Abstract
Expression of the btuB gene encoding the outer membrane cobalamin transporter in Escherichia coli is strongly reduced on growth with cobalamins. Previous studies have shown that this regulation occurs in response to adenosylcobalamin (Ado-Cbl) and operates primarily at the translational level. Changes in the level and stability of btuB RNA are consequences of the modulated translation initiation. To examine how Ado-Cbl affects translation, the binding of E. coli 30S ribosomal subunits to btuB RNA was investigated by using a primer extension inhibition assay. Ribosome binding to btuB RNA was much less efficient than to other RNAs and was preferentially lost when the ribosomes were subjected to a high-salt wash. Ribosome binding to btuB RNA was inhibited by Ado-Cbl but not by cyanocobalamin, with half-maximal inhibition around 0.3 μM Ado-Cbl. Ribosome-binding activity was increased or decreased by mutations in the btuB leader region, which affected two predicted RNA hairpins and altered expression of btuB-lacZ reporters. Finally, the presence of Ado-Cbl elicited formation of a single primer extension-inhibition product with the same specificity and Cbl-concentration dependence as the inhibition of ribosome binding. These results indicate that btuB expression is controlled by the specific binding of Ado-Cbl to btuB RNA, which then affects access to its ribosome-binding sequence.
Although bacterial gene regulation usually occurs at the stage of transcription initiation, some genes are controlled at the posttranscriptional level through changes in RNA secondary structure, mediated by ribosome pausing or effector-responsive RNA-binding proteins (reviewed in ref. 1). Changes in RNA secondary structure can affect access of the ribosome-binding site or the stability of the RNA. Posttranscriptional control has been implicated in repression of several genes in response to adenosylcobalamin (Ado-Cbl). Expression of the btuB gene of Escherichia coli and Salmonella typhimurium, encoding the outer membrane transporter for corrinoids such as vitamin B12 [cyanocobalamin (CN-Cbl)], and of the S. typhimurium cob operon, encoding the de novo cobalamin biosynthetic pathway, is strongly reduced by addition of cobalamins (reviewed in ref. 2). The specific role of Ado-Cbl as regulatory effector was indicated by the constitutive expression of btuB in E. coli btuR mutants defective in conversion of CN-Cbl to Ado-Cbl (3). S. typhimurium cobA mutants are defective in the same gene as btuR (4) but do not show altered regulatory behavior, because additional pathways of Ado-Cbl synthesis exist in this organism (2, 5).
Analyses of transcriptional and translational lacZ fusions to the btuB and cob genes revealed similar features of their Ado-Cbl-responsive regulation, even though their 5′-leader regions possess only a few conserved regions, called B12 boxes (6–11). Their promoters are unaffected by Cbl supplementation and can be replaced by other promoters with no major effect on regulation (8). The degree of regulation is much greater for translational fusions than for transcriptional fusions, i.e., 25- to 40-fold and 1- to 5-fold, respectively. The length of the transcribed sequences needed for regulation of the different types of fusions also differs. Translational regulation of btuB requires the 241-nt leader region, but transcriptional regulation also requires the next 60–100 nucleotides of btuB coding sequence, called the translated regulatory region (8). Translational regulation involves an RNA hairpin that includes the Shine–Dalgarno sequence, as judged from the constitutive btuB expression in mutants that affect this hairpin and the restoration of regulation by compensatory mutations that restore the potential to form the hairpin (8, 9). Mutations at other sites in the leader region, including the conserved B12 box, affect translational regulation (7, 9, 11), but no regulatory proteins have been identified.
Transcriptional regulation of btuB involves attenuation and altered RNA stability but is secondary to translational control (8). Transcription is unregulated when translation operates in a constitutive manner, and btuB RNA levels are strongly reduced when translation is prevented by ablation of the start codon or by early termination codons (8). These properties focus attention on translation initiation as the primary site of control, but biochemical investigation of this regulation has been limited to a report that Cbl-dependent regulation of cob gene expression occurs in a transcription–translation system (12). The key question is how Ado-Cbl interferes with translation of btuB RNA. Perhaps the conformation of btuB RNA is altered by the action of an Ado-Cbl-responsive RNA-binding protein or by the binding of Ado-Cbl directly to the RNA or to the ribosome. Here we describe the ability of Ado-Cbl to inhibit the binding of btuB RNA to the ribosome, perhaps by its specific binding to btuB RNA.
Materials and Methods
Bacterial Strains and Plasmids.
The E. coli K-12 strain used for all studies of btuB-lacZ expression and for preparation of plasmids is RK5173, a metE derivative of strain MC4100 [Δ(argF-lac)U169 araD139 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR22 non-9 gyrA219 metE70] (13). Strain MRE600 was used as source of ribosomes and was provided by P.-C. Tai (Georgia State University, Atlanta, GA). Plasmids are derivatives of plasmids pRS414 and pRS415 (14), which carry EcoRI-BamHI fragments that cover nucleotides −60 to + 450 of the btuB regulatory region (coordinates are relative to the transcription start site), as described previously (6). These DNA inserts generate translational and transcriptional fusions to lacZ, respectively.
DNA Isolation and Mutagenesis.
All DNA manipulations were carried out by using standard techniques, as previously described (8, 15). The mutations in the btuB regulatory region were introduced by a two-step PCR protocol, as previously described (8). All mutations were verified by nucleotide sequence determination. The sequences of all PCR primers are available on request.
DNA templates for in vitro transcription were prepared by PCR. Templates containing the btuB regulatory region from nucleotides + 1 to + 315 were used for synthesis of the RNA molecules for ribosome-binding assays. Primers T7btu, corresponding to the phage T7 φ10 late promoter sequence and the first 22 nucleotides of the btuB transcript, and XNO-6, annealing to transcript position 298 to 315, were used for PCR. The ribosome-binding sequence for the lacZ gene fragment carried on plasmid pET-15b was used as control. A DNA template containing this region was isolated by PCR, by using primers XNO-71, containing T7 promoter sequence, and XNO-77, combining the sequences in XNO-6 and a region annealing to the multiple-cloning site of plasmid pET-15b (Novagen) and pET15b DNA as template. In vitro transcription by T7 RNA polymerase by using the PCR products as template was performed according to the instructions of the manufacturer (BMB–Roche Diagnostics).
Ribosome Preparation.
High-salt-washed 30S ribosomal subunits were isolated from MRE600 cells as described by Spedding (16). Washed cells in buffer A [20 mM Tris⋅HCl, pH 7.5 at 4°C/10.5 mM magnesium acetate/100 mM NH4Cl/0.5 mM EDTA/3 mM 2-mercaptoethanol (2ME)] were lysed by two passes through a French press at 12,000 psi. After removal of debris by centrifugation at 30,000 × g for 45 min, ribosomes were twice precipitated by centrifugation at 100,000 × g for 10 h through an equal volume of 1.1 M sucrose in buffer B (buffer B is buffer A plus 400 mM NH4Cl). The washed ribosomes were suspended in buffer F (10 mM Tris⋅HCl/1.1 mM magnesium acetate/60 mM NH4Cl/0.1 mM EDTA/2 mM 2ME) and isolated by centrifugation at 43,000 × g for 15 h in a gradient of 10 to 30% (wt/vol) sucrose in buffer F. Fractions containing 30S ribosomal subunits were mixed with an equal volume of buffer E (10 mM Tris⋅HCl/10 mM magnesium acetate/60 mM NH4Cl/3 mM 2ME), pelleted by centrifugation at 150,000 × g for 10 h, suspended in buffer E, and stored at −70°C. The quality of ribosome preparations was assessed by the A260/A280 ratio and tricine-SDS/PAGE. Low-salt-washed ribosomes were prepared in an identical manner, except that buffer B was replaced by buffer A.
Ribosome-Binding Assay.
Binding of 30S subunits to RNA was determined by primer extension inhibition, as described by Hartz et al. (17). About 50 fmols of RNA and a similar amount of 32P-labeled primer XNO-6 were annealed in SB buffer [10 mM Tris-acetate, pH 7.4/60 mM NH4Cl/6 mM 2ME] at 60°C for 3 min, then at −70°C for 1 min, and then placed on ice. The annealing mixture was supplemented to 10 mM magnesium acetate, and dNTP, tRNAfMet, and 30S ribosomal subunits were added. Unless specified, final concentrations were 5 nM RNA, 5 nM primer, 10 mM Mg2+, 200 μM dNTP, 1 μM tRNAfMet, and 100 nM 30S ribosomal subunits, in a volume of 10 μl. Binding was allowed to proceed for 10 min at 37°C, followed by the addition of 1 unit of AMV reverse transcriptase (BMB–Roche Diagnostics). After 10 min, the primer extension reaction was terminated by addition of 20 μl of formamide loading buffer, and the products were resolved by electrophoresis on a 6% sequencing gel (15). Radioactivity was quantified by using PhosphorImager and ImageQuant (Molecular Dynamics) analysis or by autoradiography.
β-Galactosidase Assay.
Expression of β-galactosidase was measured as previously described (8). Cells were grown in the absence and presence of 5 μM Ado-Cbl (Sigma).
Results
Cobalamin Repression Occurs in Vitro.
As a preliminary test for in vitro regulation of btuB expression, commercial transcription–translation kits were used with plasmid templates encoding btuB-lacZ transcriptional and translational fusions. Incorporation of [35S]methionine into polypeptides with the sizes of β-galactosidase or the BtuB-LacZ fusion polypeptide was found (data not shown). Synthesis of the BtuB-LacZ translational fusion product was strongly reduced by the presence of 5 μM Ado-Cbl, but synthesis of LacZ encoded by the transcriptional fusion was only modestly reduced. These results, although not quantified, are consistent with the regulation seen in cells, confirming that Cbl-dependent regulation of btuB operates mainly at the translational level, as was reported for the cob operon (12).
Ribosome Binding to btuB RNA Is Inhibited by Ado-Cbl.
To dissect this regulatory process, a toe-print or primer extension-inhibition assay (17) was used to detect the binding of ribosomes to btuB mRNA. This assay measures the ability of a bound ribosome to block extension by reverse transcriptase of a primer hybridized to btuB RNA downstream of the ribosome-binding site. The btuB regulatory region (residues 1 to 315) and the templates and primers used in this assay are presented in Fig. 1A. RNA substrates for ribosome binding were synthesized with T7 RNA polymerase, and lacZ (the fragment carried on pET15b) RNA was included as control for ribosome activity. Ribosomes were prepared by sucrose gradient centrifugation as described (16), except without the usual high-salt-wash step.
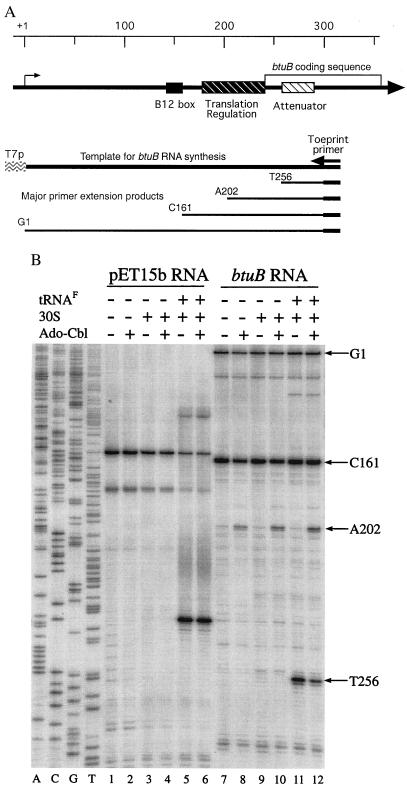
Figure 1.
(A Upper) Schematic structure of the btuB regulatory region. The positions of regulatory elements are indicated relative to the transcription start site and the start of the btuB coding sequence. The region from +1 to +315 is carried on the DNA template for synthesis of btuB RNA, with transcription from the T7 φ10 late promoter; the region for primer hybridization in the toe-print assay is indicated (Right). (Lower) The major primer extension products are identified by the nucleotide position of their 3′ end. (B Lower) Ribosome binding to lac and btuB RNA. RNA containing the translation initiation region from lacZ and btuB genes were synthesized by using T7 RNA polymerase and used in the primer extension inhibition assay. Incubation in the presence of tRNAfMet, 20 nM 30S ribosomal subunits, or 5 μM Ado-Cbl, as indicated (Upper), was for 10 min; primer extension was then carried out for 10 min after addition of AMV reverse transcriptase. The four lanes (Left) (A,C,G,T) are sequencing ladders for the btuB region; (Right) the positions of the last base of the primer extension fragments.
In the absence of ribosomes, primer extension on btuB RNA yielded the full-length run-off product, which is designated G1 to reflect the template position at the end of the fragment (Fig. 1B, lanes 7 to 12). A major product whose formation was independent of ribosomes ended at position C161 in the B12 box and is different from the 165-nt RNaseIII-dependent endonucleolytic product made in vivo (8). Some minor fragments were produced whose abundance increased with the age of the transcript and that could result from endonucleolytic cleavage or blockage of reverse transcriptase by RNA secondary structures.
In the presence of 30S ribosomal subunits and tRNAfMet, a termination product was produced at position T256 (Fig. 1B, lane 11). This fragment ends 15 nucleotides downstream of the AUG start codon at position 241, as expected for the binding of an initiating ribosome (17). Formation of this ribosomal toe-print product required both 30S subunits and initiator tRNAfMet. Weak ribosome binding to btuB RNA also occurred further upstream to the AUG at position 72, which is preceded by a purine-rich Shine–Dalgarno sequence and encodes a 19-residue peptide.
The presence of 5 μM Ado-Cbl caused reduced binding of 30S subunits to the btuB start site and a marked increase of a primer-extension product ending at position A202 (Fig. 1B, lane 12). Formation of this A202 product was independent of the presence of ribosomal subunits or tRNAfMet (lanes 8, 10, and 12). Once formed in the absence of Ado-Cbl, the btuB RNA-ribosome complex was not affected by subsequent addition of Ado-Cbl (not shown). When lacZ RNA transcribed from plasmid pET15b was used, a single toe-print product was formed in the presence of 30S subunits and tRNAfMet and was invariably unaffected by Ado-Cbl (Fig. 1B, lanes 1 to 6).
Binding of Ribosomes to btuB RNA Requires a Ribosome-Associated Factor(s).
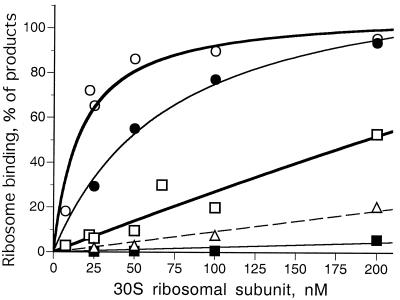
Ribosome subunits prepared with the usual high-salt-wash step were markedly and specifically impaired in their ability to bind btuB RNA. The ability of low- and high-salt-washed 30S subunits to bind to btuB and lacZ RNA was quantified as the percentage of the major extension products bound to the start codon. Low-salt-washed subunits bound lacZ RNA efficiently, with half-maximal binding at 15 to 20 nM 30S subunits (Fig. 2, open circles). In contrast, binding to btuB RNA (open squares) required at least a 10-fold higher concentration of subunits. This weaker binding to btuB RNA could reflect differences in the strength of the ribosome-binding site, the sequestration of the ribosome-binding region, or a property of the ribosomes themselves.
Figure 2.
Dependence of lac and btuB RNA binding on concentration of 30S ribosomal subunits. Ribosome binding is measured as the relative amount of the T256 product, expressed as a percentage of the major primer extension products. Binding of unwashed (open symbols) and high-salt-washed (filled symbols) ribosomes to lac (circles) and btuB (squares) RNA. Binding of unwashed ribosomes to btuB RNA in the presence of 5 μM Ado-Cbl is indicated (triangles). Curves are fit to a hyperbolic binding process.
Ribosomes washed with 0.5 M NH4Cl showed a moderate <3-fold reduction in binding to lacZ RNA (Fig. 2, filled circles) but at least 20-fold reduction in binding to btuB RNA (filled squares). When the material released by salt-washing was added back to washed ribosomes, the ability to form the ribosome toe-print T256 product was extensively restored (data not shown), suggesting that some ribosome-associated factor allows ribosomes to bind btuB RNA.
Inhibition of Ribosome Binding Is Cobalamin Specific.
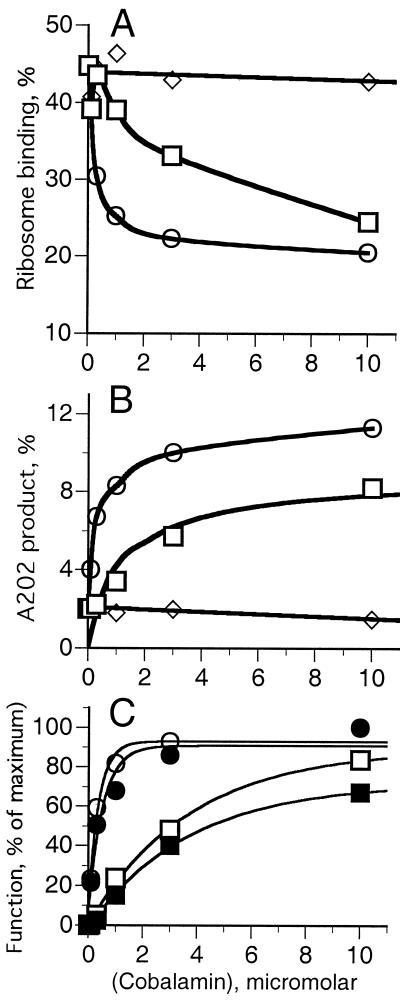
The effect of cobA/btuR mutations on btuB expression indicated that Ado-Cbl, but not CN-Cbl, was the regulatory signal (3). Hence, the toe-print assay was carried out in the presence of Ado-Cbl, methyl-Cbl, or CN-Cbl at 0.1 to 30 μM (Fig. 3). The amounts of the T256 toe-print product and the Cbl-dependent A202 product were measured as a percentage of the major primer-extension products. The binding of 30S subunits to the btuB start site was inhibited by Ado-Cbl, with half-maximal inhibition at around 0.3 μM (Fig. 3A, circles). Binding was inhibited by about 10-fold higher concentration of methyl-Cbl (squares), but 30 μM CN-Cbl (diamonds), aquo-Cbl, or dicyanocobinamide (data not shown) had no effect. This cobalamin specificity for inhibition of ribosome binding mirrors the genetic studies.
Figure 3.
Effect of cobalamins on the amount of btuB primer extension inhibition products. Binding reactions were carried out in the presence of the indicated concentrations of Ado-Cbl (circles), methyl-Cbl (squares), and CN-Cbl (diamonds), with 200 nM 30S subunits and 1 μM tRNAFMet. (A) Ribosome binding to the translation initiation site to generate fragment T256. (B) Formation of cobalamin-dependent A202 product. (C) Data from A (open symbols) and B (closed symbols) are plotted as the percent of the maximal effect to compare the effect of Ado-Cbl (circles) and methyl-Cbl (squares) on inhibition of ribosome binding (open symbols) and formation of fragment A202 (closed symbols) vs. cobalamin concentration.
Production of the A202 product showed a similar dependence on cobalamin type and concentration as the inhibition of ribosome binding (Fig. 3B). Ado-Cbl elicited formation of this product with half-maximal effect in the range of 0.3 μM. CN-Cbl did not stimulate its formation, and methyl-Cbl had an intermediate effect. Plotting the data for blockage of ribosome binding (Fig. 3C, open symbols) and formation of the A202 product (Fig. 3C, closed symbols) as a percentage of the maximal effect shows that both processes have a similar Cbl-concentration dependence, suggesting that both result from the same Cbl-binding event.
Mutations in the Leader Region Affect Ribosome Binding.
Mutations in the btuB leader region affect regulation (7). The effect of some mutations on expression of btuB-lacZ transcriptional and translational fusions and on ribosome binding in the toe-print assay were compared (Table 1).
Table 1.
Regulation of expression and ribosome-binding activity of variant btuB RNAs
| RNA sequence* | β-Galactosidase activity†
|
Primer extension products, %‡
|
||||||
|---|---|---|---|---|---|---|---|---|
| TRX fusion
|
TRL fusion
|
T256
|
A202
|
|||||
| Ado-Cbl− | Ado-Cbl+ | Ado-Cbl− | Ado-Cbl+ | Ado-Cbl− | Ado-Cbl+ | Ado-Cbl− | Ado-Cbl+ | |
| lac | 86.2 | 85.3 (1.0) | ||||||
| btuB wild type | 2,200 | 340 (6.5) | 405 | 3 | 32.5 | 14.1 (2.3) | 4.6 | 21.0 |
| Δ(B12 box) | 100 | 90 (1.1) | 3 | 4 | 10.7 | 9.9 (1.1) | <2 | <2 |
| Hairpin-2 | ||||||||
| M26 | 2,700 | 2400 (1.1) | 590 | 430 | 75.9 | 74.3 (1.0) | 2.4 | 11.6 |
| M27 | 2,870 | 2470 (1.1) | 720 | 560 | 58.7 | 31.9 (1.8) | 2.9 | 16.9 |
| M26.27 | 1,610 | 315 (5.1) | 220 | 17 | 53.8 | 25.8 (2.1) | 2.8 | 14.6 |
| Hairpin-1 | ||||||||
| M31 | 530 | 280 (1.9) | 26 | 5 | 16.3 | 11.5 (1.4) | 1.5 | 1.5 |
| M35 | 390 | 310 (1.3) | 22 | 3 | 13.5 | 13.3 (1.0) | 1.5 | 1.7 |
| M31.35 | 1,600 | 550 (2.9) | 95 | 1 | 28.4 | 11.8 (2.4) | 2.1 | 10.3 |
| M36 | 330 | 390 (0.8) | 13 | 6 | ND | ND | ||
| M31.36 | 650 | 420 (1.5) | 15 | 12 | ND | ND | ||
The sequence changes in the variant btuB leader regions are presented in Fig. 4.
β-Galactosidase activity was measured in cells carrying the variant btuB leader sequences present in btuB-lacZ transcriptional fusions expressed in plasmid pRS415 (TRX fusion) or in btuB-lacZ translational fusions expressed in plasmid pRS414 (TRL fusion). The insert in both plasmids carried the btuB sequences from positions −60 to +450. Cells were grown in the absence and presence of 5 μM Ado-Cbl, as indicated, and the repression ratio (−Ado-Cbl/+Ado-Cbl) is shown in parentheses for the transcriptional fusions.
The levels of the T256 and A202 products are presented as a percentage of the amounts of the major primer extension products. The results are average values of three experiments, one shown in Fig. 5. The value −Ado-Cbl/value +Ado-Cbl is shown in parentheses for the T256 ribosome-binding assay.
The B12 box (residues 141 to 162) in the btuB transcript is conserved among Cbl-regulated leader regions (2, 10). Its deletion results in marked reduction in btuB expression, loss of Cbl-dependent regulation, and decreased stability of btuB RNA in vivo (refs. 6 and 8; Table 1). The Δ(B12 box) deletion RNA showed reduced binding to 30S subunits, and the remaining binding was scarcely affected by Ado-Cbl (Table 1). This mutant RNA showed greatly reduced abundance of the C161 product, which ends within sequences deleted in this mutant, and the Cbl-dependent A202 product was not made (data not shown). Thus, the presence of the B12 box is essential for the Ado-Cbl-dependent processes, which lead to formation of the A202 product and to blockage of ribosome binding.
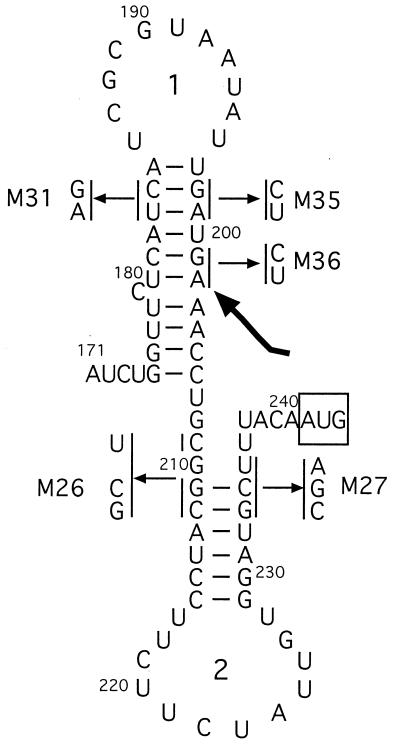
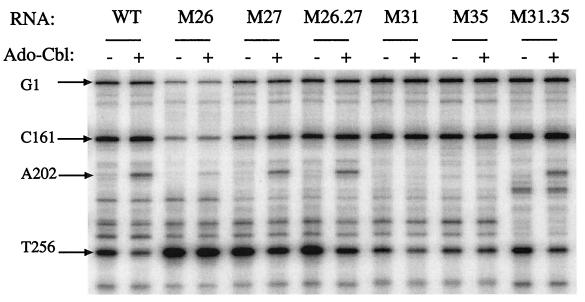
The rnafold structure-prediction program (GCG suite) indicates the potential for formation of two stem-loop structures in the btuB transcript between positions + 180 and + 240, termed hairpins 1 and 2 (Fig. 4). We showed previously that mutations affecting hairpin-2, which sequesters the Shine–Dalgarno sequence, interfere with Cbl-dependent regulation, and that compensatory changes that restore the potential for hairpin formation allowed near-normal regulation (8). Ribosome binding to these mutant RNAs was measured in the primer-extension assay (Fig. 5 and Table 1). Mutation M26 affecting the upstream anti-Shine–Dalgarno stem resulted in greatly increased ribosome binding, close to the level seen with lac RNA and not inhibited by Ado-Cbl. Mutation M27 in the downstream Shine–Dalgarno stem resulted in ribosome binding that was greater than the wild type but less than M26 RNA. Ribosome binding to M27 RNA was inhibited by Ado-Cbl but less than in wild type. RNA carrying either of these mutations was bound by high-salt-washed ribosomes, consistent with the role of a ribosome-associated factor in increasing access to the ribosome-binding site. RNA carrying the compensatory M26.27 double changes exhibited near-normal regulation of expression and showed ribosome binding that was still greater in extent than wild type but near normal in its reduction by Ado-Cbl. With all three RNAs, the A202 product was formed in the presence of Ado-Cbl but in reduced amounts because of the extensive blockage of reverse transcriptase by the ribosome.
Figure 4.
Predicted secondary structure of btuB RNA in the region from nucleotides 171 to 243. The sequence changes of mutants described in this study are indicated, along with the positions of hairpins 1 and 2. The start codon is boxed and the position of the 3′ end of the A202 product is indicated by the arrow.
Figure 5.
Ribosome binding to variant btuB RNAs. Primer extension assay was carried out with RNAs carrying the indicated mutations (described in Fig. 4). Ribosomal 30S subunits were present at 200 nM, and 5 μM Ado-Cbl was present as indicated. The identity of the primer extension products are indicated on the sides of the figure.
To evaluate the role of RNA hairpin-1 in btuB expression, the M31 and M35 mutations were designed to disrupt the stem (Fig. 4). Expression of btuB-lacZ transcriptional fusions carrying the M31 or M35 mutations was reduced by >75% and was comparable to the repressed wild type (Table 1). Regulation of β-galactosidase by Ado-Cbl was <2-fold. The M31 and M35 mutations reduced expression of btuB-lacZ translational fusions to about 5% of the wild-type basal level, and this activity was further reduced by Ado-Cbl down to the repressed wild-type level. RNAs carrying the M31 or M35 changes had low and unregulated ribosome binding, comparable to that of the repressed wild type (Fig. 5). In contrast, the compensatory M31.35 double mutant showed nearly complete restoration of btuB-lacZ expression and regulation, and near wild-type levels of ribosome binding and response to Ado-Cbl. The Ado-Cbl-dependent formation of the A202 product was completely lost in the M31 and M35 variants and was restored in the double mutant.
As a test of the specificity of the compensatory mutations, variant M36 was constructed carrying the same changes as the M35 mutation but 3 nucleotides downstream in the repeated sequence UGAUGA (Fig. 4). The M36 variant conferred the same regulatory phenotype as the M35 variant, but the M31.36 double mutation showed no restoration of btuB expression or regulation, confirming the importance of the base-pairing shown in Fig. 4.
Discussion
The cobalamin-dependent modulation of the cobalamin transport and biosynthetic genes operates by a complex process. The major site of regulation is at translation initiation, and the control of RNA levels through attenuation and changes in RNA stability is a consequence of the translational control (8, 9). The importance of two elements in the long and diverse leader regions of the Cbl-regulated genes, the B12 box and hairpin-2 that sequesters the Shine–Dalgarno sequence, has been shown (8–11).
The demonstration here that Ado-Cbl inhibits the binding of 30S ribosomal subunits to btuB RNA begins a biochemical dissection of cobalamin-dependent regulation. Our efforts to demonstrate ribosome binding to btuB RNA were routinely negative before the discovery that the usual salt-washing step for ribosome purification resulted in specific loss of ability to bind btuB RNA. The ability of the eluted material to restore btuB RNA-binding activity indicates the requirement for some ribosome-associated factor. Substoichiometric levels of this factor could account for the reduced binding of btuB RNA relative to lacZ RNA. The most likely situation is that the btuB Shine–Dalgarno sequence is sequestered in a hairpin, and the factor unfolds this structure to allow ribosome binding. The btuB Shine-Dalgarno sequence is intrinsically effective, as shown by the strong ribosome binding to M26 RNA, altered in the stem complementary to the Shine–Dalgarno sequence. A similar situation was seen for ribosome binding to the cbiA transcript from the cob operon (11). Although the HF-1 protein, encoded by the hfq gene, increases translation of rpoS RNA during stationary phase or osmotic stress by melting the RNA to increase ribosome access (18, 19), hfq mutants show normal btuB-lac expression (8). The identity of the ribosome-associated factor for btuB control is under study; preliminary results indicate that it is heat stable and RNase sensitive (X.N., unpublished results).
Ribosome binding to btuB RNA is inhibited 2- to 3-fold by Ado-Cbl. Half-maximal inhibition occurred in the range of 0.3 μM Ado-Cbl, which is very close to the concentration for inhibition of in vitro expression of a cbi-lac fusion protein (12). CN-Cbl, aquo-Cbl, and cobinamide had no effect, in agreement with the requirement for the cobA/btuR gene, which forms Ado-Cbl from CN-Cbl, for control of btuB-lacZ expression by CN-Cbl, but not by Ado-Cbl (3). Methyl-Cbl inhibited ribosome binding at a 10-fold higher concentration. The inhibition of ribosome binding by methyl-Cbl indicates that the axial adenosyl moiety is not required for signaling. Electrophoretic analysis of the methyl-Cbl sample revealed the presence of <1% Ado-Cbl (C. Bradbeer, personal communication). This specificity of regulation suggests a direct interaction of btuB RNA with Ado-Cbl. Consistent with the potential for specific RNA-Cbl recognition, Lorsch and Szostak (20) isolated RNA aptamers that bind CN-Cbl but not Ado-Cbl by repeated enrichment in the Selex procedure.
Although Ado-Cbl elicits a >25-fold modulation of expression of btuB translational fusions in vivo, its effect on ribosome binding was only 2- to 3-fold. Even accounting for the 3- to 5-fold decrease in the cellular level and stability of btuB RNA in response to Ado-Cbl, the inhibition of ribosome binding is less than expected for translation-level control. Substantial ribosome binding was also present with mutant btuB RNAs that have very low levels of expression, such as the Δ(B12 box) or the M31 and M35 mutants. These discrepancies can be explained by kinetic considerations if Ado-Cbl inhibits the steps that allow ribosome binding but does not affect the ribosome–RNA complex once formed. The instantaneous inhibition by Ado-Cbl in the toe-print assay may be comparable to the in vivo effect but appears to be less pronounced because of the trapping of the complex during the time periods for ribosome binding and primer extension.
Cbl derivatives that inhibit ribosome binding to btuB RNA also elicit the appearance of the A202 primer extension product with similar concentration dependence and specificity, indicating that both processes ensue from the same Cbl-binding event. The formation of the A202 product occurred in the absence of ribosomes or other bacterial proteins, indicating that Ado-Cbl must bind directly to btuB RNA. We cannot demonstrate binding of Ado-[57Co]Cbl to btuB RNA and there are no major changes in RNA secondary structure elicited by Ado-Cbl, as probed with conformation-dependent nucleases (X.N., unpublished observations). We propose that Ado-Cbl affects the contact of pseudoknots formed between RNA loops rather than causing a major rearrangement in the pairing of stem-loop structures. It is noteworthy that the A202 product arises from blockage between the two hairpins shown in Fig. 4.
The existence of those two hairpin structures was supported by the effect of mutations designed to affect formation of their stems. Disruption of hairpin-1 resulted in reduced transcription and translation of the btuB-lacZ fusions, greatly reduced ribosome binding to btuB RNA, and absence of the A202 product. This behavior is the same as that of the Δ(B12 box) variant. The compensatory M31.35 double mutation restored all activities almost to wild-type levels, showing that formation of this hairpin is crucial for btuB expression. Hairpin-2 sequesters the Shine–Dalgarno sequence and plays a different role, because mutations that disrupt this hairpin resulted in elevated and constitutive expression of btuB-lacZ fusions (8) and increased ribosome binding. The Ado-Cbl-dependent A202 product was formed in all mutants affecting hairpin-2, indicating that its formation requires integrity of hairpin-1 but not of hairpin-2. The compensatory double mutations in both hairpins did not quite restore wild-type behavior, indicating the importance of both the sequence and the secondary structure in this region.
These results indicate the operation of a mode of gene regulation in which the btuB leader RNA binds the specific effector molecule Ado-Cbl. This binding of Ado-Cbl inhibits binding of ribosomes to the RNA, probably by interfering with a conformation change in the RNA that depends on a ribosome-associated factor and allows access of the ribosome-binding region. Future experiments test the specific predictions of this model, such as the effect of Ado-Cbl on RNA structure and the identity of the ribosome-associated factor.
Acknowledgments
This work was supported by research grant GM19078 from the National Institute of General Medical Sciences and by funds from the University of Virginia. We appreciate strains and advice for ribosome preparation from Phang-Chen Tai.
Abbreviations
- Ado-Cbl
adenosylcobalamin
- CN-Cbl
cyanocobalamin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.130013897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.130013897
References
- 1.Landick R, Turnbough J, C L, Yanofsky C. In: Escherichia coli and Salmonella. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasonik B, Reznikoff W S, Riley M, Schaechfer M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol. Press; 1996. pp. 1263–1286. [Google Scholar]
- 2.Roth J R, Lawrence J G, Bobik T A. Annu Rev Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 3.Lundrigan M D, Kadner R J. J Bacteriol. 1989;171:154–161. doi: 10.1128/jb.171.1.154-161.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escalante-Semerena J C, Suh S-J, Roth J R. J Bacteriol. 1990;172:273–280. doi: 10.1128/jb.172.1.273-280.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ailion M, Roth J R. J Bacteriol. 1997;179:6084–6091. doi: 10.1128/jb.179.19.6084-6091.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franklund C V, Kadner R J. J Bacteriol. 1997;179:4039–4042. doi: 10.1128/jb.179.12.4039-4042.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lundrigan M D, Köster W, Kadner R J. Proc Natl Acad Sci USA. 1991;88:1479–1483. doi: 10.1073/pnas.88.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nou X, Kadner R J. J Bacteriol. 1998;180:6719–6728. doi: 10.1128/jb.180.24.6719-6728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravnum S, Andersson D I. Mol Microbiol. 1997;23:35–42. doi: 10.1046/j.1365-2958.1997.1761543.x. [DOI] [PubMed] [Google Scholar]
- 10.Richter-Dahlfors A A, Andersson D I. Mol Microbiol. 1992;6:743–749. doi: 10.1111/j.1365-2958.1992.tb01524.x. [DOI] [PubMed] [Google Scholar]
- 11.Richter-Dahlfors A A, Ravnum S, Andersson D I. Mol Microbiol. 1994;13:541–553. doi: 10.1111/j.1365-2958.1994.tb00449.x. [DOI] [PubMed] [Google Scholar]
- 12.Andersson D. FEMS Mircobiol Lett. 1995;125:89–94. doi: 10.1111/j.1574-6968.1995.tb07340.x. [DOI] [PubMed] [Google Scholar]
- 13.Silhavy T J, Berman M L, Enquist L W. Experiments with Gene Fusions. Plainview, NY: Cold Spring Harbor Lab. Press; 1984. [Google Scholar]
- 14.Simons R W, Houman F, Kleckner N. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 16.Spedding G. Isolation and Analysis of Ribosomes from Prokaryotes, Eukaryotes, and Organelles. Oxford: IRL; 1990. pp. 1–9. [Google Scholar]
- 17.Hartz D, McPheeters D S, Traut R, Gold L. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 18.Brown L, Elliott T. J Bacteriol. 1996;178:3763–3770. doi: 10.1128/jb.178.13.3763-3770.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muffler A, Fischer D, Hengge-Aronis R. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 20.Lorsch J R, Szostak J W. Biochemistry. 1994;33:973–982. doi: 10.1021/bi00170a016. [DOI] [PubMed] [Google Scholar]