies of the survival and transmission of these bacteria in the poultry, pig, and cow industries (see references 41 and 106 for reviews). For example, Salmonella has been detected in several locations within cow farms, pig farms, and slaughterhouses, both before and after sacrifice (5, 48, 65); the same Salmonella clone has been recovered in a cow herd and in ground meat products following processing (65). Investigation of a piggery in Denmark revealed the presence of Salmonella in samples from the animals, their environment, and their feed collected over a 2-year period (5). Again, all isolates originated from a single clone, suggesting that the same population persisted in the farmhouse environment. Waste slurry from infected pigs, which is used as an agricultural fertilizer, remained positive for culturable Salmonella up to 21 days after it was spread onto farmland outside the contaminated area (5). These studies indicate that Salmonella was continually introduced into the pig population through a cycle of animal shedding and short-term survival in the farm environment rather than from an external source (5). Long-term contamination of farms appears to be a widespread phenomenon as Salmonella persistence has been documented for more than 1 year in a poultry house, despite disinfections (24).
Birds and flies are important vectors for rapid, widespread dissemination of Salmonella in the environment (25). Infection of wild bird populations with salmonellae has been correlated with proximity to farms and/or the incidence of human salmonellosis in the same area (66). Likewise, adult muscoid flies (including the common housefly, Musca domestica) can carry Salmonella, particularly on dairy farms (infection rate, 67%) and poultry farms (infection rate, 13%). Salmonella survives in flies for up to 4 weeks, which is the life span of the flies (64). Salmonella has been observed to survive in infected blowfly pupae for 18 days at 5°C (52), despite physicochemical conditions that are dramatically different from those that exist in vertebrate hosts. Moreover, germfree adult houseflies inoculated with Salmonella show total defecation outputs of as much as 107 CFU (37). Thus, flies that come in contact with contaminated materials (i.e., manure, food, and water) are capable of transmitting Salmonella (64).
Rapid dissemination of Salmonella between hosts also occurs by passage of bacteria from infected farm animals to vegetables as a result of field fertilization with raw, contaminated manure. Experimental inoculation of manure has revealed seasonal patterns of Salmonella survival in fertilized soil and transmission to crops (70). Detection of Salmonella on vegetables is correlated with warm, moist summer conditions (average daily temperature, >20°C), as no vegetable contamination was observed when manure was applied during the late fall (average daily temperature, <10°C) or following repeated freezing-thawing cycles (70). Contact of Salmonella with the flowers, stems, or fruits of tomato plants leads to infiltration and colonization of plant tissues (42, 43), and tomato plants grown hydroponically have been shown to take up Salmonella (44). In summary, the high nutrient content of sewage results in a good fertilizer, but the ability of Salmonella to survive in the fertilized soil may help perpetuate the presence of Salmonella species outside animal hosts (79).
Transmission of Salmonella between hosts is not limited to natural nonhost environments, as the presence of bacteria on household surfaces has important health consequences. A striking study of the survival of Salmonella in bathrooms and toilets following bacterium-induced illness demonstrated that there were high contamination levels for up to several weeks (7). Persistence was enhanced by adhesion and biofilm formation. Toilet seeding experiments designed to mimic conditions of acute diarrhea resulted in isolation of Salmonella from air samples immediately following flushing and in toilet bowl biofilm samples for up to 50 days (7). Inhalation has been found to be a potential alternate route of Salmonella infection in pigs, given their rooting behavior (30). The presence of aerosolized salmonellae in bathrooms raises the possibility that inhalation is an alternative route of human infection.
SALMONELLA SURVIVES IN WATER AND SOIL
Salmonella is frequently isolated from water sources (21), which serve as bacterial reservoirs and may aid transmission between hosts (32). Like E. coli, Salmonella is constantly released into the environment from infected humans, farm animals, pets, and wildlife (8). Despite efforts to contain and sanitize human waste, Salmonella can survive for 10 to 15 days in a septic system (73), whereas E. coli has a negative growth rate in this environment (36). Seepage from septic tanks and sewage injection well fields, as well as sewage and storm runoff, facilitates bacterial passage into surface waters (8, 74). Detection of Salmonella correlates with proximity to the sewage discharge area (3, 8). For example, Salmonella was isolated from 65% of the water samples collected along the Peavine Creek in Decatur, Ga., which flows through urban industrial and residential areas (21). In addition, Salmonella was detected within 350 feet of the subsurface origin of a small stream in the same area, suggesting that contamination occurs rapidly once water emerges from the earth (21).
Compared to other bacteria, Salmonella has high survival rates in aquatic environments (20); it outlives both Staphylococcus aureus and the waterborne Vibrio cholerae in groundwater and in heavily eutrophied river water (27). The presence of Salmonella in marine environments does not vary seasonally and is independent of water temperature (3). Furthermore, Salmonella has a high survival rate following mixing of sewage effluent with brackish water, a transition that causes a dramatic increase in salinity (63), whereas osmotic stress has been implicated in the apparent death of E. coli in seawater (13). Thus, compared to E. coli, Salmonella appears to withstand a wider variety of stresses associated with environmental fluctuations and may persist in water environments for some time.
Salmonella can be widely disseminated in soil and sediment, even in the absence of active fertilization, as a result of water currents, underground springs, and rain runoff carrying contaminated material (1, 20). Percolation of wastewater through soil filters out bacteria that become trapped in this environment (20). S. enterica serovar Adelaide exhibited extended longevity in moist or saturated sand proximal to underground septic tanks (73). Salmonella has been detected frequently in environmental soil samples collected from both agricultural and recreational areas (1, 103-105). In contrast to E. coli, which has a low survival rate in soil (i.e., an average half-life of 3 days) (13, 101), Salmonella can survive and multiply for at least 1 year in this ecosystem (25, 104).
Soil and sediment particles are believed to function as microecological niches in which bacterial species can survive and perhaps replicate (14). Association with soil particles can provide bacteria with both high concentrations of nutrients, due to the release of organic molecules from attached algal cells, and protection against predation, by providing shelter against grazing protozoans (31). Adhesion of Salmonella cells to soil particles correlates with cell surface hydrophobicity (99), which is typically manifested by modification of the bacterial outer membrane in response to changes in environmental conditions. The adhesive properties of Salmonella, as well as the nutrient-rich growth conditions provided by association with soil, may account for the fact that nearly 100% of the Salmonella-positive samples taken downstream of a sewage treatment plant over a 1-year period were samples obtained from the bottom sediment rather than samples obtained from the surface water (45). Salmonella has exhibited fourfold-greater adherence to some mineral particles than E. coli (99), although E. coli can survive in estuarine sediment environments (14). The ubiquitous nature and high rate of survival of Salmonella in soil and sediment may represent a critical adaptation to its cyclic lifestyle involving host and nonhost environments.
IS E. COLI A VALID INDICATOR SPECIES FOR SALMONELLA?
E. coli is typically used as the indicator species for environmental fecal contamination. However, this species serves as a relevant indicator only if its survival rate in a given environment is equal to or slightly higher than that of the bacterium of interest. Because the majority of nonhost environments appear to be dead ends for E. coli but a means to a new host for Salmonella, this premise may not always be valid. For example, Salmonella survives longer than E. coli in many nonhost environments, such as a brackish lagoon following discharge from a sewage treatment plant stabilization pond (63) and during exposure to polar aquatic conditions (93). Likewise, Salmonella was more resistant to killing by biotic factors (microbial predators or competing organisms) than E. coli in three drinking water sources in Sierra Leone (108), perhaps reflecting a difference in adhesion to protective particles. The rate of survival of Salmonella in estuarine water at temperatures below 10°C is significantly higher than that of E. coli (80). Furthermore, Salmonella grows better than E. coli on alfalfa sprouts, which are frequently irrigated; this may be due to the 1,000-fold-higher level of attachment of Salmonella to vegetation (6). In summary, the different rates of survival of Salmonella and E. coli in nonhost environments suggest that E. coli may not be an appropriate indicator of Salmonella contamination.
SPECIES-SPECIFIC GENES MEDIATING SURVIVAL IN NONHOST ENVIRONMENTS
A pairwise comparison of the Salmonella and E. coli genomes revealed that more than 1,100 Salmonella genes are absent from the E. coli genome and more than 800 E. coli genes are not present in the Salmonella genome (62). Furthermore, there are more than 1,000 genes in both genomes whose functions remain unknown and which could contribute to proliferation in nonhost environments, such as soil and water. While a comprehensive genomic analysis of these two organisms is beyond the scope of this paper, below we discuss a few examples of genes that may contribute to the different abilities of Salmonella and E. coli to survive in nonhost environments.
The majority of nonhost environments are characterized by thermal variability, high osmolarity, pH fluctuations, and low nutrient availability, suggesting that a stress response is activated in Salmonella and E. coli in such environments. One regulatory protein involved in such a response is the stationary-phase alternative sigma factor RpoS (28, 56), which is expressed when Salmonella and E. coli are present in seawater (69). There was a 1,000-fold-greater decrease in the number of culturable cells for an E. coli strain lacking the rpoS gene than for the isogenic wild-type strain following 8 days of exposure to seawater (69). In contrast, inactivation of the rpoS gene decreased the number of culturable Salmonella cells only 10-fold (69), suggesting that Salmonella may employ additional genetic mechanisms to respond to stresses in this environment.
Salmonella harbors several genes that, although absent from related enteric species, are not required for virulence, suggesting that they may be involved in survival in nonhost environments (Table 1). These genes encode products involved in nutrient acquisition and utilization, motility, and transcriptional regulation, as well as genes having unknown functions. For example, Salmonella has a periplasmic d-Ala-d-Ala dipeptidase, termed PcgL, that allows growth on d-Ala-d-Ala (46), which originates during peptidoglycan remodeling and is also released from the cell walls of dead bacteria. Inactivation of the pcgL gene renders Salmonella hypervirulent in mice but defective for survival in nutrient-poor conditions (68), such as those that it might encounter outside an animal host. This has led to the hypothesis that despite the virulence-attenuating effects of the pcgL gene, Salmonella has retained this gene because of the contribution that it makes to fitness in nonhost environments (68).
TABLE 1.
Salmonella-specific genes not implicated in virulence
| Gene | Putative function | Reference(s) |
|---|---|---|
| cob | Vitamin B12 biosynthesis | 39, 62 |
| fljA | Repressor of phase 1 flagellin gene | 39, 62 |
| fljB | Flagellar subunit | 39, 62 |
| iicA | Unknown | 75 |
| iviXXII | Unknown | 23 |
| iviXVII | Unknown | 23 |
| iviXIX | Unknown | 23 |
| mgtB | Magnesium transport | 96 |
| nanH | Sialidase | 39, 62 |
| pcgL | d-Ala-d-Ala dipeptidase | 46, 68 |
| phoN | Phosphatase | 40, 51 |
| sinR | Transcriptional regulator | 39, 62 |
The E. coli genome also encodes a d-Ala-d-Ala dipeptidase, termed DdpX, that localizes to the cytoplasm (58). Although the PcgL and DdpX proteins exhibit a low level of amino acid identity (40%), they have almost identical enzymatic activities (46, 58). However, these proteins differ in their sites of action; the Salmonella d-Ala-d-Ala dipeptidase encounters its substrate in the periplasm (46), whereas the E. coli enzyme encounters its substrate in the cytoplasm following uptake by a d-Ala-d-Ala transporter encoded in the same operon as the ddpX gene (58). This suggests that Salmonella and E. coli evolved different solutions to the same problem: the utilization of peptidoglycan-derived fragments in nutrient-limited environments (46, 58, 68). Transcription of the Salmonella pcgL gene is regulated by the PhoP-PhoQ two-component system, which is activated in response to low-magnesium conditions (98), and does not require the RpoS protein. On the other hand, expression of the E. coli ddpX gene is regulated by RpoS under nutrient-rich conditions (58) and by the NtrC-NtrB two-component system in nitrogen-poor environments (109). The differences in regulation of the Salmonella and E. coli d-Ala-d-Ala dipeptidases imply that these enzymes are used in different environments.
The phoN gene of Salmonella encodes a periplasmic nonspecific acid phosphatase (40, 51) that exhibits sequence similarity to an acid phosphatase from Citrobacter sp. strain N14, which has been implicated in detoxification of heavy metals that accumulate in metal-contaminated soil (67). E. coli lacks the phoN gene but contains a periplasmic alkaline phosphatase (termed PhoA), which is absent from Salmonella (62). Like the d-Ala-d-Ala dipeptidases described above, the phosphatases of E. coli and Salmonella are regulated differently: the E. coli phoA gene is transcriptionally induced under phosphate starvation conditions by the PhoB-PhoR regulatory system (107), whereas the Salmonella phoN gene is transcriptionally regulated by the PhoP-PhoQ system (38, 51). Together with the different pH optima, this suggests that Salmonella and E. coli use their periplasmic phosphatases in different milieus.
Mg2+ is the most abundant divalent cation in living organisms, and bacteria are equipped with systems that sense and transport Mg2+ (94). Both Salmonella and E. coli contain the constitutive Mg2+ transporter CorA (50), the low-Mg2+-induced Mg2+ transporter MgtA (11, 62, 95), and the Mg2+-responding PhoP-PhoQ two-component regulatory system (38). On the other hand, Salmonella, but not E. coli, contains the mgtCB operon, which encodes a third Mg2+ transporter termed MgtB (95, 96) and the MgtC membrane protein, which is required for normal growth under Mg2+-limiting conditions (10). Salmonella can grow in the presence of lower Mg2+ concentrations than E. coli, and introduction of the mgtC gene is sufficient to enhance E. coli growth under Mg2+-limiting conditions (10). In contrast to the mgtC gene, the mgtB gene is not required for virulence, suggesting that it may enhance Salmonella survival in nonhost environments.
CONCLUSIONS
E. coli populations appear to exist in two distinct states outside the host. On the one hand, E. coli cells that are shed from a mammalian host find themselves at a dead end in the external environment because of their inability to survive environmental biotic and abiotic antagonists and because of the low probability of replacing the resident strain of another host (33, 34, 86, 89, 90). On the other hand, E. coli is a member of the normal flora in certain tropical ecosystems, possibly due to the high nutrient concentrations, constant temperatures, and moist conditions of the environments (49), which mimic the mammalian colon. At this time it is not known how similar the E. coli strains that live in our guts are to the strains that live in tropical ecosystems.
One way in which Salmonella differs from E. coli is that its enhanced survival in the external environment promotes transmission to a new host. Detection of Salmonella in the environment correlates with human or animal activity. Because Salmonella is able to infect a large number of animal species, including flies, identification of the source of environmental contamination may not always be possible. Nevertheless, survival of Salmonella in the soil, in water, and on a variety of surfaces provides the bacterium with an increased probability of infecting a new host.
The differences between Salmonella and E. coli in terms of survival outside animal hosts result from genes that are specific to each of these species. However, other genetic mechanisms, such as disparate regulation and/or allelic differences between homologous genes, may also contribute to the distinct lifestyles of these enteric microorganisms.
Acknowledgments
We thank J. Bijlsma, Y. Shotland, and C. Mouslim for comments on the manuscript.
Our research is supported in part by grants AI42236 and AI49561 from the National Institutes of Health to E.A.G., who is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Abdel-Monem, M. H. A. A., and A. Dowidar. 1990. Recoveries of Salmonella from soil in eastern region of Saudi Arabia Kingdom. J. Egypt. Public Health Assoc. 65:61-75. [PubMed] [Google Scholar]
- 2.Adlerberth, I., F. Jalil, B. Carlsson, L. Mellander, L. A. Hanson, P. Larsson, K. Khalil, and A. E. Wold. 1998. High turnover rate of Escherichia coli strains in the intestinal flora of infants in Pakistan. Epidemiol. Infect. 121:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso, J. L., M. S. Botella, I. Amoros, and A. Rambach. 1992. Salmonella detection in marine waters using a short standard method. Water Res. 26:973-978. [Google Scholar]
- 4.Asakura, H., M. Watarai, T. Shirahata, and S. Makino. 2002. Viable but nonculturable Salmonella species recovery and systemic infection in morphine-treated mice. J. Infect. Dis. 186:1526-1529. [DOI] [PubMed] [Google Scholar]
- 5.Baloda, S. B., L. Christensen, and S. Trajcevska. 2001. Persistence of a Salmonella enterica serovar Typhimurium DT12 clone in a piggery and in agricultural soil amended with Salmonella-contaminated slurry. Appl. Environ. Microbiol. 67:2859-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barak, J. D., L. C. Whitehand, and A. O. Charkowski. 2002. Differences in attachment of Salmonella enterica serovars and Escherichia coli O157:H7 to alfalfa sprouts. Appl. Environ. Microbiol. 68:4758-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barker, J., and S. F. Bloomfield. 2000. Survival of Salmonella in bathrooms and toilets in domestic homes following salmonellosis. J. Appl. Microbiol. 89:137-144. [DOI] [PubMed] [Google Scholar]
- 8.Baudart, J., K. Lemarchand, A. Brisabois, and P. Lebaron. 2000. Diversity of Salmonella strains isolated from the aquatic environment as determined by serotyping and amplification of the ribosomal DNA spacer regions. Appl. Environ. Microbiol. 66:1544-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bettelheim, K. A. 1997. Escherichia coli in the normal flora of humans and animals, p. 85-109. In M. Sussman (ed.), Escherichia coli: mechanisms of virulence. Cambridge University Press, Cambridge, United Kingdom.
- 10.Blanc-Potard, A. B., and E. A. Groisman. 1997. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 16:5376-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 12.Bogosian, G., P. J. Morris, and J. P. O'Neil. 1998. A mixed culture recovery method indicates that enteric bacteria do not enter the viable but nonculturable state. Appl. Environ. Microbiol. 64:1736-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bogosian, G., L. E. Sammons, P. J. Morris, J. P. O'Neil, M. A. Heitkamp, and D. B. Weber. 1996. Death of the Escherichia coli K-12 strain W3110 in soil and water. Appl. Environ. Microbiol. 62:4114-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brettar, I., and M. G. Hofle. 1992. Influence of ecosystematic factors on survival of Escherichia coli after large-scale release into lake water mesocosms. Appl. Environ. Microbiol. 58:2201-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Button, D. K., F. Schut, P. Quang, R. Martin, and B. Robertson. 1993. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl. Environ. Microbiol. 59:881-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caro, A., P. Got, J. Lesne, S. Binard, and B. Baleux. 1999. Viability and virulence of experimentally stressed nonculturable Salmonella typhimurium. Appl. Environ. Microbiol. 65:3229-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrillo, M., E. Estrada, and T. C. Hazen. 1985. Survival and enumeration of the fecal indicators Bifidobacterium adolescentis and Escherichia coli in a tropical rain forest watershed. Appl. Environ. Microbiol. 50:468-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caugant, D. A., B. R. Levin, and R. K. Selander. 1984. Distribution of multilocus genotypes of Escherichia coli within and between host families. J. Hyg. Camb. 92:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caugant, D. A., B. R. Levin, and R. K. Selander. 1981. Genetic diversity and temporal variation in the E. coli population of a human host. Genetics 98:467-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chao, W., R. Ding, and R. Chen. 1987. Survival of pathogenic bacteria in environmental microcosms. Chinese J. Microbial Immunol. (Taipei) 20:339-348. [PubMed]
- 21.Cherry, W. B., J. B. Hanks, B. M. Thomason, A. M. Murlin, J. W. Biddle, and J. M. Croom. 1972. Salmonellae as an index of pollution of surface waters. Appl. Microbiol. 24:334-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung, Y. C., Y. S. Kim, A. Shadchehr, A. Garrido, I. L. Macgregor, and M. H. Sleisenger. 1979. Protein digestion and absorption in human small intestine. Gastroenterology 76:1415-1421. [PubMed] [Google Scholar]
- 23.Conner, C. P., D. M. Heithoff, S. M. Julio, R. L. Sinsheimer, and M. J. Mahan. 1998. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc. Natl. Acad. Sci. USA 95:4641-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies, R. H., and C. Wray. 1995. Observations on disinfection regimens used on Salmonella enteritidis infected poultry units. Poultry Sci. 74:638-647. [DOI] [PubMed] [Google Scholar]
- 25.Davies, R. H., and C. Wray. 1996. Seasonal variations in the isolation of Salmonella typhimurium, Salmonella enteritidis, Bacillus cereus and Clostridium perfringens from environmental samples. J. Vet. Med. Ser. B 43:119-127. [DOI] [PubMed] [Google Scholar]
- 26.Desmarais, T. R., H. M. Solo-Gabriele, and C. J. Palmer. 2002. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl. Environ. Microbiol. 68:1165-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiRita, V. J. 2001. Molecular basis of Vibrio cholerae pathogenesis, p. 457-508. In E. A. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, San Diego, Calif.
- 28.Fang, F. C., S. J. Libby, N. A. Buchmeier, P. C. Loewen, J. Switala, J. Harwood, and D. G. Guiney. 1992. The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc. Natl. Acad. Sci. USA 89:11978-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faust, M. A., A. E. Aotaky, and M. T. Hargadon. 1975. Effect of physical parameters on the in situ survival of Escherichia coli MC-6 in an estuarine environment. Appl. Microbiol. 30:800-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fedorka-Cray, P. J., L. C. Kelley, T. J. Stabel, J. T. Gray, and J. A. Laufer. 1995. Alternate routes of invasion may affect pathogenesis of Salmonella typhimurium in swine. Infect. Immun. 63:2658-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fish, J. T., and G. W. Pettibone. 1995. Influence of freshwater sediment on the survival of Escherichia coli and Salmonella sp. as measured by three methods of enumeration. Lett. Appl. Microbiol. 20:277-281. [DOI] [PubMed] [Google Scholar]
- 32.Foltz, V. D. 1969. Salmonella ecology. J. Am. Oil Chem. Soc. 46:222-224. [DOI] [PubMed] [Google Scholar]
- 33.Formal, S. B., and R. B. Hornick. 1978. Invasive Escherichia coli. J. Infect. Dis. 137:641-644. [DOI] [PubMed] [Google Scholar]
- 34.Freter, R., H. Brickner, J. Fekete, M. M. Vickerman, and K. E. Carey. 1983. Survival and implantation of Escherichia coli in the intestinal tract. Infect. Immun. 39:686-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerba, C. P., and J. S. McLeod. 1976. Effect of sediments on the survival of Escherichia coli in marine waters. Appl. Environ. Microbiol. 32:114-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon, D. M., S. Bauer, and J. R. Johnson. 2002. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology 148:1513-1522. [DOI] [PubMed] [Google Scholar]
- 37.Greenberg, B., and M. Klowden. 1972. Enteric bacterial interactions in insects. Am. J. Clin. Nutr. 25:1459-1466. [DOI] [PubMed] [Google Scholar]
- 38.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Groisman, E. A., and H. Ochman. 1994. How to become a pathogen. Trends Microbiol. 2:289-294. [DOI] [PubMed] [Google Scholar]
- 40.Groisman, E. A., M. H. Saier, Jr., and H. Ochman. 1992. Horizontal transfer of a phosphatase gene as evidence for mosaic structure of the Salmonella genome. EMBO J. 11:1309-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guard-Petter, J. 2001. The chicken, the egg and Salmonella enteritidis. Environ. Microbiol. 3:421-430. [DOI] [PubMed] [Google Scholar]
- 42.Guo, X., J. Chen, R. E. Brackett, and L. R. Beuchat. 2002. Survival of Salmonella on tomatoes stored at high relative humidity, in soil, and on tomatoes in contact with soil. J. Food Prot. 65:274-279. [DOI] [PubMed] [Google Scholar]
- 43.Guo, X., J. Chen, R. E. Brackett, and L. R. Beuchat. 2001. Survival of salmonellae on and in tomato plants from the time of inoculation at flowering and early stages of fruit development through fruit ripening. Appl. Environ. Microbiol. 67:4760-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo, X., M. W. van Iersel, J. Chen, R. E. Brackett, and L. R. Beuchat. 2002. Evidence of association of salmonellae with tomato plants grown hydroponically in inoculated nutrient solution. Appl. Environ. Microbiol. 68:3639-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hendricks, C. W. 1971. Increased recovery rate of salmonellae from stream bottom sediments versus surface waters. Appl. Microbiol. 21:379-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hilbert, F., F. G. del Portillo, and E. A. Groisman. 1999. A periplasmic d-alanyl-d-alanine dipeptidase in the gram-negative bacterium Salmonella enterica. J. Bacteriol. 181:2158-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hood, M. A., and G. E. Ness. 1982. Survival of Vibrio cholerae and Escherichia coli in estuarine waters and sediments. Appl. Environ. Microbiol. 43:578-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hurd, H. S., J. D. McKean, R. W. Griffith, I. V. Wesley, and M. H. Rostagno. 2002. Salmonella enterica infections in market swine with and without transport and holding. Appl. Environ. Microbiol. 68:2376-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jimenez, L., I. Muniz, G. A. Toranzos, and T. C. Hazen. 1989. Survival and activity of Salmonella typhimurium and Escherichia coli in tropical freshwater. J. Appl. Bacteriol. 67:61-69. [DOI] [PubMed] [Google Scholar]
- 50.Kehres, D. G., C. H. Lawyer, and M. E. Maguire. 1998. The CorA magnesium transporter gene family. Microb. Comp. Genomics 3:151-169. [DOI] [PubMed] [Google Scholar]
- 51.Kier, L. D., R. M. Weppelman, and B. N. Ames. 1979. Regulation of nonspecific acid phosphatase in Salmonella: phoN and phoP genes. J. Bacteriol. 138:155-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knuckles, J. L. 1972. Survival of enteric pathogens in the pupae of Phormia regina (Meigen). J. Med. Entomol. 9:9-12. [DOI] [PubMed] [Google Scholar]
- 53.Kourany, M., C. W. Myers, and C. R. Schneider. 1970. Panamanian amphibians and reptiles as carriers of Salmonella. Am. J. Trop. Med. Hyg. 19:632-638. [DOI] [PubMed] [Google Scholar]
- 54.Kourany, M., and S. R. Telford. 1981. Lizards in the ecology of salmonellosis in Panama. Appl. Environ. Microbiol. 41:1248-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuhn, I., K. Tullus, and R. Mollby. 1986. Colonization and persistence of Escherichia coli phenotypes in the intestines of children aged 0 to 18 months. Infection 14:7-12. [DOI] [PubMed] [Google Scholar]
- 56.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5:49-59. [DOI] [PubMed] [Google Scholar]
- 57.Lawrence, J. G., and H. Ochman. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA 95:9413-9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lessard, I. A., S. D. Pratt, D. G. McCafferty, D. E. Bussiere, C. Hutchins, B. L. Wanner, L. Katz, and C. T. Walsh. 1998. Homologs of the vancomycin resistance d-Ala-d-Ala dipeptidase VanX in Streptomyces toyocaensis, Escherichia coli and Synechocystis: attributes of catalytic efficiency, stereoselectivity and regulation with implications for function. Chem. Biol. 5:489-504. [DOI] [PubMed] [Google Scholar]
- 59.Lim, C. H., and K. P. Flint. 1989. The effects of nutrients on the survival of Escherichia coli in lake water. J. Appl. Bacteriol. 66:559-569. [DOI] [PubMed] [Google Scholar]
- 60.Lopez-Torres, A. J., T. C. Hazen, and G. A. Toranzos. 1987. Distribution and in situ survival and activity of Klebsiella pneumoniae and Escherichia coli in a tropical rain forest watershed. Curr. Microbiol. 15:213-218. [Google Scholar]
- 61.Marshall, K. C. 1980. Adsorption of microorganisms to soils and sediments, p. 317-329. In G. Britton and K. C. Marshall (ed.), Adsorption of microorganisms to surfaces. Wiley, New York, N.Y.
- 62.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 63.Mezrioui, N., B. Baleux, and M. Trousselier. 1995. A microcosm study of the survival of Escherichia coli and Salmonella typhimurium in brackish water. Water Res. 29:459-465. [Google Scholar]
- 64.Mian, L. S., H. Maag, and J. V. Tacal. 2002. Isolation of Salmonella from muscoid flies at commercial animal establishments in San Bernardino County, California. J. Vector Ecol. 27:82-85. [PubMed] [Google Scholar]
- 65.Millemann, Y., S. Gaubert, D. Remy, and C. Colmin. 2000. Evaluation of IS200-PCR and comparison with other molecular markers to trace Salmonella enterica subsp. enterica serotype Typhimurium bovine isolates from farm to meat. J. Clin. Microbiol. 38:2204-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Monaghan, P., C. B. Shedden, K. Ensor, C. R. Fricker, and R. W. A. Girdwood. 1985. Salmonella carriage by herring gulls in the Clyde area of Scotland in relation to their feeding ecology. J. Appl. Ecol. 22:669-680. [Google Scholar]
- 67.Montgomery, D. M., A. C. Dean, P. Wiffen, and L. E. Macaskie. 1995. Phosphatase production and activity in Citrobacter freundii and a naturally occurring, heavy-metal-accumulating Citrobacter sp. Microbiology 141:2433-2441. [DOI] [PubMed] [Google Scholar]
- 68.Mouslim, C., F. Hilbert, H. Huang, and E. A. Groisman. 2002. Conflicting needs for a Salmonella hypervirulence gene in host and non-host environments. Mol. Microbiol. 45:1019-1027. [DOI] [PubMed] [Google Scholar]
- 69.Munro, P. M., G. N. Flatau, R. L. Clement, and M. J. Gauthier. 1995. Influence of the RpoS (KatF) sigma factor on maintenance of viability and culturability of Escherichia coli and Salmonella typhimurium in seawater. Appl. Environ. Microbiol. 61:1853-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Natvig, E. E., S. C. Ingham, B. H. Ingham, L. R. Cooperband, and T. R. Roper. 2002. Salmonella enterica serovar Typhimurium and Escherichia coli contamination of root and leaf vegetables grown in soils with incorporated bovine manure. Appl. Environ. Microbiol. 68:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nixon, S. R., and G. E. Mawer. 1970. The digestion and absorption of protein in man. 2. The form in which digested protein is absorbed. Br. J. Nutr. 24:241-258. [DOI] [PubMed] [Google Scholar]
- 72.Pang, T., Z. A. Bhutta, B. B. Finlay, and M. Altwegg. 1995. Typhoid fever and other salmonellosis: a continuing challenge. Trends Microbiol. 3:253-255. [DOI] [PubMed] [Google Scholar]
- 73.Parker, W. F., and B. J. Mee. 1982. Survival of Salmonella adelaide and fecal coliforms in coarse sands of the swan costal plain, Western Australia. Appl. Environ. Microbiol. 43:981-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paul, J. H., J. B. Rose, S. Jiang, C. Kellogg, and E. A. Shinn. 1995. Occurrence of fecal indicator bacteria in surface waters and the subsurface aquifer in Key Largo, Florida. Appl. Environ. Microbiol. 61:2235-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pfeifer, C. G., S. L. Marcus, O. Steele-Mortimer, L. A. Knodler, and B. B. Finlay. 1999. Salmonella typhimurium virulence genes are induced upon bacterial invasion into phagocytic and nonphagocytic cells. Infect. Immun. 67:5690-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pommepuy, M., M. Butin, A. Derrien, M. Gourmelon, R. R. Colwell, and M. Cormier. 1996. Retention of enteropathogenicity by viable but nonculturable Escherichia coli exposed to seawater and sunlight. Appl. Environ. Microbiol. 62:4621-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Puente, J. L., and B. B. Finlay. 2001. Pathogenic Escherichia coli, p. 387-456. In E. A. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, San Diego, Calif.
- 78.Ravel, J., I. T. Knight, C. E. Monahan, R. T. Hill, and R. R. Colwell. 1995. Temperature-induced recovery of Vibrio cholerae from the viable but nonculturable state: growth or resuscitation? Microbiology 141:377-383. [DOI] [PubMed] [Google Scholar]
- 79.Renner, R. 2002. From flush to farm. Sewage is a great fertilizer, but is it a health hazard? Sci. Am. 287:21-22. [PubMed] [Google Scholar]
- 80.Rhodes, M. W., and H. Kator. 1988. Survival of Escherichia coli and Salmonella spp. in estuarine environments. Appl. Environ. Microbiol. 54:2902-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rivera, S. C., T. C. Hazen, and G. A. Toranzos. 1988. Isolation of fecal coliforms from pristine sites in a tropical rain forest. Appl. Environ. Microbiol. 54:513-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roszak, D. B., D. J. Grimes, and R. R. Colwell. 1984. Viable but nonrecoverable stage of Salmonella enteritidis in aquatic systems. Can. J. Microbiol. 30:334-338. [DOI] [PubMed] [Google Scholar]
- 83.Rozen, Y., and S. Belkin. 2001. Survival of enteric bacteria in seawater. FEMS Microbiol. Rev. 25:513-529. [DOI] [PubMed] [Google Scholar]
- 84.Sanyal, D., T. Douglas, and R. Roberts. 1997. Salmonella infection acquired from reptilian pets. Arch. Dis. Child. 77:345-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Savageau, M. A. 1998. Demand theory of gene regulation. II. Quantitative application to the lactose and maltose operons of Escherichia coli. Genetics 149:1677-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Savageau, M. A. 1983. Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am. Nat. 122:732-744. [Google Scholar]
- 87.Savageau, M. A. 1974. Genetic regulatory mechanisms and the ecological niche of Escherichia coli. Proc. Natl. Acad. Sci. USA 71:2453-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Scherer, C. A., and S. I. Miller. 2001. Molecular pathogenesis of salmonellae, p. 265-333. In E. A. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, San Diego, Calif.
- 89.Sears, H. J., and I. Brownlee. 1952. Further observations on the persistence of individual strains of Escherichia coli in the intestinal tract of man. J. Bacteriol. 63:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sears, H. J., I. Brownlee, and J. K. Uchiyama. 1950. Persistence of individual strains of Escherichia coli in the intestinal tract of man. J. Bacteriol. 59:293-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sears, H. J., H. Jones, R. Saloum, I. Brownlee, and L. F. Lamoreaux. 1956. Persistence of individual strains of Escherichia coli in man and dog under varying conditions. J. Bacteriol. 71:370-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smith, H. W. 1965. Observations on the flora of the alimentary tract of animals and factors affecting its composition. J. Pathol. Bacteriol. 89:95-122. [PubMed] [Google Scholar]
- 93.Smith, J. J., J. P. Howington, and G. A. McFeters. 1994. Survival, physiological response, and recovery of enteric bacteria exposed to a polar marine environment. Appl. Environ. Microbiol. 60:2977-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smith, R. L., and M. E. Maguire. 1998. Microbial magnesium transport: unusual transporters searching for identity. Mol. Microbiol. 28:217-226. [DOI] [PubMed] [Google Scholar]
- 95.Snavely, M. D., S. A. Gravina, T. T. Cheung, C. G. Miller, and M. E. Maguire. 1991. Magnesium transport in Salmonella typhimurium. Regulation of mgtA and mgtB expression. J. Biol. Chem. 266:824-829. [PubMed] [Google Scholar]
- 96.Snavely, M. D., C. G. Miller, and M. E. Maguire. 1991. The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J. Biol. Chem. 266:815-823. [PubMed] [Google Scholar]
- 97.Solo-Gabriele, H. M., M. A. Wolfert, T. R. Desmarais, and C. J. Palmer. 2000. Sources of Escherichia coli in a coastal subtropical environment. Appl. Environ. Microbiol. 66:230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Soncini, F. C., E. Garcia Vescovi, F. Solomon, and E. A. Groisman. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178:5092-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stenstrom, T. A. 1989. Bacterial hydrophobicity, an overall parameter for the measurement of adhesion potential to soil particles. Appl. Environ. Microbiol. 55:142-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tauxe, R. V. 1997. Emerging foodborne diseases: an evolving public health challenge. Emerg. Infect. Dis. 3:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Temple, K. L., A. K. Camper, and G. A. McFeters. 1980. Survival of two enterobacteria in feces buried in soil under field conditions. Appl. Environ. Microbiol. 40:794-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Thomas, S. J. 1954. The number of bacilli harboured by enteric carriers. J. Hyg. Camb. 52:67-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thomason, B. M., J. W. Biddle, and W. B. Cherry. 1975. Detection of salmonellae in the environment. Appl. Microbiol. 30:764-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Thomason, B. M., D. J. Dodd, and W. B. Cherry. 1977. Increased recovery of salmonellae from environmental samples enriched with buffered peptone water. Appl. Environ. Microbiol. 34:270-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thomason, B. M., G. A. Hebert, and W. B. Cherry. 1975. Evaluation of a semiautomated system for direct fluorescent antibody detection of salmonellae. Appl. Microbiol. 30:557-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thorns, C. J. 2000. Bacterial food-borne zoonoses. Rev. Sci. Tech. 19:226-239. [DOI] [PubMed] [Google Scholar]
- 107.Torriani-Gorini, A. 1994. Regulation of phosphate metabolism and transport, p. 1-4. In A. Torriani-Gorini, E. Yagil, and S. Silver (ed.), Phosphate in microorganisms. ASM Press, Washington, D.C.
- 108.Wright, R. C. 1989. The survival patterns of selected faecal bacteria in tropical fresh waters. Epidemiol. Infect. 103:603-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 97:14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]