Abstract
16S rRNA-targeted oligonucleotide probes were designed for butyrate-producing bacteria from human feces. Three new cluster-specific probes detected bacteria related to Roseburia intestinalis, Faecalibacterium prausnitzii, and Eubacterium hallii at mean populations of 2.3, 3.8, and 0.6%, respectively, in samples from 10 individuals. Additional species-level probes accounted for no more than 1%, with a mean of 7.7%, of the total human fecal microbiota identified as butyrate producers in this study. Bacteria related to E. hallii and the genera Roseburia and Faecalibacterium are therefore among the most abundant known butyrate-producing bacteria in human feces.
The microbiota of the gastrointestinal tract of humans has been studied extensively because of the role played by gut bacteria both in disease and in the maintenance of gut health (7, 17, 27). One important activity of the large intestinal microbiota is to break down substrates, such as resistant starch and plant cell wall polysaccharides. The main fermentation products are the short-chain fatty acids acetate, propionate, and butyrate. Of these, butyrate is known to play an important role in the metabolic welfare of colonocytes (19, 20) and is also implicated in providing protection against cancer and ulcerative colitis (3-5). Despite this prominent role, the taxonomy, population structure, and dynamics of predominant butyrate-producing bacteria in the human intestinal tract are poorly understood.
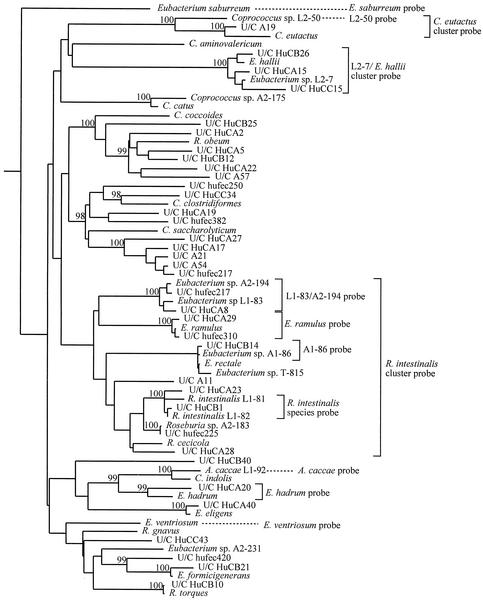
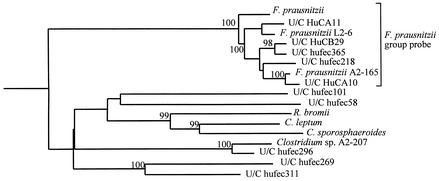
There is no simple way to selectively isolate butyrate-producing bacteria, and the majority of those recovered from nonselective isolation have proved to be highly oxygen sensitive (2). The purpose of the present study was, therefore, to design 16S rRNA-targeted oligonucleotide probes for butyrate-producing bacteria, including recent isolates from the human gut, many of which represent new species (8, 9, 21). The majority of these butyrate-producing isolates belong to the clusters XIVa and IV of clostridia (6) (Fig. 1 and 2), which account for a significant proportion of total bacterial diversity in the human large intestine on the basis of 16S rRNA sequence analyses (12, 25).
FIG. 1.
Phylogenetic tree showing the coverage of the newly designed probes within Clostridium cluster XIVa. The tree was constructed by using neighbor-joining analysis of a distance matrix obtained from a multiple-sequence alignment. Bootstrap values (expressed as percentages of 1,000 replications) are shown at branch points; values of 97% or more were considered significant.
FIG. 2.
Phylogenetic tree showing the coverage of the newly designed F. prausnitzii probe within Clostridium cluster IV. The tree was constructed by using neighbor-joining analysis of a distance matrix obtained from a multiple-sequence alignment. Bootstrap values (expressed as percentages of 1,000 replications) are shown at branch points; values of 97% or more were considered significant.
Four broad-specificity probes were designed to target the small subunit rRNA of bacteria related to Eubacterium hallii, the recently reclassified Faecalibacterium prausnitzii (formerly Fusobacterium prausnitzii) (9), Coprococcus eutactus, and Roseburia intestinalis clusters (Table 1 and Fig. 1 and 2). The latter probe is predicted to recognize R. intestinalis, Eubacterium rectale, and Eubacterium ramulus as well as the butyrate-producing Eubacterium isolates A2-194 and L1-83 and the Roseburia isolate A2-183 from the study of Barcenilla et al. (2). In addition, six more specific probes were designed to recognize R. intestinalis (8), Anaerostipes caccae (21), the Eubacterium isolates L1-83 and A2-194, Coprococcus isolate L2-50, and E. rectale isolate A1-86. The new probes were designed with the ARB (16) software package, checked against the Ribosomal Database Project (RDP) and EMBL databases, and named according to the nomenclature suggested by the Oligonucleotide Probe Database (OPD) (1). The probe sequences have also been deposited in the ProbeBase data bank (15). The specificity of the newly designed probes was tested by whole-cell in situ hybridization against a panel of 120 reference strains derived from the human and animal gastrointestinal tract as described by Schwiertz et al. (22) and also against the new target butyrate-producing strains. Hybridization temperatures (TH) are given in Table 1. All newly designed probes hybridized only to the respective target organisms but not to any of the other organisms tested. The exception was the L1-83 probe, which showed weak cross-reactivity with Eubacterium sp. strain A2-194.
TABLE 1.
Probes used during the study
| Probe name | OPD namea | Target species with zero mismatches | Probe sequence 5′-3′ | TH (°C) | Reference or source |
|---|---|---|---|---|---|
| Species-specific probes | |||||
| A. caccae | S-S-Acac-0194-a-A-18 | A. caccae | CTA TAC TGC CAG GGC TTT | 46 | This study |
| R. intestinalis | S-S-Rint-1102-a-A-18 | R. intestinalis | GCT TAC CCG CTG GCT ACT | 46 | This study |
| Eubacterium sp. strain L1-83b | S-St-xxxx-0576-a-A-18 | Eubacterium sp. strains L1-83 and A2-194 | AGC CTT CCG CCT GCG CTC | 58 | This study |
| Coprococcus sp. strain L2-50b | S-St-xxxx-0060-a-A-18 | Coprococcus sp. strain L2-50 | CAC CGA TCT TCT CTC GTT | 54 | This study |
| E. rectale sp. strain A1-86b | S-S-Erec-0207-a-A-18 | E. rectale strain A1-86 | GGT GGT GTA CAA GAC CCG | 52 | This study |
| E. barkeri | S-S-Ebar-1237-a-A-18 | E. barkeri, E. aggregans | CCT TTG TCC CAA CCC ATT | 51 | 22 |
| E. biforme | S-S-Ebif-0462-a-A-18 | E. biforme | CAC TCA CTC ATC ATT CCC | 51 | 22 |
| E. cylindroides | S-St-Ecyl-0461-a-A-18 | E. cylindroides | ACC CAC GGA TCA TTC CCT | 51 | 22 |
| E. cylindroides | S-St-Ecyl-0466-a-A-18 | E. cylindroides | CCG TCA CCC ACA TAG CAT | 51 | 22 |
| E. hadrum | S-*-Ehad-0579-a-A-20 | E. hadrum | GAC TTG CCA TAC CAC CTA CG | 54 | 22 |
| E. limosum | S-*-Elim-1433-a-A-18 | E. callanderi, E. limosum | GCG GTT CTC TCA CAG GCT | 51 | 22 |
| E. moniliforme | S-S-Emon-0084-a-A-18 | E. moniliforme | CCG CTA ATC CAT TTC CCG | 51 | 23 |
| E. multiforme | S-S-Emul-0183-a-A-18 | E. multiforme | GTT CCT TCA TGC GAA GGT | 51 | 23 |
| E. ramulus | S-S-Eram-0997-a-A-18 | E. ramulus | ACA TGT TCT GTC ACC GGG | 46 | 24 |
| E. saburreum | S-S-Esab-1467-a-A-18 | E. saburreum | AGT TAT CCT CCC TGC CTT | 48 | 23 |
| E. ventriosum | S-S-Even-0066-a-A-18 | E. ventriosum | TCT GTC CAA GGT GCT TCG | 55 | 22 |
| Group-and cluster-specific probes | |||||
| E. hallii L2-7/ E. hallii | S-*-Ehal-0578-a-A-18 | E. hallii L2-7, E. hallii | TTG CAC TGC CAC CTA CGC | 58 | This study |
| E. cylindroides cluster | S-*-Ecyl-0387-a-A-18 | C. innocuum, E. biforme, E. cylindroides, E. dolichum, E. tortuosum, Streptococcus pleomorphus | CGC GGC ATT GCT CGT TCA | 46 | 11 |
| Ruminococcus-Eubacterium-Clostridium cluster | S-*-Erec-0482-a-A-19 | For details see Franks et al. (10) | GCT TCT TAG TCA RGT ACC G | 50 | 10 |
| C. eutactus | S-*-Ceut-0705-a-A-21 | C. eutactus, Coprococcus sp. strain L2-50 | GTC AGT AGC AGT CCA GTA AGT | 54 | This study |
| F. prausnitzii | S-*-Fpra-0655-a-A-18 | F. prausnitzii A2-165 and L2-6 | CGC CTA CCT CTG CAC TAC | 58 | This study |
| R. intestinalis subcluster | S-*-Rint-0623-a-A-18 | Eubacterium sp. strains L1-83 and A2-194, E. rectale sp. strains A1-86, T1-815, and Roseburig sp. strain A2-183, and R. cecicola, R. intestinalis, E. rectale, E. ramulus | TTC CAA TGC AGT ACC GGG | 54 | This study |
| C. histolyticum | S-*-Chis150-a-A-23 | For details see Franks et al. (10) | TTA TGC GGT ATT AAT CTY CCT TT | 50 | 10 |
| C. lituseburense | S-*-Clit135-a-A-19 | For details see Franks et al. (10) | GTT ATC CGT GTG TAC AGG G | 50 | 10 |
Standardized probe name in accordance with the OPD (1).
No valid systematic name was available.
Fresh fecal samples from 10 healthy volunteers of both sexes, aged 28 to 56, who had consumed a Western diet and had not received any antibiotic treatment at least 3 months prior to the study, were collected and fixed as described elsewhere (24). Hybridization and enumeration were performed as described previously (22), with the lower limit of detection of 107 cells g−1 of dry feces. In addition to analysis with the newly designed probes described above, the fecal samples were analyzed with 11 Eubacterium species-specific probes described previously (22-24). Broad-specificity probes or probe mixes that targeted all eubacteria were applied (14), along with the Ruminococcus-Eubacterium-Clostridium cluster probe (Erec482) (10), the Clostridium lituseburense group probe (Clit135) (13), the Clostridium histolyticum group probe (Chis150) (10), and the Eubacterium cylindroides group probe (Ecyl387) (11).
Cell counts for the target organisms in the 10 subjects are summarized in Table 2. Each subject harbored at least three groups of butyrate producers, with a mean of 7.7% of the total fecal microbiota identified as butyrate producers in this study. The Fpra655 (F. prausnitzii) probe detected between 1.4 and 5.9% (mean, 3.8%) of the total fecal microbiota in all 10 subjects, which is in agreement with previous evidence, by using a different probe, indicating that this is one of the most abundant species in human feces (26). Also found in all 10 subjects was the R. intestinalis cluster (by using the Rint603 probe), which accounted for 0.9 to 5.0% (mean, 2.3%) of the total microbiota. Thus, the R. intestinalis and F. prausnitzii groups, which are likely to consist largely if not wholly of butyrate-producing strains, together accounted for not less than 3% and up to 10.9% (mean, 6.1%) of total eubacterial cells in the subjects studied. Organisms detected by the Ehal578 (E. hallii) probe were also widespread, being found in nine subjects and accounting for up to 2.4% (average of 0.6%) of the total microbiota. Recent work by Harmsen et al. (11) with a different probe showed that E. hallii and its close relatives can account for up to 3.6% of the total human fecal microbiota.
TABLE 2.
Quantification of the various bacterial components known to produce butyric acid within the human fecal microbiotaa
| Target | Probeb | No. of cells/g (dry wt) and percentage of the total microbiota in volunteer no.b
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| All eubacteria | Universal | 11.7 | 11.8 | 11.6 | 11.8 | 12.2 | 11.7 | 12 | 11.7 | 11.5 | 11.9 |
| Ruminococcus-Eubacterium-Clostridium cluster | Erec482 | 10.7 (5.2) | 10.7 (8.06) | 11 (26.4) | 10.5 (5.2) | 10.9 (5.3) | 10.9 (15) | 10.8 (6.3) | 10.6 (8.4) | 10.8 (19.7) | 10.9 (8.8) |
| E. hallii cluster | Ehal578 | 8.8 (0.13) | 10.1 (2) | 8.3 (0.03) | 9.1 (0.1) | 9 (0.17) | 10.4 (2.4) | 8.8 (0.12) | 9.2 (0.48) | 9 (0.12) | |
| R. intestinalis cluster | Rint603 | 9.6 (0.87) | 10.4 (3.88) | 10 (2.6) | 10 (1.66) | 10.6 (2.2) | 10 (2.07) | 10.4 (2.74) | 9.8 (1.22) | 10.2 (4.98) | 10 (1.16) |
| R. intestinalis | Rint1102 | 8.4 (0.05) | 8.4 (0.03) | 9.5 (0.44) | 9.5 (0.22) | 9.4 (0.47) | 8.5 (0.06) | 8.8 (0.2) | 9.2 (0.2) | ||
| Eubacterium sp. strains L1-83 and A2-194 | 8.4 (0.03) | 10.1 (1.28) | 9.1 (0.24) | 9.4 (0.51) | 8.8 (0.2) | 9.4 (0.26) | |||||
| E. rectale sp. strain A1-86 | Erec207 | 8.8 (0.12) | 8.5 (0.06) | 8.5 (0.04) | 9.5 (0.34) | ||||||
| E. ramulus | Eram997 | 8.5 (0.05) | 8.1 (0.03) | 8.9 (0.05) | 8.9 (0.01) | 8 (0.02) | 8.6 (0.05) | ||||
| Coprococcus sp. strain L2-50 | 8.8 (0.035) | ||||||||||
| E. hadrum | Ehad579 | 9.1 (0.4) | 9.1 (0.2) | 8.5 (0.1) | 9 (0.14) | 9.3 (0.01) | 9.5 (0.56) | 9.2 (0.17) | 9.2 (0.2) | ||
| E. ventriosum | Event66 | 10.1 (1.7) | 10.2 (1.7) | ||||||||
| E. cylindroides group | Ecyl387 | 10.2 (2.5) | 9.9 (1.7) | ||||||||
| E. biforme | Ebif462 | 10.2 (2.5) | 9.9 (1.7) | ||||||||
| F. prausnitzii cluster | Fpra655 | 10.1 (2.7) | 10.5 (4.6) | 10.2 (4.1) | 10.4 (3.75) | 10.8 (3.6) | 10.5 (5.53) | 10.1 (1.35) | 10.5 (5.89) | 10.2 (4.9) | 10.2 (1.8) |
All probes were negative for E. barkeri, E. multiforme, E. moniliforme, E. saburreum, E. cylindroides, E. dolichum, E. limosum, A. caccae, C. lituseburense, and C. histolyticum.
Counts of bacteria are expressed as numbers of organisms log10 per gram of feces (dry weight). Coefficient of variation due to assay error of the fluorescent in situ hybridization method was 0.18 (13). The percentage of the total microbiota was calculated by using counts from the universal eubacterial probe (1H).
The species-specific probes for R. intestinalis and Eubacterium hadrum were positive for eight subjects. Bacteria related to Eubacterium sp. strains L1-83 and A2-194 and E. ramulus were each found in six subjects, while relatives of E. rectale sp. strain A1-86 were detected in four subjects. Eubacterium biforme and Eubacterium ventriosum were detected in only two subjects, and Coprococcus sp. strain L2-50 was detected in only one subject. In a PCR-based analysis the Coprococcus cluster was shown to account for up to 8% of total bacterial diversity in one human individual (25). Species probes for Anaerostipes caccae, Eubacterium barkeri, E. cylindroides, Eubacterium dolichum, Eubacterium saburreum, Eubacterium limosum, Eubacterium moniliforme, and Eubacterium multiforme failed to detect cells above the limit of detection in samples from any of the 10 subjects. Negative results were also obtained with the cluster probes Chis150 and Clit135, but the Ecyl387 group probe detected significant numbers in two individuals.
The Erec482 probe used for the detection of the whole Ruminococcus-Eubacterium-Clostridium cluster (cluster XIVa) detected between 5.2 and 26.4% of total fecal bacteria in the 10 volunteers tested here. These numbers are essentially in agreement with previously published data (10, 22, 23). Nonoverlapping probes designed to recognize butyrate-producing species within the Erec482 cluster, namely Rint603, Ehal578, Ehad579, and Event66, together accounted for between 10.2 and 85% (mean, 43%) of the Erec482 signal. Thus, in some subjects (subjects 2, 4, and 7) almost all of the Erec482 representatives were closely related to known butyrate producers, while in others the remaining Erec482 diversity may correspond to species that do not produce butyrate (e.g., Ruminococcus sp.) but might also include groups of butyrate producers that have yet to be targeted.
We have now designed and validated probes that target most of the presently known butyrate-producing species or groups from the human gut within the Clostridium clusters IV and XIVa. The main conclusion is that butyrate producers accounted for, on average, 7.7% of the bacteria in the 10 subjects studied, with the most abundant groups by far being R. intestinalis and F. prausnitzii. Interestingly, the proportion of bacterial cells belonging to Clostridium cluster XIVa was lower in this set of volunteers than for those of previously published data sets (10, 18). This highlights interindividual differences possibly due to diet or geographic location. Also, the fact that the narrower strain or species-specific probes tested here did not detect bacteria in all fecal samples further emphasizes the diversity of the colonic microbiota at the strain level.
Acknowledgments
This work was supported in part by SEERAD (Scottish Executive Environment and Rural Affairs Department).
We are grateful to Jennifer Martin and Sylvia Duncan for supplying many of the bacterial isolates used in this work. This work was also carried out with financial support from the Commission of the European Communities, specific RTD program “Quality of Life and Management of Living Resources,” QLK1-2000-108, “Microbe Diagnostics.” It does not necessarily reflect its views and in no way anticipates the commission’s future policy in this area.
REFERENCES
- 1.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barcenilla, A., S. E. Pryde, J. C. Martin, S. H. Duncan, C. S. Stewart, C. Henderson, and H. J. Flint. 2000. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 66:1654-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradburn, D. M., J. C. Mathers, A. Gunn, J. Burn, P. D. Chapman, and I. D. Johnston. 1993. Colonic fermentation of complex carbohydrates in patients with familial adenomatous polyposis. Gut 34:630-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman, M. A., M. F. Grahn, M. A. Boyle, M. Hutton, J. Rogers, and N. S. Williams. 1994. Butyrate oxidation is impaired in the colonic mucosa of sufferers of quiescent ulcerative colitis. Gut 35:73-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman, M. A., M. F. Grahn, M. Hutton, and N. S. Williams. 1995. Butyrate metabolism in the terminal ileal mucosa of patients with ulcerative colitis. Br. J. Surg. 82:36-38. [DOI] [PubMed] [Google Scholar]
- 6.Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. [DOI] [PubMed] [Google Scholar]
- 7.Cummings, J. H., and G. T. Macfarlane. 1997. Colonic microflora: nutrition and health. Nutrition 13:476-478. [DOI] [PubMed] [Google Scholar]
- 8.Duncan, S. H., G. L. Hold, A. Barcenilla, C. Stewart, and H. Flint. 2002. Roseburia intestinalis sp. nov., a novel saccharolytic butyrate-producing bacteria from human faeces. Int. J. Syst. E vol. Microbiol. 52:1615-1620. [DOI] [PubMed] [Google Scholar]
- 9.Duncan, S. H., G. L. Hold, H. J. Harmsen, C. Stewart, and H. Flint. 2002. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify the species into the new genus Faecalibacterium gen. nov. Int. J. Syst. E vol. Microbiol. 52:2141-2146. [DOI] [PubMed] [Google Scholar]
- 10.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harmsen, H. J., G. C. Raangs, T. He, J. E. Degener, and G. W. Welling. 2002. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 68:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hold, G. L., S. E. Pryde, V. J. Russell, E. Furrie, and H. J. Flint. 2002. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 39:33-39. [DOI] [PubMed] [Google Scholar]
- 13.Jansen, G. J., A. C. Wildeboer-Veloo, R. H. Tonk, A. H. Franks, and G. W. Welling. 1999. Development and validation of an automated, microscopy-based method for enumeration of groups of intestinal bacteria. J. Microbiol. Methods 37:215-221. [DOI] [PubMed] [Google Scholar]
- 14.Kruse, H. P., B. Kleessen, and M. Blaut. 1999. Effects of inulin on faecal bifidobacteria in human subjects. Br. J. Nutr. 82:375-382. [DOI] [PubMed] [Google Scholar]
- 15.Loy, A., M. Horn, and M. Wagner. 2003. probeBase: an online resource for rRNA-targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 17.Pryde, S. E., S. H. Duncan, G. L. Hold, C. Stewart, and H. Flint. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 17:133-139. [DOI] [PubMed] [Google Scholar]
- 18.Rigottier-Gois, L., A. G. Le Bourhis, G. Gramet, V. Rochet, and J. Dore. 2003. Fluorescent hybridisation combined with flow cytometry and hybridisation of total RNA to analyse the composition of microbial communities in human faeces using 16S rRNA probes. FEMS Microbiol. Ecol. 43:237-245. [DOI] [PubMed] [Google Scholar]
- 19.Scheppach, W. 1994. Effects of short chain fatty acids on gut morphology and function. Gut 35:S35-S38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheppach, W., H. P. Bartram, and F. Richter. 1995. Role of short-chain fatty acids in the prevention of colorectal cancer. Eur. J. Cancer 31A:1077-1080. [DOI] [PubMed] [Google Scholar]
- 21.Schwiertz, A., G. L. Hold, S. H. Duncan, B. Gruhl, M. D. Collins, P. A. Lawson, H. J. Flint, and M. Blaut. 2002. Anaerostipes caccae gen. nov., sp. nov., a new saccharolytic, acetate-utilising, butyrate-producing bacterium from human faeces. Syst. Appl. Microbiol. 25:46-51. [DOI] [PubMed] [Google Scholar]
- 22.Schwiertz, A., G. Le Blay, and M. Blaut. 2000. Quantification of different Eubacterium spp. in human fecal samples with species-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 66:375-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwiertz, A., U. Lehmann, G. Jacobasch, and M. Blaut. 2002. Influence of resistant starch on the SCFA production and cell counts of butyrate-producing Eubacterium spp. in the human intestine. J. Appl. Microbiol. 93:157-162. [DOI] [PubMed] [Google Scholar]
- 24.Simmering, R., B. Kleessen, and M. Blaut. 1999. Quantification of the flavonoid-degrading bacterium Eubacterium ramulus in human fecal samples with a species-specific oligonucleotide hybridization probe. Appl. Environ. Microbiol. 65:3705-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suau, A., V. Rochet, A. Sghir, G. Gramet, S. Brewaeys, M. Sutren, L. Rigottier-Gois, and J. Dore. 2001. Fusobacterium prausnitzii and related species represent a dominant group within the human fecal flora. Syst. Appl. Microbiol. 24:139-145. [DOI] [PubMed] [Google Scholar]
- 27.Topping, D. L., and P. M. Clifton. 2001. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 81:1031-1064. [DOI] [PubMed] [Google Scholar]