Abstract
Two-component signal transduction systems (2CSs) are widely used by bacteria to sense and adapt to changing environmental conditions. With two separate approaches, three different 2CSs were identified on the chromosome of the probiotic bacterium Bifidobacterium infantis UCC 35624. One locus was identified by means of functional complementation of an Escherichia coli mutant. Another two were identified by PCR with degenerate primers corresponding to conserved regions of one protein component of the 2CS. The complete coding regions for each gene cluster were obtained, which showed that each 2CS-encoding locus specified a histidine protein kinase and an assumed cognate response regulator. Transcriptional analysis of the 2CSs by Northern blotting and primer extension identified a number of putative promoter sequences for this organism while revealing that the expression of each 2CS was growth phase dependent. Analysis of the genetic elements involved revealed significant homology with several distinct regulatory families from other high-G+C-content bacteria. The conservation of the genetic organization of these three 2CSs in other bacteria, including a number of recently published Bifidobacterium genomes, was investigated.
Two-component regulatory systems (2CSs) are employed extensively in nature by microorganisms to modify their cellular physiology in response to alterations in environmental conditions (for a review, see references 35, 36, 37, and 51). A 2CS typically consists of a membrane-associated sensor protein or histidine protein kinase (HPK), which monitors one or more environmental parameters, and a cytoplasmic effector protein or response regulator (RR), which induces a specific cellular adaptive response. The HPK and RR each comprise two modular elements. A typical HPK contains an N-terminally located input or sensing domain and a C-terminal transmitter domain, which is autophosphorylated at a conserved histidine residue in response to fluctuations in chemical and/or physical conditions (sensed by the input domain). This phosphate group is transferred to an aspartate residue on the N-terminally positioned receiver domain of the cognate RR, which in turn alters the activity of the output domain (situated in the C-terminal region of the RR) to elicit an adaptive response (either functioning at the level of transcriptional regulation or by interacting directly with proteins). The transmitter module of the HPK contains a number of conserved residues in addition to the histidine at the site of autophosphorylation. These include an asparagine box, a glycine residue, a phenylalanine box, and a glycine-lysine motif, all located toward the C terminus of the kinase protein. The conserved receiver domain found in RRs contains a strictly conserved aspartate box and a lysine residue which are part of an acidic pocket involved in the phosphorylation event (33, 54).
2CSs have been found in over 50 prokaryotic species to date and in several lower eukaryotic organisms and plants (10, 23, 34, 38). However, both the number and the organization of these systems are diverse. The number of 2CSs in a given bacterial species range from the four HPKs and five RRs encoded by the entire genome of Haemophilus influenzae Rd (16) to the approximately 50 different 2CSs encoded in the genomes of enteric bacteria (5, 26).
HPKs have been sorted into classes on the basis of the sequence relationships of the residues surrounding the phosphorylated histidine (19). This classification has resulted in the organization of HPKs into five homology groups (groups I, II, IIIA, IIIB, and IV [15]). RRs have been classified into three major groups (classes 1, 2, and 3) based on the phylogenetic relatedness of their receiver module and DNA-binding domains, and four minor groups (classes 4 to 7) that exhibit output domains with unique amino acid sequences (33).
To date, no 2CSs have been reported for the probiotic genus Bifidobacterium. Very little is known about the molecular biology of bifidobacteria, despite the fact that they are among the most common genera in the human colon and have consistently had health-promoting properties attributed to them (for general reviews, see references 13, 14, and 52). Genetic characterization of bifidobacteria is essential to define their possible beneficial activities as part of the intestinal microflora and to explore and potentially exploit any such beneficial properties. 2CSs are of particular interest, as they may be of critical importance in the interaction between microbe and host (environment). In this communication, we report the identification and preliminary characterization of three putative 2CSs from Bifidobacterium infantis UCC 35624, a strain whose probiotic properties have been reported previously (14).
MATERIALS AND METHODS
Bacterial strains, media, chemicals, and culture conditions.
The strains and plasmids used in this study are listed in Table 1. Bifidobacteria were routinely cultured in de Man, Rogosa, and Sharpe medium (MRS [12]; Oxoid Ltd., Hampshire, England) supplemented with 0.2% (wt/vol) glucose. MRS was supplemented with 0.05% (wt/vol) cysteine-HCl, and strains were grown at 37°C under anaerobic conditions, maintained with the Anaerocult oxygen-depleting system (Merck, Darmstadt, Germany) in an anaerobic chamber. Escherichia coli strains were grown in Luria-Bertani (LB) medium at 37°C with agitation (44). Stocks of all cultures were maintained at −20°C in 40% glycerol. When necessary, antibiotics were added to the media as follows: ampicillin at 100 μg ml−1 (50 μg ml−1 in the case of plasmid pWSK29), tetracycline at 12.5 μg ml−1, and chloramphenicol at 20 μg ml−1. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and 5-bromo-4-chloro-3-indolylphosphate (X-P) were used at final concentrations of 40 μg ml−1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| Strains | ||
| E. coli XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10(Tcr)] | Stratagene Ltd. |
| E. coli ANCC22 | phoR and creC mutations | 29 |
| E. coli ANCL1 | PhoB− | 29 |
| E. coli VJS3051 | ΔnarQ251::Tn10d (Tcr) ΔnarX242 zch-2084::Ω-Cmr φ(fdnG-lacZ) | 40 |
| E. coli VJS3081 | Δ(lac-argF)U169 λφ(fdnG-lacZ) narL215::Tn10 | 41 |
| B. infantis UCC 35624 | Wild-type human isolate | UCC Culture Collectiona |
| Plasmids | ||
| pBluescript KS− | Apr αlacZb | Stratagene Ltd. |
| pWSK29 | Apr αlacZ,b low copy number | 56 |
UCC, National University of Ireland, Cork, Ireland.
αlacZ is the portion of the lacZ gene that provides α-complementation for blue and white color selection of recombinant phagemids.
DNA manipulations and sequence analysis.
Plasmid DNA was obtained from E. coli with either an alkaline lysis method (8) or the QIAprep spin plasmid miniprep kit (Qiagen GmbH, Hilden, Germany). Large-scale preparations of total DNA from B. infantis were made as described previously (32). Purified DNA was obtained by cesium chloride ultracentrifugation of this preparation, as described by Sambrook et al. (44). Restriction endonucleases, T4 DNA ligase, and calf intestinal alkaline phosphatase were purchased from Roche Diagnostics Ltd. (Lewes, East Sussex, United Kingdom) or New England Biolabs Ltd. (Hitchin, United Kingdom) and used as recommended by the manufacturers. Electroporation of plasmid DNA into E. coli was performed essentially as described previously (44).
PCRs were accomplished with either the Taq PCR Master Mix (Qiagen) or the Expand Long Template PCR system (Roche Diagnostics GmbH, Mannheim, Germany) in accordance with the manufacturer's instructions. PCRs were executed with an Omnigene thermal cycler (Hybaid Ltd., Middlesex, United Kingdom). Sequencing was performed by MWG-Biotech AG (Ebersberg, Germany). Sequence data assembly and analysis were performed with DNAStar software (DNAStar, Madison, Wis.). Database searches were performed with nonredundant sequences at the NCBI Internet site (http://www.ncbi.nlm.nih.gov) with the tBlastN, tBlastX, and BlastP programs (2, 3). Sequence alignments were performed with the Clustal method of the Megalign program of the DNAStar software package. Functional domains in deduced proteins were identified with the SMART database (46, 47) Internet site (http://smart.embl-heidelberg.de).
Phenotypic complementation and activity assays of mutant strains.
Ligation mixes were prepared essentially as described previously (30). The ligation mixes were introduced into competent E. coli ANCC22 or VJS3051 by electrotransformation (44) with the Bio-Rad gene pulser apparatus according to the manufacturer's instructions (Bio-Rad Laboratories, Richmond, Calif.). Colonies phenotypically exhibiting increased activity (alkaline phosphatase activity on X-P plates in the case of strain ANCC22 or β-galactosidase activity on X-Gal plates for strain VJS3051), as indicated by the formation of a blue colony, were selected for quantitative assay. Alkaline phosphatase activity assays were performed as described previously (1).
Degenerate PCR.
PCR was performed on B. infantis UCC 35624 chromosomal DNA with degenerate oligonucleotide primers designed specifically to correspond to conserved regions of RRs, essentially as described previously (28). Sequences of (assumed) RRs from bacteria with high G+C content were obtained from the Blast database and aligned with the Megalign program from DNAStar. Conserved residues were identified (approximately 97 amino acids apart), and degenerate primers (MWG-Biotech, Ebersburg, Germany) were designed on these. Two different forward oligonucleotides, GT(G/A/T/C)GT(G/A/T/C)GA(G/A/T/C)GA(C/T)GA and (A/C)T(G/A/T/C)GT(G/A/T/C)GA(G/A/T/C)GA(C/T)GA, corresponding to the amino acids VV(DE)D(DE) and (ILM)V(DE)D(DE), respectively, and one reverse oligonucleotide, (A/G)(A/T)A(A/G)TC(G/A/T/C)GC(G/A/T/C)CC, corresponding to the amino acid sequence GAD(IN), were designed based on conserved amino acid residues around the DD and K boxes of known RRs (28). PCR conditions were essentially as described previously (28). Fragments of the expected size (approximately 300 bp) were excised from 2% agarose gels, purified with the Concert Rapid PCR purification system (Gibco-BRL, Paisley, Scotland), and cloned into the pCR2.1-TOPO vector prior to sequencing.
Anchored PCR and Southern hybridization.
Anchored PCR was used in order to obtain the DNA sequence surrounding the cloned open reading frame (ORF) specifying the assumed HPK or RR, essentially as described previously (11). PCR products were purified and used for sequencing purposes. Restricted chromosomal DNA from B. infantis UCC 35624 was separated by agarose gel electrophoresis and transferred to nylon membranes (Hybond N+; Amersham International, Little Chalfont, Bucks, United Kingdom) by the method of Southern (48) as modified by Wahl et al. (55). DNA was labeled with the enhanced chemiluminescence gene detection system (Amersham). Probe labeling, hybridization conditions, and washing steps were completed according to the manufacturer's instructions.
RNA isolation, Northern analysis, and 5′ extension analysis.
Northern analysis was performed on aliquots of total RNA extracted by the Macaloid method (20) from bifidobacterial cultures which had been harvested at a range of optical densities at 600 nm between 0.2 and 1.4. RNA samples were treated with DNase and RNase inhibitors (Roche Diagnostics), denatured at 70°C for 10 min, and loaded with formamide-containing dye on to a 1.2% formaldehyde gel (6). RNA size standards from Promega (Madison, Wis.) were used to enable transcript size estimation. Capillary blotting to Hybond-N+ nylon membranes (Amersham) was performed essentially as described previously (44). An internal 500-bp fragment (amplified with PCR) from each ORF identified for each of the three 2CS-encoding loci was used as a probe (for primer sequences, see Table 2). The probes were radiolabeled with γ-32P with a Prime-a-Gene kit (Promega).
TABLE 2.
Primers used to amplify internal fragments of the genes described in this study to be used as probes for Northern hybridizations
| Gene | Forward primer | Reverse primer |
|---|---|---|
| gtpA | GCAACAGTCTCACGATTC | GGGGCGTTCCTCAAATAC |
| birA | AACACCATGGCGACCATC | TCCATCGGAGTGAGATTC |
| bikA | AGTCTGATTTCTGACGAC | GTGGTCACCGGGGTACGC |
| lipA | TGGGTTCCTTGGATTCGC | CACATTTGCGTCGGCATC |
| biaA | GATTGGTGCCAAGAAGGC | CGGGGTGCGTGGCCAGCC |
| biaB | GCCAAGGTCATCACCTCC | GCCTGCATCACGCAGATC |
| biaC | TTCGGCCTGCTGGCCGGC | GGAGCCGAGCACGTAGCC |
| birB | GACGTCATGCTGCCTGAC | GGTCACGTCGTGGGAGTC |
| bikB | GCCGAATTCAGCCTTGCC | GGACTGCTTGGGCTCAGG |
| bikC | TCGAGCACATGGTCGGCC | CTGCGCCAGCGTCCAGGC |
| birC | CGTGAGGGGCTGCGCGCC | TTGTGTGCGGTCGGCGAC |
| orfC (3′) | CTGCTGGCCGAAGCGGCG | GGCGCACCAGTTCGACGC |
| orfC (5′) | GAGATCCACAGCACCAGC | GAATTCAAGGACGATTAC |
Primer extension to identify the transcriptional start site was accomplished by annealing γ-33P-radiolabeled synthetic oligonucleotides to RNA as previously described (39). Primers were designed approximately 100 bp downstream of the predicted ribosome binding site of the assumed first coding sequence of each transcript, and primer extension was performed by annealing 5 pmol of γ-33P-labeled primer to 50 μg of RNA. Sequence ladders for each of the primer extension reactions were produced with the same primer used for the primer extension and with the aid of the T7 DNA polymerase sequencing kit (USB Corp.).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the three regions specifying 2CSs identified in this study are as follows: system A, AY266333; system B, AY266334; and system C, AY266335.
RESULTS
Isolation of HPK-encoding gene by functional complementation of E. coli ANCC22.
By using a complementation strategy (see Materials and Methods), 15 transformants, each carrying one or more random chromosomal fragments of B. infantis UCC 35624 cloned into the high-copy-number pBluescript vector, were shown to be capable of suppressing the E. coli ANCC22 PhoA-negative phenotype on solid medium. This phenotypic suppression strategy was also employed without success with a second mutant E. coli strain, VJS3051 (40), and a low-copy-number vector, pWSK29 (data not shown). The complementing ANCC22 clones were quantitatively assayed for increased alkaline phosphatase activity. All transformants exhibited increased alkaline phosphatase activity, ranging from 40 to 200 U, compared to a negative control of ANCC22 containing pBluescript (<5 U). Furthermore, introduction of the recombinant plasmids from the suppressed isolates into the control strain ANCL1 showed that suppression was not due to the cloning of a phosphatase or a regulator of phoA transcription, as outlined previously (29). Sequence data for the inserts (ranging from 1 to 2 kb) of each plasmid capable of phenotypic suppression revealed the presence of (various 3′ sections of) a single HPK-encoding gene, corresponding to the transmitter domain of this assumed HPK (designated bikA; see below).
Identification of two putative RR-encoding genes by degenerate PCR.
Sequence comparison of 50 independent plasmid inserts obtained with a PCR strategy (see Materials and Methods) allowed the identification of two ORFs, each displaying significant similarity with the N-terminal internal fragment of an RR-encoding gene. These assumed RR-encoding genes were designated birB and birC (see Table 3). The PCR product encoding BirB was obtained with the forward primer VV(DE)D(DE) in conjunction with the reverse primer (see above). The second RR-encoding moiety, birC, was obtained with the second degenerate primer, (ILM)V(DE)D(DE), and the reverse primer.
TABLE 3.
Classification and putative functional domains of HPKs and RRs identified in this study
| ORF | Size (amino acids) | Positiona (amino acids) or homology
|
Class or group | |||
|---|---|---|---|---|---|---|
| N-terminal domainsa
|
C-terminal domain HATPase-c or effector | |||||
| Transmembrane or receiver | HAMP | HPK-A | ||||
| BikA | 565 | 29-51 | 210-278 | 290-356 | 402-513 | IIIA |
| 172-194 | ||||||
| 207-229 | ||||||
| BikB | 448 | 51-73 | 69-121 | 134-202 | 266-413 | IIIA |
| BikC | 348 | N/A | N/A | 172-241 | 275-321 | II |
| BirA | 240 | CheY | PhoP/OmpR | 2 | ||
| BirB | 227 | CheY | OmpR/PhoB | 2 | ||
| BirC | 214 | CheY | NarL/DegU | 3 | ||
HAMP, histidine kinase, adenlyl cyclase, methyl binding protein, phosphatase domain; HPK-A, histidine kinase A motif; HATPase-c, histidine kinase-, DNA gyrase B-, phytochrome-like ATPase. N/A, not apparent.
Comparative sequence analysis of three 2CSs.
Analysis of the DNA regions surrounding bikA, birB, and birC, which were obtained by anchored PCR, showed that each gene was flanked by either an RR- or an HPK-encoding gene, revealing three complete 2CSs. Additional ORFs were identified in some cases. All identified ORFs are schematically depicted in Fig. 1 and summarized in Table 4 along with a number of their salient features. bikA was located immediately downstream of its cognate RR-encoding gene, birA (birA-bikA was designated system A). This genetic organization was also observed for birB-bikB (referred to as system B). In contrast, bikC was located immediately upstream of its cognate RR-encoding gene, birC (bikC-birC was named system C). HAMP domains (cytoplasmic helical linker domains proposed to have a role in regulation of phosphorylation of the HPK and present in many prokaryotic signaling proteins [4]), HPK A motifs (the predicted dimerization and phosphoacceptor domain), and histidine kinase-like ATPase domains involved in ATP binding were identified in each of these two HPKs (see Table 3) with the SMART database (see Materials and Methods).
FIG. 1.
Schematic representation of three 2CSs identified on the chromosome of B. infantis UCC 35624 and their surrounding ORFs. Arrows represent each ORF, with the gene name positioned above. The lengths of the transcripts identified by Northern analysis are indicated underneath each system by a thin arrow. Positions of promoter sequences deduced from primer extension and/or Northern blot analysis are indicated by bent arrows. The positions of putative transcriptional terminator structures are indicated by “lollipops.”
TABLE 4.
| ORF | Size (amino acids) | ORF with highest similarity score | Organism | Identity (%) | P | GenBank accession no. |
|---|---|---|---|---|---|---|
| System A | ||||||
| GtpA | 459 | Hypothetical protein | B. longum DJO10A | 96 | 0.0 | ZP_00121667 |
| BirA | 240 | Hypothetical protein | B. longum DJO10A | 80 | 1e-101 | ZP_00121666 |
| BikA | 565 | Hypothetical protein | B. longum DJO10A | 96 | 0.0 | ZP_00121665 |
| LipA | 467 | Hypothetical protein | B. longum NCC2705 | 93 | 0.0 | NP_695263 |
| BiaA | 324 | Probable solute-binding protein of ABC transporter system, possibly for sugars | B. longum NCC2705 | 93 | 1e-149 | NP_695264 |
| BiaB | 504 | ATP-binding protein of ABC transporter | B. longum NCC2705 | 100 | 0.0 | NP_695265 |
| BiaC | 696 | Hypothetical protein | B. longum DJO10A | 88 | 1e-138 | ZP_00121659 |
| System B | ||||||
| BirB | 227 | Hypothetical protein | B. longum DJO10A | 91 | 2e-91 | ZP_00121583 |
| BikB | 448 | Histidine kinase sensor of two-component system | B. longum NCC2705 | 96 | 0.0 | NP_695237 |
| System C | ||||||
| BikC | 348 | Sensor histidine kinase | Pseudomonas syringae pv. tomato DC3000 | 42 | 1e-20 | NP_791953 |
| BirC | 219 | Putative two-component system regulator | Streptomyces coelicolor A3(2) | 40 | 2e-17 | NP_627834 |
See also Fig. 2.
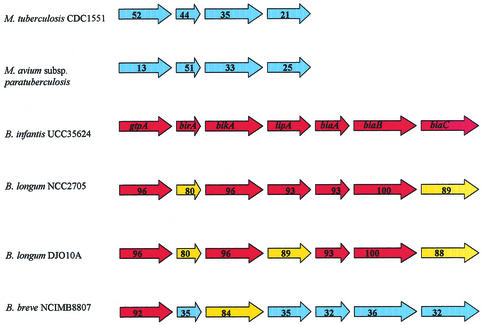
A 1,380-bp ORF was located immediately upstream of birA, and the deduced protein product of this gene, designated gtpA, displayed high similarity to a GTP-binding protein. A predicted lipoprotein-encoding ORF, designated lipA, was identified downstream of the HPK. Downstream of lipA, three genes were identified which appeared to constitute a putative ABC transport system. The gene organization of the system A operon (see below) was conserved in Bifidobacterium longum DJO10A, Bifidobacterium longum NCC2705, and Bifidobacterium breve NCIMB 8807 (Fig. 2). A partly homologous gene cluster consisting of the first four genes of this operon was found in a number of Mycobacterium spp. (Fig. 2). Interestingly, while clear homologues were found for systems A and B in other sequenced Bifidobacterium spp., this was not the case for system C. A number of other ORFs were identified immediately up- or downstream of the identified 2CSs; however, as they were not found to be transcriptionally linked to the 2CSs (see below), they will not be discussed further.
FIG. 2.
Alignment of genetic organization of system A from B. infantis UCC 35624 with corresponding loci in M. tuberculosis CDC1551 (accession no. AE007145), Mycobacterium avium subsp. paratuberculosis (AF10884), B. longum NCC2705 (AE014617), B. longum DJO10A (NZ_AABM02000022), and B. breve NCIMB8807 (unpublished data). The names of the genes are indicated within arrows for UCC 35624. The percent identities for each protein-encoding gene compared to the corresponding ORF from UCC 35624 are indicated within the arrows for each genome. The degree of amino acid identity (>90%, >80%, >70%, and <70%) is indicated by the color of the arrow (red, yellow, green, and blue, respectively).
A number of putative rho-independent transcriptional terminator structures were identified on the basis of being able to form stable stem-loop structures (ΔG < −15 kcal mol−1) and are depicted in Fig. 1. No putative hairpin structures with significant ΔG values were identified immediately downstream of lipA; however, a region rich in C and poor in G was detected (13% G over 60 bases), suggesting the involvement of a rho-dependent terminator (22).
Transcriptional regulation and 5′ extension analysis.
Northern analysis was performed to elucidate the manner in which the three 2CS-encoding loci are transcribed. All probes used for system A hybridized to a large (11-kb) transcript, indicating that all these genes are cotranscribed, comprising an 11-kb-long operon (Fig. 3a and 3b). In addition, a smaller transcript of 4 kb was observed only when the probes derived from birA, bikA, and lipA were used (Fig. 3a). This second transcript was constitutively expressed from the early exponential to the late stationary phase. On the other hand, the 11-kb transcript was evident only from the late exponential to the late stationary phase. Both gene probes obtained from bikB and birB in system B hybridized to a transcript of 3.0 kb in mRNA samples obtained from cells at the late exponential to late stationary growth phase, indicating that these genes are transiently transcribed as a dicistronic operon (Fig. 3c). Similarly, the bikC- and birC-derived probes hybridized to a single 3.0-kb mRNA transcript only from mRNA of late-exponential- to late-stationary-phase cells. A probe obtained from B. infantis orfC, encompassing DNA on the 5′ side of the putative transcriptional terminator (Fig. 1), also hybridized to a similar-sized transcript, whereas a probe consisting of DNA located at the 3′ side of this stem-loop structure did not (results not shown).
FIG. 3.
Northern analysis of systems A and B with RNA isolated from B. infantis UCC 35624 at different optical densities at 600 nm (indicated above each lane). The estimated sizes of the transcripts are indicated on the right. (a) Transcription of system A with an internal 500-bp fragment of bikA as a probe. Similar results were obtained with probes birA and lipA. (b) Transcription of system A with probe gtpA. Similar results were obtained with probes biaA, biaB, and biaC. (c) Transcription of system B with probe bikB. Similar results were obtained with probe birB. Northern blots also revealed a 3-kb transcript for system C (not shown) with probes bikC, birC, and B. infantis orfC.
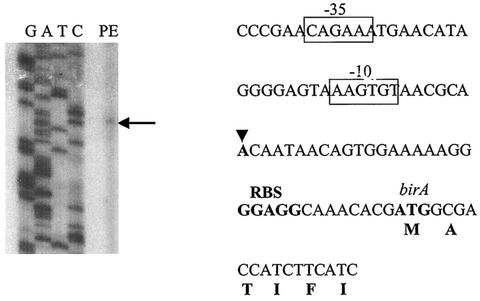
Primer extension analysis was attempted for each system to elucidate the transcriptional start site of the four identified transcripts. The transcriptional start site for the large 11-kb transcript of system A was identified as an adenine base, situated 13 bp upstream of the assumed start codon of gtpA (Fig. 4). The transcriptional start site for the putative promoter immediately proximal to birA was identified as an adenine residue, situated 33 bp upstream of the presumed translational start site of birA, as indicated in Fig. 5. No definitive sequence ladder-primer extension pair could be obtained for either system B or C despite exhaustive attempts.
FIG. 4.
Primer extension (PE) analysis of transcriptional start site of 11-kb transcript of system A. The assumed ribosome-binding site (RBS) and start codon (ATG) of gtpA are indicated in bold. The transcriptional start site is indicated by a solid triangle, and the name of the gene is indicated in italics over the initial methionine residue. The translated amino acid residues of GtpA are shown underneath the corresponding DNA sequence. The arrow indicates the position of the extension product. Proposed −10 and −35 motifs are boxed.
FIG. 5.
Primer extension (PE) analysis of transcriptional start site of 4-kb transcript of system A. The assumed ribosome-binding site (RBS) and start codon (ATG) of birA are indicated in bold. The transcriptional start site is indicated by a solid triangle, and the name of the gene is indicated in italics over the initial methionine residue. The translated amino acid residues of BirA are shown underneath the corresponding DNA sequence. The arrow indicates the position of the extension product. Proposed −10 and −35 motifs are boxed.
DISCUSSION
This report describes the identification and transcriptional analysis of three 2CSs from B. infantis. These three systems are, to our knowledge, the first fully sequenced and transcriptionally analyzed examples of such signal transduction systems in this genus. Two different methods were used to maximize identification of 2CSs on the chromosome of B. infantis UCC 35624. A complementation strategy (30, 53) resulted in the identification of a single HPK, bikA, with the E. coli mutant ANCC22. A second E. coli mutant, VJS3051, was used without success. This strain carries mutations in the class II HPK-encoding genes narX and narQ. Complementation of this mutant through cross talk of a cloned HPK would in theory lead to the activation of the cognate RR, NarL, which in turn induces transcription of an fdnG-lacZ fusion, resulting in increased β-galactosidase activity (40, 41, 50). Complementation would be expected from class II HPKs, extending the variety of HPK-encoding genes that could be identified in B. infantis.
A second, PCR-based strategy which allowed the identification of two RRs was used. This approach has been successfully applied in a number of gram-positive and gram-negative bacteria (21, 27, 28, 31). Morel-Deville et al. (28) noted that the design of the K primer employed in their study (conserved residue in the RR receiver domain around which the reverse primer was designed) was biased toward class II proteins according to the Pao and Saier (33) classification of RRs. In this study, a more specific set of degenerate primers was designed and optimized for use in gram-positive bacteria with high G+C contents. Subsequent sequence analysis with the three HPK- and RR-encoding fragments allowed the identification of three complete 2CSs.
The complementation strategy has various technical limitations, such as the particular mutant strain used, the intrinsic properties of the kinase itself, and the portion of the kinase cloned. All of these factors determine if “cross talk” or heterologous transphosphorylation is possible. These limitations are possibly exacerbated by the difference in G+C content between E. coli (typically 48 to 52% [9]) and bifidobacterial DNA (58%). BikA belongs to the group IIIA kinases (15) and would therefore be predicted to suppress the phenotypic effect of the HPK mutations in ANCC22 (also group IIIA HPKs). BikB is also a member of class IIIA of HPKs and thus would be expected to have been detected by the complementation procedure. However, when the C-terminal conserved moiety of this kinase was cloned into ANCC22, no phenotypic complementation was observed (unpublished data); thus, it may be that the specificity of BikB prevents the transmitter domain from participating in heterologous transphosphorylation in this case. The genome of B. infantis UCC 35624 would be expected to harbor more than three 2CSs, considering the frequency with which such systems occur in other bacterial species (see the introduction). It is likely that not all 2CSs have been identified with the strategies used in this study. It is interesting, for example, that the recently published sequence for B. longum NCC2705 harbors seven 2CSs (45).
All three operons appear to be typical two-component His-Asp phosphorelay systems. The HPK-RR pair of system A display significant similarity to a number of putative 2CSs from the related, high-G+C genera Corynebacterium and Mycobacterium as well as the B. longum strains DJO10A and NCC2705 (Table 4, Fig. 2) and B. breve NCIMB 8807 (D. van Sinderen and G. F. Fitzgerald, unpublished data). The genetic organization of the large 11-kb transcription unit of system A (see above) is highly conserved across the bifidobacterial genomes investigated. Comparative sequence analysis indicates that this 2CS may have a function in solute (specifically sugar) uptake by the bacterium. The lipA gene, transcriptionally linked to system A, is consistently located immediately downstream of a 2CS in the bifidobacterial genomes investigated (see Fig. 2), as well as in Mycobacterium avium subsp. paratuberculosis, Mycobacterium tuberculosis H37Rv, and Mycobacterium leprae TN (accession numbers AF410884, Z95121, and NC_002677, respectively).
The BirB-BikB and BirA-BikA 2CSs both belong to the OmpR superfamily of 2CSs (15). Homologues of system B were observed in B. longum DJO10A, B. longum NCC2705, B. breve NCIMB 8807, and M. tuberculosis CDC1551, indicating that this 2CS is widely conserved among high-G+C bacterial species. In B. longum NCC2705 in particular, a number of ORFs (not discussed in this study) surrounding the 2CS displayed significant similarity (data not shown).
The genetic organization of system C is different from that of the other two 2CSs (see Results). Notably absent in BikC are the transmembrane domains typical of the N terminus of HPKs. BikC therefore appears to be a cytoplasmic HPK and possibly responds to an intracellular signal. BikC represents a member of the group II HPKs specifically categorized in the DegS subgroup. BirC lacks the C-terminal DNA-binding motif of the OmpR family and is a member of the NarL/DegU family of RRs (class 3) (7, 15, 33). System C is of particular interest, as it does not appear to have a close homologue in B. longum NCC2705, B. longum DJO10A, or B. breve NCIMB8807 (Table 4), indicating that this 2CS may fulfill a regulatory function not present in (some) other Bifidobacterium spp.
The comparative analysis of three 2CSs from B. infantis UCC 35624 suggests that two of these have functional homologs in three partially or completely sequenced Bifidobacterium genomes (Table 4, Fig. 2). The conserved gene organization of system A and its cotranscribed genes suggest either that such genes may be targets of the 2CS (several 2CSs are located next to cotranscribed genes, in many cases encoding ABC transport systems [31]) or that they control or may be part of the signal transduction pathway itself. Also, the signals to which these 2CSs respond remain elusive (as they are for most known 2CSs).
The HPKs encoded by systems A and B are predicted to be associated with the cytoplasmic membrane and are therefore expected to respond to extracellular stimuli. In contrast, the protein specified by bikC does not appear to contain a membrane-spanning input domain and may therefore respond to an intracellular signal. All three systems incorporate an RR protein that contains an effector domain with a DNA binding motif, suggesting that these systems act to respond to their stimulus by adjusting gene expression.
This is one of the first reported studies of Northern blotting being used in the transcriptional analysis of genes from Bifidobacterium spp. Each of the 2CSs appears to be growth phase regulated, a feature which is common in such systems throughout the bacterial kingdom (see references 17 and 31 for specific reviews on Streptomyces and Lactococcus spp., respectively). It is an observed phenomenon in many bacterial species that promoter elements have higher A+T contents than intragenic DNA. The only experimentally mapped bifidobacterial promoter regions, i.e., the β-gal1 and lactose permease genes of B. infantis, have a relatively high A+T content (66% and 73%, respectively [18]). Our results are in accordance with this observation, as the sequences immediately upstream of the transcriptional start sites of gtpA and birB have an A+T content of 48%, and 50% in the case of birA.
There is only one other report to date of the primer extension technique being used in the determination of a transcriptional start site in Bifidobacterium spp. (42). If the vegetative B. infantis RNA polymerase recognizes promoter sequences similar to those from other bacteria (i.e., −10 TATAAT and −35 TTGACA), putative promoter motifs (Fig. 4 and 5) may be proposed upon inspection of the DNA sequence immediately upstream of the transcriptional start site (which concurs with previously reported motifs for this genus [25, 42]). However, further experimentation will be required to establish the validity of this assumption. No definitive consensus sequence can be determined from these motifs, which may be due to the fact that these RNA polymerase recognition sites can tolerate a significant amount of degeneracy or that the sequences investigated in this study are not representative of typical bifidobacterial −10 and −35 hexamers. It is also possible that the recognition sites of the vegetative RNA polymerase in Bifidobacterium spp. are dissimilar to those previously reported for a variety of bacterial species.
Previous research has demonstrated the beneficial effects of administering probiotic combinations of Lactobacillus salivarius subsp. salivarius UCC 118 and B. infantis UCC 35624 (14). The mechanisms of action of these probiotic bacteria remain to be elucidated; however, a number of putative modes have been proposed (for a review, see references 13 and 49). Genetic investigation of Bifidobacterium species has been very limited due to a paucity of genetic tools and a relatively low electrotransformation efficiency (generally reported as approximately 104 to 105 cells per μg of DNA [24, 43]). The low transformation frequency of B. infantis UCC 35624 (unpublished results) does not allow single-crossover recombination for the purpose of gene knockouts. Thus, it is not possible at this time to attribute in vivo phenotypic characteristics to the mutation of any of these systems, and functionality can be proposed only as a result of homology studies. Any information on genetic organization and regulation, particularly on systems which act at the interface between host and bacterium, such as two-component systems, will be invaluable for understanding of the probiotic properties attributed to these bacteria.
Acknowledgments
We are grateful to Takeshi Mizuno for providing E. coli ANCC22 and ANCL1 and to Valley Stewart for providing E. coli VJS3051 and VJS3081.
This work was supported by Enterprise Ireland (grant no. BR/1998/202) and the Higher Education Authority Programme for Research in Third Level Institutions (HEA PRTLI/HEA PRTLIII).
REFERENCES
- 1.Aiba, H., M. Nagaya, and T. Mizuno. 1993. Sensor and regulator proteins from the cyanobacterium Synechococcus species PCC7942 that belong to the bacterial signal-transduction protein families: implication in the adaptive response to phosphate limitation. Mol. Microbiol. 8:81-91. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tools. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schafer, J. Hang, Z. Hang, W. Miller, and D. J. Lipman. 1997. Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 5.Arthur, M., F. Depardieu, and P. Courvalin. 1999. Regulated interactions between partner and nonpartner sensors and response regulators that control glycopeptide resistance gene expression in enterococci. Microbiology 145:1849-1858. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
- 7.Baikalov, I., I. Schroder, M. Kaczor-Grzeskowiak, K. Grzeskowiak, R. P. Gunsalus, and R. E. Dickerson. 1996. Structure of the Escherichia coli response regulator NarL. Biochemistry 35:11053-11061. [DOI] [PubMed] [Google Scholar]
- 8.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brock T. D.,. 1997. Chapter 16. Prokaryotic diversity: bacteria, p.635-740. In M. T. Madigan, J. M. Martinko, and J. Parker (ed.), Biology of microorganisms, 8th ed. Prentice-Hall, Inc., Trenton, New Jersey.
- 10.Chang, C., and E. M. Meyerowitz. 1995. The ethylene hormone response in Arabidopsis: a eukaryotic two-component signalling system. Proc. Natl. Acad. Sci. USA 92:4129-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cintas, L. M., P. Casaus, H. Holo, P. E. Hernandez, I. F. Nes, and L. S. Håvarstein. 1998. Enterocins L50A and L50B, two novel bacteriocins from Enterococcus faecium L50, are related to staphylococcal hemolysins. J. Bacteriol. 180:1988-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Man, J., M. Rogosa, and M. E. Sharpe. 1960. A medium for the culture of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 13.Dunne, C. 2001. Adaptation of bacteria to the intestinal niche: probiotics and gut disorder. Inflamm. Bowel Dis. 7:136-145. [DOI] [PubMed] [Google Scholar]
- 14.Dunne, C., L. Murphy, S. Flynn, L. O'Mahony, S. O'Halloran, M. Feeney, D. Morrissey, G. Thornton, G. Fitzgerald, C. Daly, B. Kiely, E. M. Quigley, G. C. O'Sullivan, F. Shanahan, and J. K. Collins. 1999. Probiotics: from myth to reality. Demonstration of functionality in animal models of disease and in human clinical trials. Antonie Van Leeuwenhoek 76:279-292. [PubMed] [Google Scholar]
- 15.Fabret, C., V. A. Feher, and J. A. Hoch. 1999. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol. 181:1975-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. G. Sutton, W. FitzHugh, C. A. Fields, J. D. Gocayne, J. D. Scott, R. Shirley, L. I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and C. J. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 17.Huang, J., C. J. Lih, K. H. Pan, and S. N. Cohen. 2001. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor with DNA microarrays. Genes. Dev. 15:3183-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hung, M.-N., Z. Xia, N.-T. Hu, and B. H. Lee. 2001. Molecular and biochemical analysis of two β-galactosidaseactosidases from Bifidobacterium longum HL96. Appl. Environ. Microbiol. 67:4256-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, D.-J., and S. Forst. 2001. Genomic analysis of the histidine kinase family in bacteria and archaea. Microbiology 147:1197-1212. [DOI] [PubMed] [Google Scholar]
- 20.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1993. Characterisation of the Nisin gene cluster nisABTCIPR of Lactococcus lactis, requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 21.Lee, P. J., and A. M. Stock. 1996. Characterisation of the genes and proteins of a two-component system from the hyperthermophilic bacterium Thermotoga maritima. J. Bacteriol. 178:5579-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewin, B. 1995. Control of prokaryotic gene expression, p. 375-523. In Genes V. Oxford University Press, Oxford, United Kingdom.
- 23.Loomis, W. F., G. Shaulsky, and N. Wang. 1997. Histidine kinases in signal transduction pathways of eukaryotes. J. Cell Sci. 110:1141-1145. [DOI] [PubMed] [Google Scholar]
- 24.Matsumura, H., Takeuchi, A., and Y. Kano. 1997. Construction of an Escherichia coli-Bifidobacterium longum shuttle vector transforming B. longum 105-A and 108-A. Biosci. Biotechnol. Biochem. 61:1211-1212. [DOI] [PubMed] [Google Scholar]
- 25.Minowa, T., S. Iwata, H. Sakai, H. Masaki, and T. Otha. 1989. Sequence and characteristics of the Bifidobacterium longum gene encoding L-lactate dehydrogenase and the primary structure of the enzyme: a new feature of the allosteric site. Gene 85:161-168. [DOI] [PubMed] [Google Scholar]
- 26.Mizuno, T. 1997. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of E. coli. DNA Res. 4:161-168. [DOI] [PubMed] [Google Scholar]
- 27.Morel-Deville, F., F. Fauvel, and P. Morel. 1998. Two-component signal-transducting systems involved in stress response and vancomycin susceptibility in Lactobacillus sakei. Microbiology 144:2873-2883. [DOI] [PubMed] [Google Scholar]
- 28.Morel-Deville, F., S. D. Ehrlich, and P. Morel. 1997. Identification by PCR of genes encoding multiple response regulators. Microbiology 143:1513-1520. [DOI] [PubMed] [Google Scholar]
- 29.Nagasawa, S., K. Ishige, and T. Mizuno. 1993. Novel members of the two-component signal transduction genes in E. coli. J. Biochem. 114:350-357. [DOI] [PubMed] [Google Scholar]
- 30.O'Connell-Motherway, M., G. F. Fitzgerald, and D. van Sinderen. 1997. Cloning and sequence analysis of putative histidine protein kinases isolated from Lactococcus lactis MG1363. Appl. Environ. Microbiol. 63:2454-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Connell-Motherway, M., D. van Sinderen, F. Morel-Deville, G. F. Fitzgerald, S. Dusko Ehrlich, and P. Morel. 2000. Six putative two-component regulatory systems isolated from Lactococcus lactis subsp. cremoris MG1363. Microbiology 146:935-947. [DOI] [PubMed] [Google Scholar]
- 32.O'Riordan, K. 1998. Studies on antimicrobial activity and genetic diversity of Bifidobacterium species: molecular characterisation of a 5.75 kb plasmid and a chromosomally encoded recA gene homologue from Bifidobacterium breve. Ph.D. thesis. Department of Microbiology, National University of Ireland, Cork, Ireland.
- 33.Pao, G. M., and M. H. Saier, Jr. 1995. Response regulators in bacterial transduction systems: selective domain shuffling during evolution. J. Mol. E. 40:136-154. [DOI] [PubMed] [Google Scholar]
- 34.Pao, G. M., and M. H. Saier, Jr. 1997. Nonplastid eucaryotic response regulators have a monophyletic origin and evolved from their bacterial precursors in parallel with their cognate sensor kinases. J. Mol. E. 44:605-613. [DOI] [PubMed] [Google Scholar]
- 35.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signalling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 36.Parkinson, J. S. 1993. Signal transduction schemes in bacteria. Cell 73:857-871. [DOI] [PubMed] [Google Scholar]
- 37.Parkinson, J. S. 1995. Genetic approaches for signalling pathways and proteins, p. 9-24. In J. Hoch and T. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 38.Phalip, V., J. H. Li, and C. C. Zhang. 2001. HstK, a cyanobacterial protein with both a serine/threonine kinase domain and a histidine kinase domain: implication for the mechanism of signal transduction. Biochem. J. 360:639-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pujic, P., R. Dervyn, A. Sorokin, and S. D. Ehrlich. 1998. The kdgRKAT operon of Bacillus subtilis: detection of the transcript and regulation by the kdgR and ccpA genes. Microbiology 144:3111-3118. [DOI] [PubMed] [Google Scholar]
- 40.Rabin, R. S., and V. Stewart. 1992. Either of two functionally redundant sensor proteins, NarX and NarQ, is sufficient for nitrate regulation in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 89:8419-8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rabin, R. S., and V. Stewart. 1993. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J. Bacteriol. 175:3259-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossi, M., L. Altomare, A. Gonzalez Vara y Rodriguez, P. Bridigi, and D. Matteuzzi. 2000. Nucleotide sequence, expression and transcriptional analysis of the Bifidobacterium longum MB219 lacZ gene. Arch. Microbiol. 174:74-80. [DOI] [PubMed] [Google Scholar]
- 43.Rossi, M., P. Bridigi, and D. Matteuzzi. 1997. An efficient transformation system for Bifidobacterium spp. Lett. Appl. Microbiol. 24:33-36. [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 45.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M.-C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schultz, J., R. R. Copley, T. Doerks, C. P. Ponting, and P. Bork. 2000. SMART: a web based tool for the study of genetically mobile domains. Nucleic Acids Res. 28:231-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 49.Stanton, C., G. Gardiner, H. Meehan, K. Collins, G. Fitzgerald, P. B. Lynch, and R. P. Ross. 2001. Market potential for probiotics. Am. J. Clin. Nutr. 73:476S-483S. [DOI] [PubMed]
- 50.Stewart, V., J. Parales, Jr., and S. M. Merkel. 1989. Structure of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J. Bacteriol. 171:2229-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stock, J. B., M. G. Surette, M. Levit, and P. Park. 1995. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis, p. 25-52. In J. A. Hock and T. J Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 52.Tannock, G. W. 1999. Probiotics: a critical review. Horizon Scientific Press, London, England.
- 53.Utusumi, R., S. Katayama, M. Taniguchi, T. Horie, M. Ikeda, S. Igaki, H. Nakagawa, A. Miwa, H. Tanabe, and M. Noda. 1994. Newly identified genes involved in the signal transduction of E. coli K-12. Gene 140:73-77. [DOI] [PubMed] [Google Scholar]
- 54.Volz, K. 1995. Structural and functional conservation in response regulators, p. 53-64. In J. A. Hock and T. J Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 55.Wahl, G., M. Stern, and G. R. Stark. 1979. Efficient transfer of large DNA fragments from agarose gels to diazobenzylomethyl paper and rapid hybridization with dextran sulfate. Proc. Natl. Acad. Sci. USA 76:3683-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]