Abstract
Over a 3-year period, a total of 646 fecal samples from pigs in 22 indoor and outdoor herds from Western Australia were screened for Cryptosporidium spp. by microscopy. Results revealed that 39 of 646 samples (6.03%) were positive for Cryptosporidium. Cryptosporidium was much more common in outdoor herds (17.2%) than in indoor herds (0.5%) and was more common in animals between the ages of 5 and 8 weeks (69.2%) than in younger animals (P < 0.0001). Molecular characterization of the positive samples at the 18S ribosomal DNA locus identified two distinct genotypes of Cryptosporidium: the previously identified pig genotype I and a novel pig genotype (pig genotype II), both of which warrant species status.
Preweaning diarrhea, caused by a complex of protozoan organisms including Isospora suis and Cryptosporidium spp., remains a major problem in pigs worldwide (2, 3, 6). Yet, due to the inadequacies of conventional diagnostics, little is known about the prevalence and significance of Cryptosporidium in pigs. As a consequence, it has not been possible to adequately determine the contribution of Cryptosporidium to porcine preweaning diarrhea. Evidence suggests not only that Cryptosporidium is prevalent in domestic pigs but also that Cryptosporidium infection can occur concurrently with Isospora infection, as well as in older animals (7, 9-11, 14, 16, 17).
Recent genetic characterization studies have revealed that pigs are infected with a genetically distinct and apparently host-adapted form of Cryptosporidium (Cryptosporidium “pig” genotype) (5, 8, 9, 13). Pigs can also be infected with the zoonotic Cryptosporidium parvum “cattle” genotype, indicating that they can potentially play a role as reservoirs of infection for humans and other animals (9). In this study, in order to gain a better understanding of the prevalence and significance of Cryptosporidium in pigs, fecal samples were collected over a 3-year period and screened for the presence of Cryptosporidium by morphological and molecular analyses.
MATERIALS AND METHODS
Sources of parasite isolates.
Fecal samples were collected between August 1999 and July 2001 from indoor and outdoor piggeries in Western Australia. A total of 646 fecal samples from pigs below the age of 9 weeks in 22 (17 indoor and 5 outdoor) herds were screened for the presence of Eimeria spp., I. suis, Giardia spp., and Cryptosporidium spp. by direct smear, saturated ZnSO4, and malachite green staining (for the identification of Cryptosporidium) (4).
DNA extraction, 18S rDNA gene amplification, and sequencing.
DNA was extracted from fecal samples by using the QiAmp stool detection kit (Qiagen, Hilden, Germany). A two-step nested PCR protocol for the 18S ribosomal DNA (rDNA) gene was performed as previously described (19).
PCR products were purified by using Qiagen spin columns and sequenced by using an ABI Prism Dye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, Calif.) according to the manufacturer's instructions, except that the annealing temperature was raised to 58°C. Sequences were analyzed using SeqEd (version 1.0.3; Applied Biosystems).
Phylogenetic analyses of 18S rDNA.
Nucleotide sequences obtained from this study were aligned against each other and those obtained previously by using Clustal X (15). Plasmodium falciparum was used as an outgroup (GenBank accession no. M19172). Distance-based analyses were conducted using Tamura-Nei distance estimates, and trees were constructed using the neighbor-joining algorithm (D. L. Swofford, PAUP, 1999). Phylograms were drawn by using the TreeView program (12).
Statistical analysis.
Prevalences in different groups of pigs (indoor versus outdoor; different age groups) were compared with a chi-square test for independence.
Nucleotide sequence accession number.
A pig genotype II sequence has been submitted to GenBank under accession no. AY271721.
RESULTS
Microscopic analysis.
Cryptosporidium was detected in 39 of 646 pigs from both indoor and outdoor herds in Western Australia, a prevalence of 6.03% (Table 1). Cryptosporidium was significantly more common in outdoor herds (37 of 216 [17.2%]) than in indoor herds (2 of 430 [0.5%]), where only two low-level positive samples were found (P < 0.0001). Cryptosporidium was found in 1- to 8-week-old pigs but was significantly more common (69.2%) in older animals (5 to 8 weeks old) than in pigs below the age of 5 weeks (P < 0.0001). This contrasts with bovine hosts, where cryptosporidiosis is most common in calves 2 weeks old or younger (11).
TABLE 1.
Cryptosporidium isolates used in this study
| Isolate | Age of host (wks) | I/O herda | Toltrazuril treatmentb | No. of oocysts/g of feces | Date collected (day/mo/yr) | Genotype at 18S rDNA locusc |
|---|---|---|---|---|---|---|
| PW 1 | 4-6 | O | Y | <1,000 | 16/09/99 | ND |
| PW 2 | 6-8 | O | Y | <1,000 | 16/09/99 | Pig genotype II |
| PW 3 | 6 | O | Y | <1,000 | 16/09/99 | Pig genotype II |
| PW 4 | 5 | O | Y | <1,000 | 16/09/99 | Pig genotype II |
| PM3-2 | 4 | I | N | <1,000 | 21/09/99 | Pig genotype II |
| PM3-3 | 4 | I | N | <1,000 | 21/09/99 | ND |
| PGR- 1 | 4 | O | N | <1,000 | 23/12/99 | ND |
| PGR- 2 | 4 | O | N | <1,000 | 23/12/99 | ND |
| PGI 1 | 1 | O | N | 1.1 × 106 | 25/07/99 | Pig genotype II |
| PGI 2 | 2 | O | N | <1,000 | 25/07/99 | ND |
| PGI 5 | 2 | O | Y | 7 × 105 | 25/07/00 | Pig genotype I |
| PGI 7 | 2 | O | Y | 9.6 × 106 | 25/07/00 | ND |
| PGI 8 | 2 | O | Y | 7 × 106 | 25/07/00 | Pig genotype I |
| PGI 9 | 4 | O | N | <1,000 | 25/07/00 | Pig genotype I |
| PGI 11 | 5 | O | N | <1,000 | 25/07/00 | ND |
| PGI 13 | 5 | O | U | <1,000 | 25/07/00 | Pig genotype I |
| PGI 14 | 5 | O | U | 6.8 × 105 | 25/07/00 | ND |
| PGI 16 | 5 | O | U | <1,000 | 25/07/00 | Pig genotype I |
| PGI 18 | 6 | O | U | ≈1,000 | 25/07/00 | Pig genotype II |
| PGI 19 | 6 | O | U | 1.2 × 105 | 25/07/00 | Pig genotype II |
| PGI 20 | 6 | O | U | 2 × 105 | 25/07/00 | Pig genotype II |
| PGI 22 | 6 | O | U | <1,000 | 25/07/00 | Pig genotype II |
| PGI 23 | 6 | O | U | 2 × 104 | 25/07/00 | Pig genotype I |
| PGI 24 | 6 | O | U | 6 × 105 | 25/07/00 | Pig genotype I |
| PGI 25 | 6 | O | U | <1,000 | 25/07/00 | Pig genotype II |
| PGI 26 | 6 | O | U | <1,000 | 25/07/00 | Pig genotype II |
| PGI 28 | 8 | O | U | <1,000 | 25/07/00 | Pig genotype II |
| PGI 29 | 8 | O | U | <1,000 | 25/07/00 | ND |
| PGI 31 | 8 | O | U | 1.5 × 106 | 25/07/00 | Pig genotype II |
| POR10 | 6 | O | N | <1,000 | 25/07/00 | Pig genotype I |
| POR12 | 5 | O | N | <1,000 | 25/07/00 | ND |
| POR21 | 8 | O | N | <1,000 | 25/07/00 | Pig genotype II |
| POR32 | 2 | O | N | <1,000 | 25/07/00 | Pig genotype I |
| POR33 | 2 | O | N | <1,000 | 25/07/00 | Pig genotype I |
| POR 3 | 6 | O | N | <1,000 | 31/07/01 | Pig genotype I |
| PMa 4 | 7 | O | N | <1,000 | 31/07/01 | ND |
| PMa12 | 7 | O | N | <1,000 | 31/07/01 | Pig genotype I |
| PMa23 | 7 | O | N | <1,000 | 31/07/01 | Pig genotype I |
| PMa28 | 7 | O | N | <1,000 | 31/07/01 | Pig genotype I |
I/O, indoor or outdoor.
Y, yes; N, no; U, unknown.
ND, not determined.
In the majority of samples the number of oocysts detected was low (<1,000/g of feces), with the exception of four samples, all from the same herd (PGl 1, PGl 7, PG1 8, and PGl 31), where oocyst numbers of >106/g of feces were detected (Table 1). A number of other parasites were detected, including I. suis (15%), a Giardia sp. (0.8%), Ascaris suum (0.6%), and Eimeria spp. (18%). In some cases multiple parasite infections were detected; for example, isolates PGl 22 to 25 were positive for Cryptosporidium, a Giardia sp., and Eimeria spp.
Sequencing and phylogenetic analysis of the 18S ribosomal DNA (rDNA) gene.
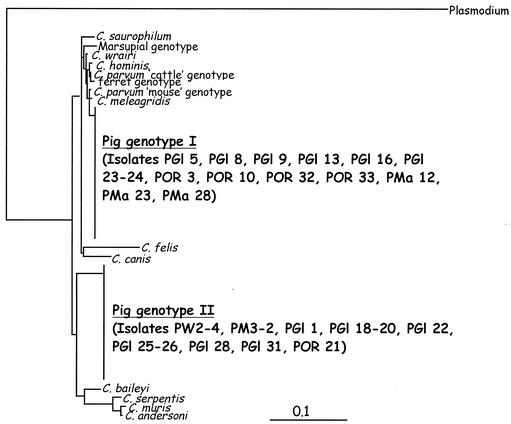
Partial sequences were obtained for 28 of 39 pig-derived Cryptosporidium isolates (Table 1). The remaining 11 isolates either could not be amplified or produced mixed chromatograms, which could not be read. Sequencing and phylogenetic analysis of the 28 pig-derived isolates and a range of Cryptosporidium 18S sequences obtained from GenBank identified two distinct genotypes in pigs: the previously identified Cryptosporidium “pig” genotype (9) and a novel genotype, which we have called Cryptosporidium “pig” genotype II (Fig. 1 and 2). In the samples that could be genotyped, pig genotype I and pig genotype II appeared to be present in equal numbers (i.e., each was present in 14 of 28 samples [50%]). There was no difference in age between pigs infected with genotypes I and II. Some pigs infected with pig genotype II did appear to shed high numbers of oocysts, but more-intensive sampling is required to confirm this.
FIG. 1.
Clustal X sequence alignment of partial 18S rDNA sequences of pig genotype I and pig genotype II.
FIG. 2.
Evolutionary relationships of Cryptosporidium isolates inferred by neighbor-joining analysis of Tamura-Nei distances calculated from pairwise comparisons of the 18S rDNA sequences.
DISCUSSION
Microscopic analysis of fecal samples from indoor and outdoor piggeries in Western Australia identified Cryptosporidium in 39 of 646 pigs tested (6.03%); most of these samples were not associated with diarrhea. A previous study of indoor and outdoor piggeries in Western Australia detected the presence of both the pig genotype and the C. parvum cattle genotype (9). That study concluded that “two genetically and biologically differing strains of Cryptosporidium appeared to be associated with acute diarrhea, chronic ill thrift and death in pigs” (9). However, it was not possible to determine whether Cryptosporidium was the primary or the secondary pathogen, because no screening for viruses and enteric bacteria was performed. Similarly, in the present study, it was not possible to determine if Cryptosporidium was the primary pathogen, but in most cases, infection with Cryptosporidium was not associated with diarrhea, suggesting that both pig genotype I and pig genotype II are host adapted. Further studies are required to confirm this hypothesis.
Few studies have been conducted in recent years to determine the prevalence of Cryptosporidium in pigs. A study in Japan reported a much higher prevalence of Cryptosporidium in pigs, with 77 (33.2%) 1- to 3-month-old weaned piglets positive for Cryptosporidium from four out of eight stock-raising farms located in Kanagawa Prefecture (7). In another study, in Germany, Cryptosporidium was detected in 1.4% of piglets, all of which exhibited diarrhea (16). More-extensive surveys need to be performed, but it is clear that cryptosporidial infection, perhaps in combination with other enteric pathogens, may be an emerging problem for the pig industry.
In the present study, the prophylactic use of toltrazuril at the age of 3 to 5 days was widespread (384 of 646 samples), particularly in the indoor intensive farms. This is interesting in light of the fact that Cryptosporidium was much more common in outdoor herds (37 of 216 [17.2%]) than in indoor herds (2 of 430 [0.5%]). A study by Armson et al. (1) has shown that toltrazuril exhibited limited efficacy against the C. parvum cattle genotype in vitro, but it has not been tested in vivo. It is possible that the extensive prophylactic use of toltrazuril in indoor herds inhibited the establishment of Cryptosporidium infections in these pigs. However, high numbers of oocysts were detected in two isolates from an outdoor herd which had been prophylactically treated with toltrazuril (Table 1). Further studies are required to confirm the effect, if any, of toltrazuril on Cryptosporidium.
A more likely explanation is that the opportunities for transmission of Cryptosporidium are much greater in outdoor herds through the contamination of the environment to which the pigs have access. In the indoor herds the pigs were maintained in pens with slatted flooring that allowed fecal matter to fall through. The slats were rinsed off daily, thus limiting any potential exposure to contaminated feces. This theory is supported by a previous study on the prevalence of Cryptosporidium in two Ohio pig farms with different management systems (18). That study reported that a farm with porous concrete floors had a significantly higher Cryptosporidium infection rate in pigs than a farm with slotted and wire floors (18).
Sequencing and phylogenetic analysis of 18S rDNA identified two distinct genotypes of Cryptosporidium: the previously identified pig genotype (9) and a novel pig genotype (pig genotype II). The previous study by Morgan et al. (9) reported the presence in pigs of the C. parvum cattle genotype, which is associated with zoonotic transmission. However, in the present study, the cattle genotype was not detected, which argues against the previous speculation that pigs may be important reservoirs of zoonotic Cryptosporidium in Australia (9).
Phylogenetic analysis of the 18S locus revealed that Cryptosporidium pig genotype I and pig genotype II are genetically distinct. Pig genotype I shared only 97.3% similarity with the C. parvum cattle genotype and C. hominis. This is less than the similarities among C. meleagridis, the C. parvum cattle genotype, and C. hominis (99 to 98.6%) and between the C. parvum cattle genotype and C. hominis (99.2%). Cryptosporidium pig genotype II is genetically very distinct and did not group within the C. parvum genotypes or closely related species but formed a separate clade on its own. Cryptosporidium pig genotype II shared only 93% similarity with pig genotype I and only 92.2 to 92.8% similarity with the C. parvum cattle genotype, the C. parvum “mouse” genotype, C. hominis, and C. meleagridis. This is similar to the level of genetic similarity between the C. parvum group and C. baileyi (93.7%) and between the C. parvum group and C. muris, C. serpentis, and C. andersoni (87.7 to 89%), which are the Cryptosporidium species most distantly related to the C. parvum group
Both pig genotypes appear to be widespread, since both were first identified in pig-derived isolates from Switzerland (8; U. M. Ryan, unpublished data) and have since been identified in other locations in Europe (5; L. Xiao et al., unpublished data). The genetically distinct nature of both these genotypes, combined with their apparently host-adapted nature, strongly suggests that both pig genotype I and the novel pig genotype II warrant species status.
Acknowledgments
This project was funded by the Australian Pig Research and Development Corporation (now Australian Pork Limited).
REFERENCES
- 1.Armson, A., B. P. Meloni, J. A. Reynoldson, and R. C. A. Thompson. 1999. Assessment of drugs against Cryptosporidium parvum using a simple in vitro screening method. FEMS Microbiol. Lett. 178:227-233. [DOI] [PubMed] [Google Scholar]
- 2.Blagburn, B. L., T. R. Boosinger, and T. A. Powe. 1991. Experimental Isospora suis infections in miniature swine. Vet. Parasitol. 38:343-347. [DOI] [PubMed] [Google Scholar]
- 3.Driesen, S. J., P. G. Cartland, and V. A. Fahy. 1993. Studies on pre-weaning piglet diarrhoea. Aust. Vet. J. 70:259-262. [DOI] [PubMed] [Google Scholar]
- 4.Elliot, A., U. M. Morgan, and R. C. A. Thompson. 1999. Improved staining method for detecting Cryptosporidium oocysts in stools using Malachite Green. J. Gen. Appl. Microbiol. 45:139-142. [DOI] [PubMed] [Google Scholar]
- 5.Enemark, H. L., V. Bille-Hansen, P. Lind, P. M. H. Heegaard, H. Vigre, P. Ahrens, and S. M. Thamsborg. 2003. Pathogenicity of Cryptosporidium parvum—evaluation of an animal infection. Vet. Parasitol. 113:35-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eysker, M. 1995. The prevalence of Isospora suis and Strongyloides ransomi in suckling piglets in the Netherlands. Pig News Infect. 16:116. [DOI] [PubMed] [Google Scholar]
- 7.Izumiyama, S., I. Furukawa, T. Kuroki, S. Yamai, H. Sugiyama, K. Yagita, and T. Endo. 2001. Prevalence of Cryptosporidium parvum infections in weaned piglets and fattening porkers in Kanagawa Prefecture, Japan. Jpn. J. Infect. Dis. 54:23-26. [PubMed] [Google Scholar]
- 8.Morgan, U. M., K. D. Sargent, P. Deplazes, D. A. Forbes, F. Spano, H. Hertzberg, A. Elliot, and R. C. A. Thompson. 1998. Molecular characterisation of Cryptosporidium from various hosts. Parasitology 117:31-37. [DOI] [PubMed] [Google Scholar]
- 9.Morgan, U. M., R. Buddle, A. Armson, and R. C. A. Thompson. 1999. Molecular and biological characterisation of Cryptosporidium in pigs. Aust. Vet. J. 77:44-47. [DOI] [PubMed] [Google Scholar]
- 10.Nagy, B. 1995. Epidemiologic data on Cryptosporidium parvum infection of mammalian domestic animals in Hungary. Magyar Allatorvosok Lapja 50:139-144. [Google Scholar]
- 11.Olson, M. E., C. L. Thorlakson, L. Deselliers, D. W. Morck, and T. A. McAllister. 1997. Giardia and Cryptosporidium in Canadian farm animals. Vet. Parasitol. 68:375-381. [DOI] [PubMed] [Google Scholar]
- 12.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 13.Pereira, M. G., E. R. Atwill, M. R. Crawford, and R. B. Lefebvre. 1998. DNA sequence similarity between California isolates of Cryptosporidium parvum. Appl. Environ. Microbiol. 64:1584-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quilez, J., C. Sanchez-Acedo, A. Clavel, E. del Cacho, and F. Lopez-Bernad. 1996. Prevalence of Cryptosporidium infections in pigs in Aragon (North-Eastern Spain). Vet. Parasitol. 67:3-88. [DOI] [PubMed] [Google Scholar]
- 15.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The Clustal X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wieler, L. H., A. Ilieff, W. Herbst, C. Bauer, E. Vieler, R. Bauerfeind, K. Failing, H. Klos, D. Wengert, G. Baljer, and H. Zahner. 2001. Prevalence of enteropathogens in suckling and weaned piglets with diarrhea in southern Germany. J. Vet. Med. B 48:151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao, L., and R. P. Herd. 1994. Infection pattern of Cryptosporidium and Giardia in calves. Vet. Parasitol. 55:257-262. [DOI] [PubMed] [Google Scholar]
- 18.Xiao, L., R. P. Herd, and G. L. Bowman. 1994. Prevalence of Cryptosporidium and Giardia infections on two Ohio pig farms with different management systems. Vet. Parasitol. 52:331-336. [DOI] [PubMed] [Google Scholar]
- 19.Xiao, L., L. Escalante, C. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]