Abstract
The lantibiotic (i.e., lanthionine-containing antibiotic) mersacidin is an antimicrobial peptide of 20 amino acids which is produced by Bacillus sp. strain HIL Y-85,54728. Mersacidin inhibits bacterial cell wall biosynthesis by binding to the precursor molecule lipid II. The structural gene of mersacidin (mrsA) and the genes for the enzymes of the biosynthesis pathway, dedicated transporters, producer self-protection proteins, and regulatory factors are organized in a biosynthetic gene cluster. For site-directed mutagenesis of lantibiotics, the engineered genes must be expressed in an expression system that contains all of the factors necessary for biosynthesis, export, and producer self-protection. In order to express engineered mersacidin peptides, a system in which the engineered gene replaces the wild-type gene on the chromosome was constructed. To test the expression system, three mutants were constructed. In S16I mersacidin, the didehydroalanine residue (Dha) at position 16 was replaced with the Ile residue found in the closely related lantibiotic actagardine. S16I mersacidin was produced only in small amounts. The purified peptide had markedly reduced antimicrobial activity, indicating an essential role for Dha16 in biosynthesis and biological activity of mersacidin. Similarly, Glu17, which is thought to be an essential structure in mersacidin, was exchanged for alanine. E17A mersacidin was obtained in good yields but also showed markedly reduced activity, thus confirming the importance of the carboxylic acid function at position 17 in the biological activity of mersacidin. Finally, the exchange of an aromatic for an aliphatic hydrophobic residue at position 3 resulted in the mutant peptide F3L mersacidin; this peptide showed only moderately reduced activity.
Mersacidin belongs to a group of antibacterial peptides which are designated lantibiotics because they contain the rare amino acids lanthionine and/or methyllanthionine (8). In contrast to peptide antibiotics produced by nonribosomal biosynthesis, lantibiotic precursors are encoded by structural genes and the rare amino acids are synthesized by posttranslational modification of the prepeptides. The various enzymes of the biosynthetic pathway, dedicated product transporters, producer self-protection proteins, and regulatory factors are encoded in biosynthetic gene clusters, which are found either on the chromosome or on mobile elements such as plasmids or transposons (25). Recently it has been shown that the biosynthetic gene cluster of mersacidin is located on the chromosome of the producer strain Bacillus sp. strain HIL Y-85,54728 (1).
Since lantibiotics are gene-encoded peptides, the exchange of single amino acids by site-directed mutagenesis is possible. However, in the case of lantibiotics it is not sufficient to simply express a single engineered gene in an Escherichia coli expression system, since the modification machinery is indispensable for the biosynthesis of the lanthionines and other modified amino acids. In addition, lantibiotics are bacteriocins, and therefore the producer strain has to be protected against its own product by the self-protection (or immunity) factors. Systems that meet these requirements have been successfully constructed for the expression of the lantibiotics subtilin (20), nisin (11, 12, 17, 19), Pep5 (4), epidermin (22), and mutacin II (9).
The activity spectrum of mersacidin includes several gram-positive bacteria, with staphylococci among the most sensitive organisms. The peptide acts by inhibition of bacterial cell wall biosynthesis via interaction with lipid II, the membrane-bound monomeric cell wall precursor (6, 7). Both the crystal and solution structures of mersacidin have been solved (24, 28). Mersacidin is a compact globular molecule. In methanol as well as in the crystal, the N terminus folds back in order to interact with the carboxyl group of the glutamic acid residue at position 17. Other lantibiotics that resemble mersacidin with respect to their mode of action are actagardine and ala(0)-actagardine, which are produced by Actinoplanes gabardinensis and Actinoplanes liguriae (23, 30). Moreover, with the exception of an exchange of didehydroalanine (Dha) for Ile at position 10 in actagardine and position 16 in mersacidin, the amino acid sequence of the third ring of mersacidin and the second ring of actagardine is conserved and has been suggested to constitute the binding pocket for lipid II (33). However the activity spectrum of actagardine is markedly different from that of mersacidin; actagardine is most active against streptococci and displays only low activity against staphylococcal species.
Here we describe the development of an expression system that enables the production of engineered mersacidin peptides, as well as the properties of the first mersacidin variants.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used for construction of the expression system are described in Table 1. The E. coli strains used for mutagenesis were grown on Luria-Bertani agar, Staphylococcus carnosus was maintained on blood agar or tryptone soy agar, and Bacillus strains were cultured on nutrient broth or tryptone soy agar, always in the presence of the appropriate antibiotics. All strains were stored at −70°C as glycerol cultures.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain(s) or plasmid | Characteristics | Reference or source |
|---|---|---|
| Bacillus sp. strain HIL Y-85,54728 TT | Producer strain of mersacidin | 8 |
| M. luteus ATCC4698 | Indicator strain | ATCC strain collection |
| S. pyogenes I-12662 | Indicator strain | This study |
| S. carnosus TM300 | Cloning host | 27 |
| E. coli JM 83 | Cloning host | 31 |
| E. coli 71/18, JM 109, HMS 174 | Cloning hosts | 26 |
| E. coli 71/18 BHM mutS | Cloning host for site directed mutagenesis | 32 |
| pTV0 | Temperature-sensitive gram-positive vector, CmR | 1 |
| pTV0[MCS] | pTV0 carrying a multiple cloning site | 15 |
| pS16I14 | pTV0 carrying S16ImrsA in its PvuII site | This study |
| pE17A14 | pTV0 carrying E17AmrsA in its PvuII site | This study |
| pF3L14 | pTV0[MCS] carrying F3LmrsA | This study |
Site-directed mutagenesis.
Site-directed mutagenesis was performed with a commercial phagemid system (Altered Sites in vitro mutagenesis system; Serva, Heidelberg, Germany) according to the instructions of the manufacturer. The 1.14-kb EcoRI-KpnI fragment containing the structural gene of mersacidin (mrsA) (3) and the 5′ part of mrsR1 (1) was cloned into the multiple cloning site of the pALTER-1 vector provided in the mutagenesis kit. The mutagenic oligonucleotides employed for site-directed mutagenesis are shown in Table 2. All mutated genes were confirmed by sequencing on an A.L.F. express sequencer (Pharmacia Biotech, Freiburg, Germany) or by Sequiserve (Vaterstetten, Germany).
TABLE 2.
Primers used in this study
| Primer | Sequencea | Product or function |
|---|---|---|
| PTV01 | CACAAAAAACAGGTACAAGAAAAAC | 442-bp fragment of pTV0[MCS] |
| PTV02 | ACCCCTCTTTCCATGTATTCACT | |
| NPTV02 | AAGGAGTTAAGTATTATGACTGGG | 818-bp product in absence of the plasmid |
| Primer 5′ | GGGTATATGCGTATAAACTTATG | |
| mrsAmutS16I | 5′d(GTTTGTACTCTAACTTTAGAATGTATTTGTTAATT)3′ | Mutagenesis |
| mrsAmutE17A | 5′d(TACTCTAACTTCTGCATGTATTTGTTAATTTG)3′ | Mutagenesis |
| mrsAmutF3L | 5′d(GAAGCAGCATGTACGCTTACATTGCCTGG)3′ | Mutagenesis |
Mismatches of the oligonucleotides used for mutagenesis are underlined.
DNA was prepared by using Qiagen genomic tips, Qiaprep spin miniprep kits, or Qiagen plasmid minikits according to the recommendations of the supplier (Qiagen, Hilden, Germany). pTV0/pTV0[MCS] or pTV0/pTV0[MCS] recombinant plasmids carrying engineered mrsA genes were purified from 50 ml of S. carnosus culture by using the HiSpeed plasmid midikit (Qiagen). General protocols were used for cloning strategies and enzymatic DNA modification (26). Digested DNA fragments were eluted from agarose gels with the QIAquick gel extraction kit (Qiagen).
The Bacillus sp. and Staphylococcus carnosus TM300 were transformed by protoplast transformation (13, 14), while E. coli strains were transformed by electroporation. Clones were screened by plasmid preparations or, in the case of Bacillus, by PCR (Table 2) performed with AGS Gold DNA polymerase (Hybaid-AGS, Heidelberg, Germany) in a PCRExpress thermocycler (Hybaid) with genomic DNA as a template. For competitive nucleotide priming, a pair of Mut and Umut oligonucleotide primers was synthesized for each mutation. These primers served as backward primers, with the 3′ terminus annealing to the base which had been chosen for mutagenesis. Therefore, the sequences of the Mut and Umut primers differed only by a single base located at the 3′ end. In case of the Mut primer, this base was complementary to the mutation, whereas the Umut primer was complementary to the wild-type sequence. The two primers were included into two separate PCRs. The forward primer (primer 5′) was added to both reaction mixtures and annealed to the sequence downstream of mrsE that is present only on the chromosome and not on the plasmid. Since Taq polymerase does not possess proofreading activity, the enzyme did not work efficiently with the primer whose 3′ end was mismatched. Therefore, only the primer whose 3′ end paired correctly with that copy of the mrsA gene which was located directly downstream of mrsE yielded a PCR product.
Purification of engineered peptides.
For purification of S16I mersacidin, 400 ml of culture supernatant were applied to a 35-ml XAD Serdolite AD2 (Serva) column, the column was washed with water-50% methanol containing 50 mM potassium phosphate buffer (pH 7), and the peptide was eluted with acetonitrile containing 0.1% trifluoroacetic acid. The eluate was concentrated by evaporation and applied in several runs to a POROS R2 10 column (Applied Biosystems, Lincoln, Calif.) at 3 ml/min and eluted with the following gradient (eluent A, water-0.1% trifluoroacetic acid; eluent B, acetonitrile-0.1% trifluoroacetic acid): 0 min, 10% B; 3 min, 10% B; 15 min, 30% B; 23 min, 40% B; and 25 min, 100% B. The adsorption of the eluate was monitored at 220 nm. Since S16I mersacidin could not be identified directly by mass spectrometry (MS) of the crude lyophilized fractions, the fractions were analyzed by high-pressure liquid chromatography (HPLC)-MS. Lyophilized fractions were dissolved in 30% acetonitrile, and a 20-μl aliquot was separated on a 100- by 1-mm column of Gromsil ODS-5ST (Grom Analytik+HPLC, Herrenberg, Germany) in a continuous gradient of 10 to 90% acetonitrile containing 0.1% trifluoroacetic acid over 30 min at a constant flow rate of 60 μl/min. Eluant was fed directly into a VG Quattro II triple-quadropole mass spectrometer (MicroMass) fitted with an electrospray ionization interface, using air as the carrier gas. The mass spectrometer was operated in positive mode, and data were collected and analyzed with the software provided by the manufacturer (MassLynx). The fractions containing S16I mersacidin were rechromatographed in order to obtain sufficient material for the determination of MICs. After lyophilization, the mass of the peptide was analyzed by MS as described above.
E17A mersacidin and F3L mersacidin were purified from 100 ml of culture supernatant, which was sterilized by filtration, lyophilized, dissolved in about 30 ml of 30% acetonitrile-0.1% trifluoroacetic acid, and applied in several runs to a POROS R2 20 column. The mass was analyzed with an API 2000 Triple Quad MS (Applied Biosystems).
For the agar diffusion assay, the culture supernatant was sterilized by filtration, and aliquots were pipetted into wells in a blood agar plate that had been overlaid with soft agar containing Micrococcus luteus as indicator strain.
Determination of the MICs of mersacidin and engineered mersacidin peptides was performed by use of a microtiter plate assay with half-concentrated Mueller-Hinton broth and an inoculum of 105 cells of M. luteus per well, which was read after 16 h of incubation at 37°C.
RESULTS
Construction of an expression system for variant mersacidin peptides.
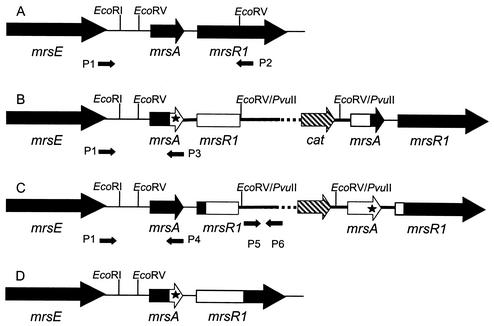
For expression of mutated peptides, we constructed a system that relies on replacement of the wild-type gene by the engineered gene on the chromosome. To this end, the EcoRV fragment carrying the engineered mrsA gene was ligated into the temperature-sensitive vector pTV0 or its derivative pTV0[MCS], both of which carry a chloramphenicol resistance gene, and was subcloned in S. carnosus at 30°C. The recombinant plasmid was then transformed into the wild-type producer strain (Fig. 1A), which was also cultured at 30°C. Since pTV0 and pTV0[MCS] replicate at a very low copy number, the colonies obtained on regeneration agar were screened by PCR with primers pTV01 and pTV02 (Table 2; Fig. 1). Subsequently, colonies which had integrated the plasmid into their chromosome were selected by a temperature shift to 45°C and growth on agar containing chloramphenicol. These colonies carried two copies of mrsA, the wild-type gene and the mutagenized gene, which are separated by the vector. It has to be kept in mind that the mutagenized mrsA gene differs from the wild-type gene by only a single or double base exchange, i.e., a point mutation. Therefore, two different kinds of mutant strains could be expected from the integration of the plasmid into the chromosome of the producer strain by a single-crossover event. When the crossover event takes place upstream of the point mutation, the mutant gene will be located downstream of mrsE, as shown in Fig. 1B. On the other hand, the wild-type gene will remain downstream of mrsE when the crossover event takes place downstream of the point mutation, as shown in Fig. 1C. The locations of the mutant and wild-type genes were checked by competitive nucleotide priming with Taq polymerase. However, these clones did not show any mersacidin production, irrespective of orientation of the two mrsA genes. The reason for this phenomenon was not investigated, but it might be the result of a polar effect caused by the insertion of the plasmid into the biosynthetic gene cluster.
FIG. 1.
Construction of clones that are produced by integration and subsequent excision of a plasmid carrying an engineered mrsA gene and the N terminus of mrsR1 (white open reading frames, thick line) into the chromosome (black open reading frames, thin line). The mutation is marked by an asterisk. (A) Wild type. (B and C) The plasmid (thick line, cat resistance gene) has been integrated into the chromosome, and the crossover took place either upstream of the mutation (B) or downstream of the mutation (C). (D) The plasmid has been excised by the second homologous recombination step, which took place either downstream (B) or upstream (C) of the mutation in mrsA, resulting in an exchange of the wild-type mrsA gene for the engineered gene. The primers are shown by arrows: P1, primer 5′; P2, NPTV0; P3, Mut; P4, Umut; P5, PTV01; P6, PTV02.
In order to obtain production of engineered peptides, we cultured the clones for about 100 generations in the absence of chloramphenicol and screened for colonies that had lost the chloramphenicol resistance by a second crossover but had kept the engineered mrsA gene (Fig. 1D). Colonies that had not grown on agar containing chloramphenicol after 2 days of incubation were screened by PCR with primers specific for the vector (PTV01 and PTV02), primers which anneal on the chromosome upstream and downstream of the EcoRV fragment containing mrsA (NPTV02 and primer 5′), and the specific Mut and Umut primers (Fig. 1). When the PCR indicated that the colony had integrated the engineered gene correctly into the biosynthetic gene cluster and had lost pTV0[MCS], the peptide was produced and purified as described above. The purified peptide was checked by MS for the correct modification.
Identification of amino acid residues essential for activity of mersacidin.
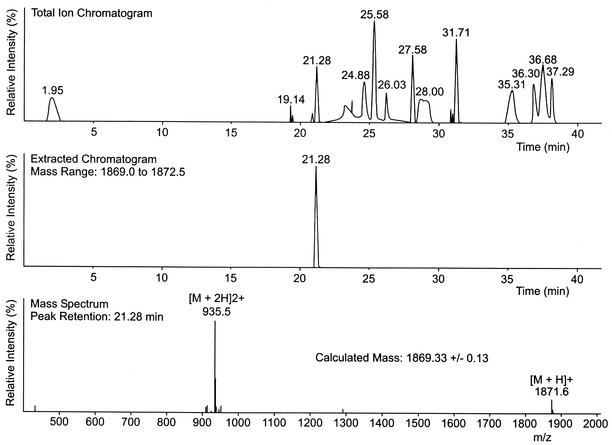
The first peptide to be produced by the method described above was S16I mersacidin, which contains the Ile residue that is found in the related lantibiotic actagardine instead of the Dha residue present in mersacidin (Fig. 2). Since no antibacterial activity and no mersacidin peak were detected in the supernatant of the correct clone after 72 h of culture, the peptide either was inactive, was produced only in small amounts, or was not synthesized at all. Loss of production has been described for some engineered nisin, epidermin, and Pep5 peptides (18). Therefore, the culture supernatant was fractionated on XAD and POROS R2 columns and the fractions eluting between 31 and 37% acetonitrile were screened by HPLC-MS (Fig. 3). The peptide with the correct mass of 1,869 Da was detected at 36% acetonitrile and thereafter was purified from 400 ml of culture supernatant with a yield lower than 0.5 mg/liter. The determination of the MIC showed that the activity of S16I mersacidin against M. luteus was nearly 1,000-fold lower than that of the wild-type peptide (mersacidin MIC, 0.1 mg/liter; S16I mersacidin MIC, 87.5 mg/liter). The antibacterial activities of HPLC fractions of culture supernatant were checked against 25 Streptococcus strains (belonging to Lancefield groups A, G, and B), two enterococci, and Micrococcus flavus, but no inhibition zones could be discerned on Mueller-Hinton agar plates. It was also inactive in a broth microdilution test against a clinical Streptococcus pyogenes isolate (I-12662) with good sensitivity to wild-type mersacidin (mersacidin MIC, 0.78 mg/liter; S16I mersacidin MIC, >25 mg/liter).
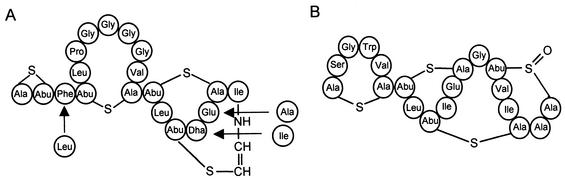
FIG. 2.
(A) Structure of mersacidin (8) and exchanges that were introduced by site-directed mutagenesis. (B) Structure of the closely related lantibiotic actagardine (34).
FIG. 3.
HPLC-electrospray ionization-MS of S16I mersacidin. (A) Total ion chromatogram obtained after separation of the fraction containing S16I mersacidin. (B) Extracted chromatogram for peaks with masses in the range of 1869.0 to 1872.5 Da. (C) Mass determined for the peak detected at 21.28 min (expected mass for S16I mersacidin, 1,869.32 Da).
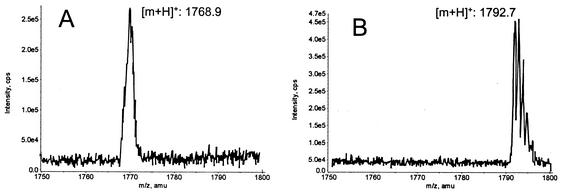
E17A mersacidin was constructed in order to assess the importance of the glutamic acid residue at position 17 for the antimicrobial activity of mersacidin. Culture supernatant of the E17A mersacidin producer did not show antimicrobial activity against M. luteus. However, the peptide could be identified by double detection at 220 and 266 nm during HPLC runs, since due to the NH-CH=CH-S structure in the C terminus of mersacidin, the extinction at 266 nm is about half of that at 220 nm. E17A mersacidin was produced in greater amounts than S16I mersacidin, and the purification yielded 0.41 mg of pure peptide from 100 ml of culture supernatant. The peptide with the correct mass (1,767 Da) (Fig. 4A) was inactive against M. luteus, with an MIC that was higher than 175 mg/liter (wild-type mersacidin MIC, 0.195 mg/liter). The yield of purified F3L mersacidin was 3.4 mg/liter of culture. The MIC of this peptide with the correct mass of 1,791 Da (Fig. 4B) against M. luteus was 12.5 mg/liter (wild-type mersacidin MIC, 0.195 mg/liter).
FIG. 4.
MS of E17A mersacidin (expected mass, 1767.2 Da) (A) and F3L mersacidin (expected mass, 1791.2 Da) (B).
DISCUSSION
For successful production of a mutagenized lantibiotic, the peptide must be expressed and correctly modified to contain the rare amino acids, e.g., methyllanthionine, didehydroalanine, and S-aminovinylmethylcysteine for mersacidin. The modification reactions are catalyzed by the modifying enzymes MrsM (dehydration and thioether formation) and MrsD (oxidative decarboxylation of Cys20) (1). In the study presented here, a gene replacement strategy was successfully employed for production of mutated mersacidin peptides. Compared to a complementation approach, it is advantageous in that the relative gene dosage of the structural gene versus the modification-export machinery (MrsD, MrsM, and MrsT) is kept at the wild-type level. Similarly, in the case of nisin yields were also higher when a gene replacement system was employed than when a complementation approach was used (11). In the present study, the rates of production of the engineered mersacidin peptides varied, but all three peptides were produced at lower rates than wild-type mersacidin, where yields of 5 to 10 mg/liter are expected. The yield was especially low for S16I mersacidin; in this peptide, the serine residue at position 16 had been exchanged for isoleucine, thereby preventing the modification reaction of Ser16 to dehydroalanine. It is possible that this exchange and the hydrophobic side chain of the Ile residue introduced interfere with the dehydration of other residues and/or thioether formation. Therefore, the lower production rates, especially of S16I mersacidin, may be caused by inhibition of the enzymes of the modification machinery. Similar observations have also been described for other lantibiotics, especially after exchange of hydroxy amino acids; e.g., T20A Pep5 was not completely modified and was produced only at low rates (5). Furthermore, some engineered peptides that had lost hydroxy amino acids (S3N epidermin and T23A nisin A) were not produced at all (18, 22). Incomplete dehydration, indicated by the presence of peptides with molecular masses corresponding to hydrated variants (having an additional 18 Da), as found with the expression system for engineering of the lantibiotic Pep5 (4), was not seen in this system, and incompletely modified peptides were not detected. However, Bacillus is a potent producer of extracellular proteases, and one function of the ring structures in lantibiotics is stabilization of the peptides against the activity of proteases (5). An incompletely modified mersacidin peptide, lacking the stabilizing ring structures, would most probably be quickly degraded by one of the numerous extracellular proteases of the producer strain. This phenomenon may also contribute to the low yields of S16I mersacidin.
The exchanges S16I and E17A are situated in the third ring of mersacidin, which is conserved between mersacidin and the lantibiotic actagardine from A. gabardinensis (Fig. 2) (8, 34) and includes the conserved Glu residue. Actagardine also inhibits cell wall biosynthesis (29) and is able to strongly inhibit the binding of mersacidin to M. luteus cells, indicating that both lantibiotics compete for a similar binding site (6). However, actagardine and its variant Ala(0)-actagardine show a different activity spectrum from mersacidin (2, 30). Whereas mersacidin displays high antibacterial activity against M. luteus and Staphylococcus aureus, actagardine and its variant show good activity against streptococci. The three-dimensional structure of actagardine was elucidated by nuclear magnetic resonance (33) and seems to be rather rigid. The only hydrophilic amino acids of actagardine, Ser2 and Glu11, are part of a putative binding pocket that is formed by Ser2-Ala1-S-Ala6-α-aminobutyric acid 7 (Abu7)-S-Ala12-Glu11. This structure is located in the part of actagardine that is conserved compared to mersacidin, and it was speculated that these residues might form the binding site for the antibacterial target of mersacidin and actagardine, the cell wall precursor lipid II (6, 33). In the conformation of mersacidin that is adopted in methanol, the N terminus is folded back into the center of the molecule and the amino group of Ala1 interacts with the carboxylate group of Glu17 (24), and in the crystal structure an additional interaction with Gly7 was observed (28). The polar interactions of Glu17 and Ala1 or Gly7 varied in their strength in the six slightly different structures that constitute the hexameric crystal, indicating a rather loose contact. This observation led to a speculative model in which the carboxylate group interacts with Ala1 or Gly7 in the crystal but upon encountering its target shifts to binding a ligand directly upon lipid II (28). Recent nuclear magnetic resonance experiments have indeed shown that mersacidin is a flexible molecule and that the three-dimensional structure that is adopted by mersacidin in methanol-water mixtures changes upon contact with dodecylphosphocholine micelles and then shifts again upon addition of lipid II. The first shift is especially strong in Abu13, Abu15, and Dha16; the second shift is especially pronounced for Abu13, which seems to act as a hinge region, and Gly7 and Glu17 (16). From these structural data, it is most likely that Glu17 is one of the key residues in mersacidin. The loss of antibacterial activity in E17A mersacidin demonstrated here confirms this hypothesis. In contrast, the low antibacterial activity displayed by S16I mersacidin was not expected from the comparison of the primary structures of mersacidin and actagardine. However, these results indicate that in mersacidin, either Dha16 is either essential for the interaction with lipid II or the presence of the bulky Ile residue in position 16 influences the position of the neighboring essential residue Glu17. Obviously, the primary structure of actagardine cannot simply be superimposed on mersacidin even if the amino acid sequence of both rings is otherwise conserved. In contrast, the comparatively better activity of F3L mersacidin shows that exchanges seem to be more compatible with antibacterial activity in the two N-terminal rings of mersacidin.
The mode of action of mersacidin and actagardine is not covered by any other antibiotics, since both substances interact with a novel target site that is present only on lipid II and do not bind to lipid I or intracellular peptidoglycan precursors (6). Although ramoplanin also binds to lipid II, this lipoglycodepsipeptide recognizes lipid I and UDP-N-acetylmuramylpentapeptide as well (10), and vancomycin and teicoplanin target the terminal d-Ala-d-Ala residues of the peptide side chain of lipid II. The promising activities of the lantibiotics against Staphylococcus and Streptococcus make them lead structures for the design of novel antibiotics against these problematic pathogens. Chemical derivatives of actagardine have been synthesized (21, 30), but chemical modification is restricted to the C and N termini and the hydroxyl group of Ser5. In mersacidin, only the N terminus and the Glu residue represent reactive groups. The development of a system for site-directed mutagenesis of mersacidin is an important step towards the design of novel antibiotics, since it allows targeted exchange of amino acids in the structure of mersacidin.
Acknowledgments
This work was supported by the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (grant 01KI9705), the Deutsche Forschungsgemeinschaft (grants Bi 504/1-2 and Bi 504/1-3), and the BONFOR program of the Medizinische Einrichtungen, Universität Bonn.
We are grateful to Aventis Pharma GmbH (Frankfurt, Germany) for providing mersacidin and the mersacidin producer strain. We thank S. Schmitz for expert technical assistance.
REFERENCES
- 1.Altena, K., A. Guder, C. Cramer, and G. Bierbaum. 2000. Biosynthesis of the lantibiotic mersacidin: organization of a type B lantibiotic gene cluster. Appl. Environ. Microbiol. 66:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arioli, V., M. Berti, and L. G. Silvestri. 1976. Gardimycin, a new antibiotic from Actinoplanes. III. Biological properties. J. Antibiot. 29:511-515. [DOI] [PubMed] [Google Scholar]
- 3.Bierbaum, G., H. Brötz, K. P. Koller, and H. G. Sahl. 1995. Cloning, sequencing and production of the lantibiotic mersacidin. FEMS Microbiol. Lett. 127:121-126. [DOI] [PubMed] [Google Scholar]
- 4.Bierbaum, G., M. Reis, C. Szekat, and H. G. Sahl. 1994. Construction of an expression system for engineering of the lantibiotic Pep5. Appl. Environ. Microbiol. 60:4332-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bierbaum, G., C. Szekat, M. Josten, C. Heidrich, C. Kempter, G. Jung, and H. G. Sahl. 1996. Engineering of a novel thioether bridge and role of modified residues in the lantibiotic Pep5. Appl. Env. Microbiol. 62:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brötz, H., G. Bierbaum, K. Leopold, P. E. Reynolds, and H. G. Sahl. 1998. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob. Agents Chemother. 42:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brötz, H., G. Bierbaum, P. E. Reynolds, and H. G. Sahl. 1997. The lantibiotic mersacidin inhibits peptidoglycan biosynthesis at the level of transglycosylation. Eur. J. Biochem. 246:193-199. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee, S., S. J. Lad, M. S. Phansalkar, R. H. Rupp, B. N. Ganguli, H. W. Fehlhaber, and H. Kogler. 1992. Mersacidin, a new antibiotic from Bacillus. Fermentation, isolation, purification and chemical characterization. J. Antibiot. 45:832-838. [DOI] [PubMed] [Google Scholar]
- 9.Chen, P., J. Novak, M. Kirk, S. Barnes, F. Qi, and P. W. Caufield. 1998. Structure-activity study of the lantibiotic mutacin II from Streptococcus mutans T8 by a gene replacement strategy. Appl. Environ. Microbiol. 64:2335-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cudic, P., D. Behenna, J. Kranz, R. Kruger, A. Wand, Y. Veklich, J. Weisel, and D. McCafferty. 2002. Functional analysis of the lipoglycodepsipeptide antibiotic ramoplanin. Chem. Biol. 9:897. [DOI] [PubMed] [Google Scholar]
- 11.Dodd, H. M., N. Horn, C. J. Giffard, and M. J. Gasson. 1996. A gene replacement strategy for engineering nisin. Microbiology 142:47-55. [DOI] [PubMed] [Google Scholar]
- 12.Dodd, H. M., N. Horn, Z. Hao, and M. J. Gasson. 1992. A lactococcal expression system for engineered nisins. Appl. Environ. Microbiol. 58:3683-3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Götz, F., and B. Schumacher. 1987. Improvement of protoplast transformation in Staphylococcus carnosus. FEMS Microbiol. Lett. 40:285-288. [Google Scholar]
- 14.Grosch, J. C., and K. L. Wollweber. 1982. Transformation of Bacillus licheniformis and Bacillus amyloliquefaciens protoplasts by plasmid DNA, p. 97-105. In U. N. Streips, H. S. Goodgal, W. R. Guild, and G. A. Wilson. (ed.), Genetic exchange. Marcel Dekker, New York, N.Y.
- 15.Guder, A., T. Schmitter, I. Wiedemann, H. G. Sahl, and G. Bierbaum. 2002. Role of the single regulator MrsR1 and the two-component system MrsR2/K2 in the regulation of mersacidin production and immunity. Appl. Environ. Microbiol. 68:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu, S. T., E. Breukink, G. Bierbaum, H. G. Sahl, B. De Kruijff, R. Kaptein, N. A. Van Nuland, and A. M. Bonvin. 2003. NMR study of mersacidin and lipid II interaction in DPC micelles: conformational changes are a key to antimicrobial activity. J. Biol. Chem. 278:13110-13117. [DOI] [PubMed]
- 17.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 18.Kuipers, O. P., G. Bierbaum, B. Ottenwälder, H. M. Dodd, N. Horn, J. Metzger, T. Kupke, V. Gnau, R. Bongers, P. van den Bogaard, H. Kosters, H. S. Rollema, W. M. de Vos, R. J. Siezen, G. Jung, F. Götz, H. G. Sahl, and M. J. Gasson. 1996. Protein engineering of lantibiotics. Antonie Leeuwenhoek 69:161-169. [DOI] [PubMed] [Google Scholar]
- 19.Kuipers, O. P., H. S. Rollema, R. Zell, H. J. Boot, R. J. Siezen, and W. M. de Vos. 1992. Engineering dehydrated amino acid residues in the antimicrobial peptide nisin. J. Biol. Chem. 267:24340-24346. [PubMed] [Google Scholar]
- 20.Liu, W., and J. N. Hansen. 1992. Enhancement of the chemical and antimicrobial properties of subtilin by site-directed mutagenesis. J. Biol. Chem. 267:25078-25085. [PubMed] [Google Scholar]
- 21.Malabarba, A., R. Pallanza, M. Berti, and B. Cavalleri. 1990. Synthesis and biological activity of some amide derivatives of the lantibiotic actagardine. J. Antibiot. 43:1089-1097. [DOI] [PubMed] [Google Scholar]
- 22.Ottenwälder, B., T. Kupke, S. Brecht, V. Gnau, J. Metzger, G. Jung, and F. Götz. 1995. Isolation and characterization of genetically engineered gallidermin and epidermin analogs. Appl. Environ. Microbiol 61:3894-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parenti, F., H. Pagani, and G. Beretta. 1976. Gardimycin, a new antibiotic from Actinoplanes. I. Description of the producer strain and fermentation studies. J. Antibiot. 29:501-506. [DOI] [PubMed] [Google Scholar]
- 24.Prasch, T., T. Naumann, R. L. Markert, M. Sattler, W. Schubert, S. Schaal, M. Bauch, H. Kogler, and C. Griesinger. 1997. Constitution and solution conformation of the antibiotic mersacidin determined by NMR and molecular dynamics. Eur. J. Biochem. 244:501-512. [DOI] [PubMed] [Google Scholar]
- 25.Sahl, H. G., and G. Bierbaum. 1998. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from gram-positive bacteria. Annu. Rev. Microbiol. 52:41-79. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Schleifer, K. H., and U. Fischer. 2002. Description of a new species of the genus Staphylococcus: Staphylococcus carnosus. Int. J. Sys. Bacteriol. 32:153-156. [Google Scholar]
- 28.Schneider, T. R., J. Karcher, E. Pohl, P. Lubini, and G. M. Sheldrick. 2000. Ab initio structure determination of the lantibiotic mersacidin. Acta Crystallogr. D 56:705-713. [DOI] [PubMed] [Google Scholar]
- 29.Somma, S., W. Merati, and F. Parenti. 1977. Gardimycin, a new antibiotic inhibiting peptidoglycan synthesis. Antimicrob. Agents Chemother. 11:396-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vertesy, L., W. Aretz, A. Bonnefoy, E. Ehlers, M. Kurz, A. Markus, M. Schiell, M. Vogel, J. Wink, and H. Kogler. 1999. Ala(0)-actagardine, a new lantibiotic from cultures of Actinoplanes liguriae ATCC 31048. J. Antibiot. 52:730-741. [DOI] [PubMed] [Google Scholar]
- 31.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertional mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 32.Zell, R., and H. J. Fritz. 1987. DNA mismatch-repair in Escherichia coli counteracting the hydrolytic deamination of 5-methyl-cytosine residues. EMBO J. 6:1809-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zimmermann, N., and G. Jung. 1997. The three-dimensional solution structure of the lantibiotic murein-biosynthesis-inhibitor actagardine determined by NMR. Eur. J. Biochem. 246:809-819. [DOI] [PubMed] [Google Scholar]
- 34.Zimmermann, N., J. W. Metzger, and G. Jung. 1995. The tetracyclic lantibiotic actagardine. 1H-NMR and 13C-NMR assignments and revised primary structure. Eur. J. Biochem. 228:786-797. [PubMed] [Google Scholar]