Abstract
Certain rpsL (which encodes the ribosomal protein S12) mutations that confer resistance to streptomycin markedly activate the production of antibiotics in Streptomyces spp. These rpsL mutations are known to be located in the two conserved regions within the S12 protein. To understand the roles of these two regions in the activation of silent genes, we used site-directed mutagenesis to generate eight novel mutations in addition to an already known (K88E) mutation that is capable of activating antibiotic production in Streptomyces lividans. Of these mutants, two (L90K and R94G) activated antibiotic production much more than the K88E mutant. Neither the L90K nor the R94G mutation conferred an increase in the level of resistance to streptomycin and paromomycin. Our results demonstrate the efficacy of the site-directed mutagenesis technique for strain improvement.
It was previously reported that certain mutations that confer resistance to streptomycin or paromomycin can activate antibiotic production (actinorhodin and undecylprodigiosin) in Streptomyces coelicolor A3(2) and Streptomyces lividans 66 (13, 15). These mutations are located in the rpsL gene, which encodes the ribosomal protein S12, and can effectively activate antibiotic production even in the genetic background of relA and relC (11, 15), the mutations of which are known to severely inhibit production of antibiotics in wild-type cells due to a failure to produce ppGpp (2, 11). The introduction of the mutation (str) causing streptomycin resistance is also effective in enhancing antibiotic production in other bacteria, including members of the genera Bacillus and Pseudomonas (7). Recently, the str mutation was also shown to confer tolerance of organic chemicals to Pseudomonas putida (6). The level of antibiotic production depended on both the type and position of amino acid substitution in the protein. The replacement of Lys-88 by Glu was the most effective in increasing actinorhodin production of S. lividans and S. coelicolor A3(2), while a replacement of Lys-43 by Asn had no effect on antibiotic production (15). The rpsL mutations that were found in S. coelicolor A3(2) and S. lividans that confer resistance to streptomycin are K43N, K43R, K43T, K88E, K88R, and P91S (see references 5, 8, 13, and 15). Of these, only two (K88E and P91S) prominently increased antibiotic production. It thus appears that certain mutations around the K88E region may distinctively affect antibiotic production. Since most of those mutations are not likely to confer resistance to streptomycin, it would be difficult or impossible to select for such mutations by resistance to the drug. Therefore, we used site-directed mutagenesis to generate rpsL mutations that may have an effect on antibiotic production.
Plasmids were constructed and amplified in Escherichia coli strain DH5α. The strain was grown at 37°C in Luria-Bertani medium. S. lividans TK21 and its derivatives were cultured at 30°C on YEME, R2YE, TSB(10), GYM, or R4 medium (15). For selection of transformants, the media were supplemented with 50 μg of ampicillin or thiostrepton per ml. The plasmid pUC18 was purchased from Takara Shuzo. The single-copy-number plasmid pV1, an E. coli-Streptomyces shuttle plasmid, was constructed by Kawamoto et al. (9). General techniques for plasmid isolation and transformation with Streptomyces and E. coli have been previously described by Hopwood et al. (10) and Sambrook et al. (14), respectively. PCR amplification was carried out by using PE480 and PE9700 (PE Biosystems), and DNA sequencing was performed by using the DNA sequencer ABI310 (PE Biosystems).
Primers were designed by using the data obtained from the S. coelicolor genome sequence (1). Total DNA was isolated from S. lividans TK21, and a 684-bp DNA fragment containing a putative promoter region (300 bp) and a coding region for the rpsL gene was amplified with pS12BmN (5′-CGGGATCCCGTACTTCGTCCGCCACGACACGGC) and pS12BmC (5′-CCGCGGATCCCGCTTACTTCTCCTTCTTGGCGCCG) primers. The amplified fragment was inserted into the BamHI site of pUC18 to obtain the recombinant plasmid pU-TK21, which carries the rpsL gene in the same direction as the lacZ gene. pU-TK21 was used as a template for the following PCR mutagenesis experiments: the primers used for mutagenesis are listed in Table 1. PCR experiments were performed as illustrated in Fig. 1. After the second PCR, the products digested with EcoRI and HindIII were ligated into pUC18 to generate pUCmut1 through -mut9. The plasmids thus obtained were sequenced, and nine clones that had the desired mutation without a PCR error were selected. The wild-type and mutant rpsL genes were excised by digesting with BamHI and were ligated with the pV1 vector to generate plasmids, i.e., pVWT, pVK43R, pVR86V, pVV87K, pVK88G, pVD89R, pVL90K, pVG92D, pVR94G, and pVK88E. These plasmids were then used to transform S. lividans TK21. The transformants were selected with thiostrepton (50 μg/ml).
TABLE 1.
Primers used for site-directed mutagenesisa
| Primer | Sequence |
|---|---|
| pMutHin | 5′-GGCCAGTGCCTAGCTTACAT-3′ |
| pMK43R | 5′-GACCACCCCGAGGAAGCCGAACTC-3′ |
| pMR86L | 5′-GCGCGGCGGCCTTGTGAAGGACCT-3′ |
| pMV87K | 5′-GGCGGCCGTAAGAAGGACCTGC-3′ |
| pMK88E | 5′-GGCCGTGTGGAGGACCTGCCGGGTG-3′ |
| pMK88G | 5′-GGCCGTGTGGGGGACCTGCCGGGTG-3′ |
| pMD89R | 5′-CGTGTGAAGCGCCTGCCGGGTGTTC-3′ |
| pML90K | 5′-GTGTGAAGGACAAGCCGGGTGTTCG-3′ |
| pMG92D | 5′-GGACCTGCCGGACGTTCGCTAC-3′ |
| pMR94G | 5′-CTGCCGGGTGTTGGCTACAAGATCATC-3′ |
Bold and underlined letters indicate mutated bases.
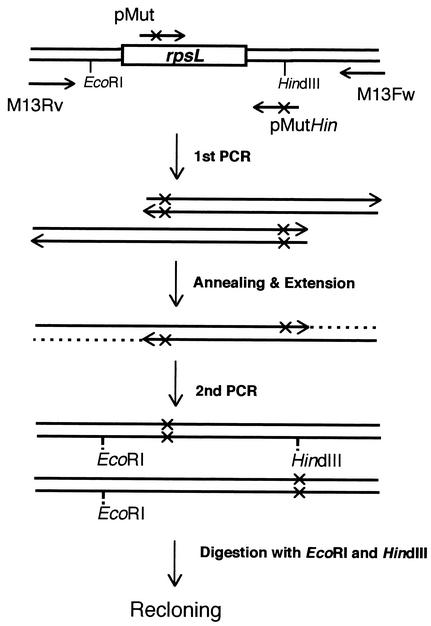
FIG. 1.
Schematic representation of the overlapping PCR strategy for introducing mutations into rpsL. pMut represents a primer containing a mutation site. In the first-step PCR, two fragments were amplified. After the overlap extension, the reconstituted mutant genes were further amplified in the second-step PCR by using primers M13-Fw and Rv.
Construction of mutation plasmid.
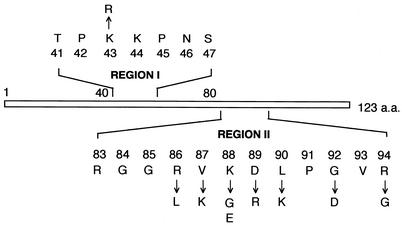
Two regions in protein S12 are highly conserved among eukaryotic and prokaryotic microorganisms; these are illustrated as region I (TPKKPNS) and region II (RVKDLPGVR) in Fig. 2. Recent X-ray crystallographic analysis of the ribosome from Thermus thermophilus demonstrated that each region consists of two independent loop structures (12, 17). rpsL mutations known to confer resistance to streptomycin in E. coli and Streptomyces spp. are all situated in either of these two regions. Interestingly, in Streptomyces spp., mutations that activate antibiotic production (K88E and P91S) are found in region II, justifying our choice of this region for the present study. Since the basic amino acid residues in region II play a role in the interaction with the rRNA phosphate backbone (3), we replaced the basic amino acids (Arg and Lys) with neutral amino acids and vice versa. Eventually, we constructed the following mutants: KO-471 (K43R), KO-472 (R86L), KO-473 (V87K), KO-474 (K88G), KO-475 (D89R), KO-476 (L90K), KO-477 (G92D), and KO-478 (R94G) (Fig. 2). The K88E mutation was also constructed as a control. The mutation K43R confers streptomycin resistance in Bacillus subtilis and markedly enhanced the production of an unidentified antibiotic in this organism (7). The G92D mutation displays a streptomycin-dependent phenotype with E. coli (16). A mutation at the position Arg-94 was also known to give rise to a streptomycin-dependent phenotype for E. coli when this residue was deleted (16). We changed Arg-94 to the smallest amino acid glycine instead of deleting it (Fig. 2).
FIG. 2.
Mutations that were constructed by site-directed mutagenesis. a.a., amino acids.
Antibiotic production by transformants.
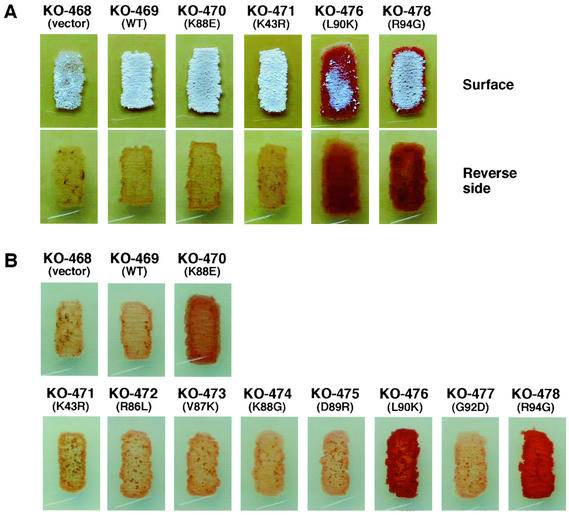
The transformants with a single-copy-number plasmid thus constructed (Table 2) were inoculated on GYM agar medium and R4 agar medium supplemented with 0.5% Casamino Acids (R4C). All transformants and the parental strain grew on GYM agar medium as shown for several representative transformants, except that transformants KO-476 (L90K) and KO-478 (R94G) displayed a somewhat retarded aerial mycelium formation (Fig. 3A). Importantly, transformants L90K and R94G produced extensive amounts of the red antibiotic undecylprodigiosin, but the transformant K88E produced only a slight amount of this antibiotic under the culture conditions used. The transformants KO-468 (vector control) and KO-469 (transformed with the wild-type rpsL gene) produced virtually no antibiotic. These results were confirmed by using the R4C medium. The transformant KO-470 (K88E) produced a considerable amount of undecylprodigosin under this set of culture conditions. Strikingly, production of the antibiotic was more pronounced in the transformants KO-476 (L90K) and KO-478 (R94G) (Fig. 3B). The amounts of undecylprodigiosin produced were 2.9-fold (KO-476) and 1.9-fold (KO-478) greater than those of the transformant KO-470 (K88E), as determined quantitatively by the method described by Kieser et al. (10). None of the other mutations tested (K43R, R86L, V87K, K88G, D89R, and G92D) were effective in activating antibiotic production. Although R4C medium is a medium suitable for production of both actinorhodin and undecylprodigiosin (15), we could not detect actinorhodin production in any transformants. This is probably because the S. lividans strains created here are hemizygous, carrying a wild type as well as a mutant copy of the rpsL gene (see below).
TABLE 2.
Strains used in this study and their level of resistance to streptomycin and paromomycin
| Strain | Description | Level of resistance (μg/ml)a to
|
Source (reference) | |
|---|---|---|---|---|
| Streptomycin | Paromomycin | |||
| E. coli DH5α | F− ΔlacU169 (φ80lacZΔM15) endA1 recA1 hsdR17 deoR supE44 thi-1 λ−gyrA96 relA1 | Invitrogen (4) | ||
| S. lividans | ||||
| TK21 | Prototroph, SLP2− SLP3− | 1.5 | 0.2 | T. Kieser (9) |
| KO-468 | TK21 harboring pV1 | 1.5 | 0.2 | This study |
| KO-469 | TK21 harboring pVWT | 1.5 | 0.2 | This study |
| KO-470 | TK21 harboring pVK88E | 40 | 0.1 | This study |
| KO-471 | TK21 harboring pVK43R | 20 | 0.1 | This study |
| KO-472 | TK21 harboring pVR86L | 1.5 | 0.1 | This study |
| KO-473 | TK21 harboring pVV87K | 1.5 | 0.1 | This study |
| KO-474 | TK21 harboring pVK88G | 10 | 0.1 | This study |
| KO-475 | TK21 harboring pVD89R | 1.5 | 0.1 | This study |
| KO-476 | TK21 harboring pVL90K | 1.5 | 0.1 | This study |
| KO-477 | TK21 harboring pVG92D | 1.5 | 0.1 | This study |
| KO-478 | TK21 harboring pVR94G | 1.5 | 0.2 | This study |
Determined after 2 days of cultivation on GYM agar.
FIG. 3.
Effect of a single copy of the mutant rpsL gene on antibiotic production. Wild-type S. lividans TK21 cells were transformed with the plasmid pV1, which expressed the mutant rpsL or wild-type rpsL. The strain KO-468 represents the vector control. The strains were grown for 4 days at 30°C on GYM (A) or R4C (B) medium. Redness represents the antibiotic undecylprodigiosin.
The ability of these transformants to exhibit resistance to streptomycin and paromomycin was further tested. The experimental system chosen in this study to evaluate the new rpsL mutations involved expressing the mutant S12 proteins from their native promoters on a single-copy-number plasmid. This means that the S. lividans strains created carry both wild-type and mutant-type rpsL genes. Wild-type copies of the rpsL gene are generally considered to encode dominant streptomycin sensitivity, and in fact we detected no streptomycin resistance when K88E transformant was incubated for 24 h on a medium containing streptomycin. However, the K88E transformant exhibited a considerable resistance (up to 40 μg/ml [Table 2]) when incubated for a longer time (48 h or more), although the resistance level is lower than that for the previously described K88E mutants that are resistant to 100 μg of streptomycin per ml (5, 8, 11, 15). Based on this fact, we evaluated the transformants with respect to the level of resistance to streptomycin. Although the transformants KO-470 (K88E), KO-471 (K43R), and KO-474 (K88G) were resistant to streptomycin, the parental strain and other transformants, including KO-476 and KO-478, showed entirely no resistance to streptomycin (1.5 μg/ml) (Table 2). It is notable that even the transformant KO-477, which has a G92D mutant-type rpsL gene that confers a streptomycin-dependent phenotype on E. coli (16), showed neither resistance to, nor dependence on, streptomycin. None of the transformants was resistant to paromomycin. Similar results were obtained when the resistance level was determined by using R4 agar medium (data not shown). Thus, unlike K88E, the mutations L90K and R94G did not confer resistance to either streptomycin or paromomycin.
Region I has been reported to interact directly with the space between the 16S rRNA 530 loop and the 1492-1493 strand of the decoding site (17). Most of the mutations in this region, found previously in E. coli and other bacteria, can increase translational accuracy. These mutations could have the effect of widening the space between the tRNA-mRNA complex and the 30S A site (17). On the other hand, only a limited number of data is available concerning the role of region II, although certain mutations found in this region can be responsible for the increased translational accuracy (16). The mutation G92D, which confers a hyperaccuracy phenotype to E. coli, did not activate antibiotic production (Fig. 3), implying that the observed activation of antibiotic production in Streptomyces spp. is not correlated with the translation accuracy. Recently, a study in our laboratory with an in vitro translation assay system raised the possibility that an increased rigidity of 70S ribosome particles caused by specific rpsL mutations may result in antibiotic production activation (T. Hosaka and K. Ochi, unpublished results). Since neither the L90K nor R94G mutation could confer resistance to streptomycin or paromomycin, it is impossible to find these mutations among the resistant isolates. In conclusion, the site-directed mutagenesis technique could be effective for improving the ribosomal protein S12 by functional modulation. Although we have no explanation for the failure to activate actinorhodin production, it is possible that the activation of actinorhodin biosynthetic genes requires more forceful metabolic modulation than that of undecylprodigiosin biosynthetic genes, as could be achieved by the presence of only the mutant-type rpsL gene.
Acknowledgments
This work was supported by a grant from the Organized Research Combination System (ORCS) of the Ministry of Education, Culture, Sports, Science and Technology, Japan.
REFERENCES
- 1.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 2.Chakraburtty, R., and M. Bibb. 1997. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J. Bacteriol. 179:5854-5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gregory, S. T., J. H. Cate, and A. E. Dahlberg. 2001. Streptomycin-resistant and streptomycin-dependent mutants of the extreme thermophile Thermus thermophilus. J. Mol. Biol. 309:333-338. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 5.Hesketh, A., and K. Ochi. 1997. A novel method for improving Streptomyces coelicolor A3(2) for production of actinorhodin by introduction of rpsL (encoding ribosomal protein S12) mutations conferring resistance to streptomycin. J. Antibiot. 50:532-535. [DOI] [PubMed] [Google Scholar]
- 6.Hosokawa, K., N.-H. Park, T. Inaoka, Y. Itoh, and K. Ochi. 2002. Streptomycin-resistant (rpsL) or rifampicin-resistant (rpoB) mutation in Pseudomonas putida KH146-2 confers enhanced tolerance to organic chemicals. Environ. Microbiol. 4:703-712. [DOI] [PubMed] [Google Scholar]
- 7.Hosoya, Y., S. Okamoto, H. Muramatsu, and K. Ochi. 1998. Acquisition of certain streptomycin-resistant (str) mutations enhances antibiotic production in bacteria. Antimicrob. Agents Chemother. 42:2041-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu, H., and K. Ochi. 2001. Novel approach for improving the productivity of antibiotic-producing strains by inducing combined resistant mutations. Appl. Environ. Microbiol. 67:1885-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawamoto, S., H. Watanabe, A. Hesketh, J. C. Ensign, and K. Ochi. 1997. Expression analysis of the ssgA gene product, associated with sporulation and cell division in Streptomyces griseus. Microbiology 143:1077-1086. [DOI] [PubMed] [Google Scholar]
- 10.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 11.Ochi, K., D. Zhang, S. Kawamoto, and A. Hesketh. 1997. Molecular and functional analysis of the ribosomal L11 and S12 protein genes (rplK and rpsL) of Streptomyces coelicolor A3(2). Mol. Gen. Genet. 256:488-498. [DOI] [PubMed] [Google Scholar]
- 12.Ogle, J. M., D. E. Brodersen, W. M. Clemons, Jr., M. J. Tarry, A. P. Carter, and V. Ramakrishnan. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292:897-902. [DOI] [PubMed] [Google Scholar]
- 13.Okamoto-Hosoya, Y., T. Sato, and K. Ochi. 2000. Resistance to paromomycin is conferred by rpsL mutations, accompanied by an enhanced antibiotic production in Streptomyces coelicolor A3(2). J. Antibiot. 53:1424-1427. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Shima, J., A. Hesketh, S. Okamoto, S. Kawamoto, and K. Ochi. 1996. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J. Bacteriol. 178:7276-7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Timms, A. R., H. Steingrimsdottir, A. R. Lehmann, and B. A. Bridges. 1992. Mutant sequences in the rpsL gene of Escherichia coli B/r: mechanistic implications for spontaneous and ultraviolet light mutagenesis. Mol. Gen. Genet. 232:89-96. [DOI] [PubMed] [Google Scholar]
- 17.Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292:883-896. [DOI] [PubMed] [Google Scholar]