Abstract
The DNA replication complex of bacteriophage T4 has been assembled as a single unit on a minicircle substrate with a replication fork that permits an independent measurement of the amount of DNA synthesis on both the leading and lagging strands. The assembled replisome consists of the T4 polymerase [gene product 43 (gp43)], clamp protein (gp45), clamp loader (gp44/62), helicase (gp41), helicase accessory factor (gp59), primase (gp61), and single-stranded DNA binding protein (gp32). We demonstrate that on the minicircle the synthesis of the leading and lagging strands are coordinated and that the C-terminal domain of the gp32 protein regulates this coordination. We show that the reconstituted replisome encompasses two coupled holoenzyme complexes and present evidence that this coupling might include a gp43 homodimer interaction.
Bacteriophage T4 is one of the more elementary replication systems using eight proteins in the formation and propagation of a replication fork. Central to the replication process, the T4 DNA polymerase [gene product 43 (gp43)] catalyzes nucleotide incorporation in a 5′ to 3′ direction whereas its 3′ to 5′ exonuclease activity serves to maintain fidelity during replication (1). Alone, the T4 polymerase will incorporate nucleotides in a distributive fashion, and, to avert quick dissociation from the DNA, interactions between the gp43 polymerase and accessory factors are essential (2, 3). These accessory factors include the gp45 protein (sliding clamp), which possesses a ring-shaped structure with an internal diameter large enough to encircle DNA, and the 44/62 protein complex, which loads the sliding clamp onto DNA (4, 5). In addition, a DNA helicase (gene product 41) is required for the ATP- or GTP-dependent unwinding of the duplex DNA to be replicated at the front of the replication fork (6). The T4 single-stranded DNA binding protein gp32 is needed to prevent the reannealing of duplex DNA at the replication fork (7). The helicase has its own accessory factor, the gene product 59 protein, which is required to load the gp41 helicase onto single-stranded DNA coated with the gp32 single-stranded DNA binding protein (8). Finally, associated with the helicase is the DNA primase (gp61), which provides the pentaribonucleotide primers needed for the initiation of lagging-strand DNA synthesis (9).
The dynamics of holoenzyme (gp43, gp45, and gp44/62) assembly have been addressed both kinetically and structurally (10). The holoenzyme assembly process proceeds in a ordered fashion in which the gp45 sliding clamp is first loaded onto the DNA by the 44/62 protein complex in an ATP-dependent process followed by the rapid association of the polymerase (gp43), with the 45 clamp mediated through the carboxyl terminus of the polymerase (11, 12). Once formed, the holoenzyme is stable with a dissociation rate constant (koff) of 0.01 sec−1.
The same basic replication strategy is shared by all DNA polymerases characterized to date in which DNA synthesis occurs by the successive addition of nucleotides to the 3′ OH of a DNA or RNA primer. Leading-strand synthesis could be replicated in principal by a single polymerase-accessory protein complex that does not dissociate until replication of the genome is complete. However, lagging-strand synthesis must proceed in a discontinuous fashion in which replication occurs through small Okazaki fragments of 1–2 kb (13). Several problems are presented to a moving replication fork as a result of the discontinuous replication of the lagging strand. For example, the lagging-strand holoenzyme complex must be stable and processive enough to replicate 1–2 kb of DNA but must expeditiously dissociate once the Okazaki fragment is complete and then recycle itself to a newly formed RNA primer to initiate synthesis of the following fragment (14).
Alberts et al. originally proposed that the lagging-strand polymerase would be held at the replication fork via protein-protein interactions to ensure the rapid and efficient recycling of the lagging-strand polymerase to the next primer after completion of an Okazaki fragment. Direct evidence for the physical coupling of two holoenzymes is scant. The most compelling experimental evidence for coupled leading- and lagging-strand holoenzyme complexes stems from the insensitivity of DNA synthesis on both strands to dilution of the reaction (15). If there are no physical protein-protein interactions, to keep the lagging-strand polymerase coupled to a moving replication fork whose rate of movement determines in part the distance on the lagging-strand template between successful initiations dilution might be expected to increase Okazaki fragment size because more time would be required to recruit a polymerase free from solution to reinitiate lagging-strand replication.
In Escherichia coli, the lagging-strand polymerase acts processively during multiple rounds of Okazaki fragment synthesis. By using extreme dilution of active replication forks, it was demonstrated that coupling of leading- and lagging-strand synthesis was mediated by the tau subunit of the holoenzyme. It was clearly demonstrated that the tau subunit was acting as a physical bridge between the two core assemblies, resulting in the retention of the lagging-strand holoenzyme at the replication fork as it is cycled from a completed Okazaki fragment to the next primer (16). Debyser et al. (17) have also demonstrated a similar coupling in the bacteriophage T7 system. However, coupling appeared to be regulated by the gene 4 helicase/primase and not by a physical coupling of two polymerases (17).
To characterize coordinated leading- and lagging-strand synthesis in a more quantitative fashion, we have used a minicircle with an annealed replication fork. Using purified bacteriophage T7 replication proteins, Lee et al. have demonstrated the utility of this template (18). The sequence of the minicircle can be controlled such that an independent measurement of both strands can be made. We describe the assembly of the bacteriophage T4 replisome using such a template. We have tested the response of this system to the presence and absence of the single-stranded DNA binding protein (gp32) and a variant gp32-A (C-terminal truncation). We present results consistent with that observed with M13 derived replication forks (19). Using the minicircle replication fork, we demonstrate that the rates of synthesis on both the leading and lagging strands are coordinated and interconnected. No reported physical data, other than binding of the 43 protein to a 43-affinity column (15), has been reported to specifically implicate a dimeric holoenzyme complex. We describe the use of an in vivo two-hybrid system based on gp43 fusions to the lambda cI repressor protein in E. coli to ascertain whether the gp43 polymerase can mediate homodimer formation (20). Specific regions within the gene 43 polymerase responsible for homodimer formation have been found, and mutations that disrupt the interaction have been identified. The response of the rates of synthesis on both the leading and lagging strands to specific mutation in this region suggests that the polymerase-polymerase contact contribute to the coordination of their travel during replication.
Materials and Methods
Reagents.
[α-32P]dGTP and [α-32P]dCTP were purchased from New England Nuclear. Unlabeled deoxynucleotides were purchased from Amersham Pharmacia (ultrapure). All oligonucleotides were synthesized by using an Expidite DNA synthesizer using standard protocols followed by purification using PAGE. The T4 exonuclease-deficient gp43 (D129A mutation) (21), gp44/62 (22), gp45 (22), gp32 (23), gp41(24), gp59 (24), and gp61 (25) proteins were purified as previously described. Gp32-A was provided by David Giedroc (Texas A & M University).
Minicicle Synthesis.
A 70-nt oligonucleotide (5′CACCATAACCTCCACCCTCCCCAATATTCACCATCAACCCTTCACCTCACTTCACTCCACTATACCACTC) was converted into a single-stranded minicircle by using a 20-nt bridging oligonucleotide (5′GGTTATGGTGGAGTGGTATA) in a manner previously described (18). The 70-mer oligonucleotide was first phosphorylated by using T4 polynucleotide kinase followed by annealing with the 20-mer bridging oligonucleotide in dilute conditions. The 20-mer is complementary to the first 5′ 10 bases and the last 3′ 10 bases of the 70-mer so that the ends of the 70-mer are brought together on annealing for subsequent ligation using T4 DNA ligase. The 70-mer circle is then purified by denaturing PAGE and subsequent electroelution. The minicircle is then annealed with a partially complementary 109-mer strand (5′AGAGGATGATATGGAGGAGAGGTATAGGAGAGAGAAGT ATGTGGAGGTTATGGTGGAGTGGTATAGTGGAGTGAAGTGAGGTGAAGGGTTGATGGTGAATATTGGGGAG) which provides the template strand for lagging-strand synthesis, a 5′ single stranded tail region for assembly of the helicase/primase complex, and two primase recognition sequences (5′-GTT-3′). The annealed template is then purified by using a 12% native polyacrylamide gel.
Minicircle DNA Synthesis Reactions.
The standard reaction conditions used consisted of 100 nM minicircle DNA, 200 nM holoenzyme (gp43, gp45 trimers, gp44/62), 60 nM primosome (gp41, gp61, gp59), and 5 μM gp32 in reaction buffer containing 150 mM KOAc, 20 mM Tris (pH7.5), 15 mM Mg(OAc)2, 1,200 μM each dATP, dCTP, dGTP, and dTTP, 4 mM ATP, and 600 μM each UTP, CTP, and GTP. [α-32P]dGTP and [α-32P]dCTP were present at 333 mCi/mmol as indicated to measure leading- and lagging-strand synthesis, respectively. The DNA polymerase holoenzyme (gp43, gp45, and gp44/62) are first preincubated with the minicircle DNA template in the presence of 2 mM ATP for 20 sec followed by the addition of the primase/helicase (gp61/41/59) and gp32 along with the dNTPs, rNTPs and 2 mM ATP to initiate strand displacement synthesis. After incubation at 30°C, aliquots were removed at the indicated time points and were quenched with an equal volume of 0.5 M EDTA. To determine the sizes of leading- and lagging-strand products, samples were electrophoresed through a 0.8% alkaline agarose gel at 1.8 V/cm for 36 h. After electrophoresis, the gels were neutralized by soaking in TBE buffer for 30 min and were dried onto DE81 paper, and the labeled strands were visualized by PhosphorImaging techniques. To quantitate the amount of DNA synthesized, samples were spotted onto DE81 filter paper, were washed three times with 0.3 M ammonium sulfate (pH 8.0) to remove any unincorporated deoxynucleotides, were air dried, and were scintillation-counted. Denaturing PAGE followed by analysis using PhosphorImaging techniques was also used to quantitate the amount of DNA synthesis. For inhibition of lagging-strand synthesis, ddCTP (600 μM) was added 75 sec after initiation of the reaction.
E. coli Two-Hybrid System.
All plasmids, lambda phage, and E. coli strains were the kind gift of Jim Hu (Texas A&M University). The plasmid pJH391 was used to construct fusions between the gene 43 polymerase and the N-terminal DNA binding domain of the lambda repressor. This ampicillin-resistant plasmid contains the Plac-promoter and a T7 terminator site. The host strain used was AG1688 (MC10G1 F′128 lacIq lacZ∷Tn5). These cells contain an F′ plasmid that carries the lacIq gene to produce enough lac repressor to turn down the expression of the fusion protein. This is important as the N-terminal domain by itself can confer immunity at high expression levels.
For the characterization of gene 43/cI N-terminal domain fusion constructs, lambda phage KH54 was used as described (26). This phage does not carry its own cI repressor gene; therefore, phage immunity will totally depend on the activity of the chimeric repressor. Bacterial cells that contained the 43/cI N-terminal-domain fusions were grown overnight in LB media containing ampicillin, 1 mM MgSO4, and 1.0% maltose. Cells were then diluted 10-fold in 2.5 ml of top agar and were plated. After 5 min, 5 μl of lambda phage kH54 (with increasing titers at 10-fold intervals from 101 to 106 plaque-forming units) were spotted on the top agar. After overnight incubation, the minimal amount of phage titer required for clearing of the lawn was determined. As a positive control, parallel experiments were done by using the bacterial strain JH370 (AG1688/pJH370). Plasmid pJH370 contains a fusion between the repressor N-terminal DNA binding domain and the leucine zipper dimerization domain of GCN4. This strain should be resistant to a high plaque-forming unit dose, 103–104, infection by lambda kH54. As a negative control, the strain JH379 (AG1688/pkH101) was used. The plasmid pkH101 expresses only the N-terminal DNA binding domain of the repressor, and this strain is sensitive to lambda kH54 infection (sensitive to 101 plaque-forming units). lambda phage imm21c was also used as a control. This phage contains a substitution of the immunity region of lambda such that the repressor fusion is not able to repress infection and should be sensitive to infection.
The protocol for mutagenic PCR was derived from standard PCR conditions with the following changes made to enhance the mutation rate. The MgCl2 concentration was increased to 7 mM to stabilize noncomplementary base pairs. MnCl2 (0.5 mM) was added to diminish the template specificity of the taq polymerase, and the concentration of dCTP and dTTP was increased to 1 mM to promote misincorporation. Finally, the amount of taq polymerase added was increased to 10 units to promote chain extension beyond positions of base mismatch. Blue/white screening was conducted by using bacterial strain JH372, which contains a chromosomal copy of the lacZ gene under the control of the lambda operator site.
Results and Discussion
Coupled Leading- and Lagging-Strand Synthesis.
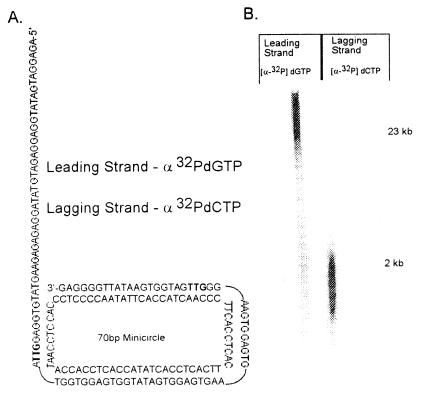
Phage M13 DNA has primarily been used as the template when studying coordinated leading- and lagging-strand synthesis. These larger DNAs depend solely on the use of a denaturing alkaline gel system to resolve the larger products of the leading strand from those made on the lagging strand, making quantitative analysis of coordinated leading- and lagging-strand synthesis difficult at best. We have used a small circular duplex molecule (70 bp) containing a 5′ single-stranded DNA tail (40 nt) (Fig. 1A). The 3′ hydroxyl terminus on the leading strand provides the primer for the assembly of the T4 holoenzyme complex (gene products 43, 45, and 44/62). The 5′ single-stranded tail provides for the loading of the 41/61 helicase/primase complex (in concert with the gene 59 and 32 proteins), which subsequently translocates in a 5′ to 3′ direction to join the leading-strand polymerase. The sequence of the lagging-strand template contains two recognition sites (5′-GTT-3′), which the primase recognizes during synthesis of the pentaribonucleotide primers required for the initiation of Okazaki fragment synthesis.
Figure 1.
(A) The structure and nucleotide sequence of the minicircle primer-template is shown. The minicircle was constructed as described in Materials and Methods. It consists of a 70-base minicircle to which a 109-base lagging-strand template is annealed to provide a 40-base 5′-single-stranded tail to which the gp41 helicase can bind. Two gp61 primase recognition sites (5′-GTT-3′), shown in bold) are included for the synthesis of the pentaribonucleotide primers required to initiate Okazaki fragment synthesis. The sequence of the minicircle (leading-strand template) is devoid of guanine residues, and the lagging-strand template (109-nt oligonucleotide) is devoid of cytosine residues, allowing leading- and lagging-strand synthesis to be independently quantitated by measuring the incorporation of dGMP and dCMP, respectively. (B) DNA synthesis on the minicircle template was performed and analyzed by alkaline gel electrophoresis as described in Materials and Methods. The reaction contained either [α-32P]dGTP or [α-32P]dCTP, as indicated, to follow the products of leading- and lagging-strand synthesis, respectively.
Lee et al. have previously shown that a small and defined minicircle template provides several advantages (18). In addition to the site-specific locations of the fork and primase sites, the minicircle can be designed such that synthesis on the leading and lagging strands can be monitored independently. The minicircle used is devoid of guanine residues in the leading-strand template, and the complementary lagging-strand template is devoid of cytosine residues. It therefore becomes possible to measure in a quantitative fashion the amount of leading- and lagging-strand synthesis independently by following either dGMP or dCMP incorporation, respectively.
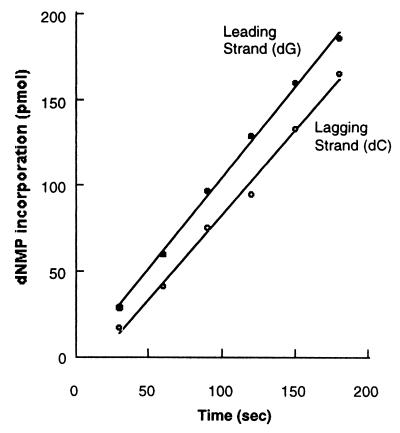
By using purified bacteriophage T4 replication proteins, the specific labeling of high molecular weight leading-strand products with [α-32P]dGMP or the smaller Okazaki fragments with [α-32P]dCMP has been demonstrated by using alkaline gel electrophoresis (Fig. 1B). Based on the sizes of the replication products as measured by gel analysis, each minicircle has been replicated thousands of times. Fig. 2 shows a comparison of the rates of leading- and lagging-strand synthesis as measured by the incorporation of dGMP or dCMP, respectively. The rates of leading- and lagging-strand synthesis are identical over the time course shown, suggesting that the additional steps (including priming and polymerase recycling) required for lagging-strand synthesis are coupled to the leading strand. The short delay in the onset of lagging-strand synthesis is most likely the result of the formation of the lagging-strand holoenzyme complex at the replication fork.
Figure 2.
Comparison of the rates of leading- and lagging-strand synthesis with the complete replication system as described in Materials and Methods. After incubation at 30°C, aliquots of the reaction were removed and quenched at the indicated time points, and the incorporation of [α-32P]dGMP or [α-32P]dCMP was quantitated as a measure of the rate of synthesis of either leading- or lagging-strand synthesis, respectively.
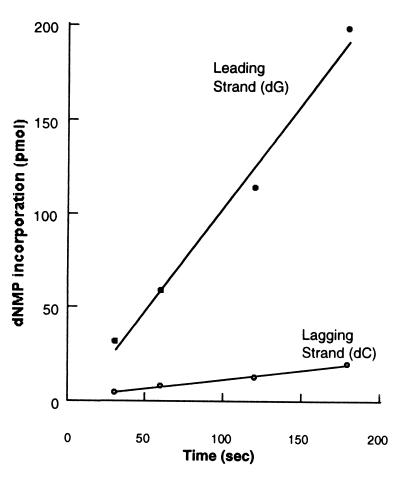
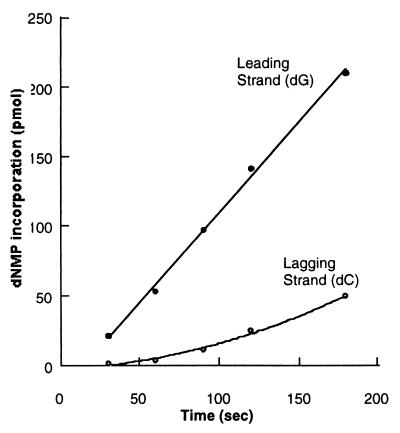
The role played by the gp32 single-stranded DNA binding protein during coordinated leading- and lagging-strand synthesis was also addressed because it served as a good test for the response of the bacteriophage T4 replication fork to the minicircle system. Earlier studies on M13 templates had implicated the importance of gp32 levels in the efficient coupling of RNA primer synthesis to initiation of Okazaki fragment synthesis (19). If gp32 is omitted from the replication reaction using the minicircle primer-template, leading- and lagging-strand synthesis becomes uncoupled, with virtually no lagging-strand synthesis (Fig. 3). We show that the requirement for gp32 is not solely attributable to its single-stranded DNA binding activity by examining the effect of substituting a C-terminally truncated derivative called gp32-A for the wild-type gp32 protein. The C-terminal A-domain has been found to mediate specific interactions with many T4 replication proteins (gp43, gp61, and gp59), and gp32-A retains DNA binding properties very similar to the intact protein (27, 28). Earlier work has shown that substituting gp32-A for intact gp32 in the T4 multienzyme complex alters the normal replication reaction in a similar fashion to omitting the gp32 protein altogether. As shown in Fig. 4, gp32-A is not able to replace wild-type 32, suggesting that protein-protein interactions between the C-terminal A-domain of gp32 and one or more of the replication proteins is required for coordinated synthesis using the minicircle replication system.
Figure 3.
Gp32 single-stranded protein is critical during coordinated leading- and lagging-strand synthesis. The reaction was performed as in Fig. 2, with the omission of gp32.
Figure 4.
Protein-protein interactions mediated by the C terminus of the gp32 protein are critical for coordinated leading- and lagging-strand synthesis using the minicircle template. The reaction was performed as in Fig. 2, replacing wild-type gp32 with gp32-A, a truncated derivative of the gene 32 protein that lacks the C-terminal A domain.
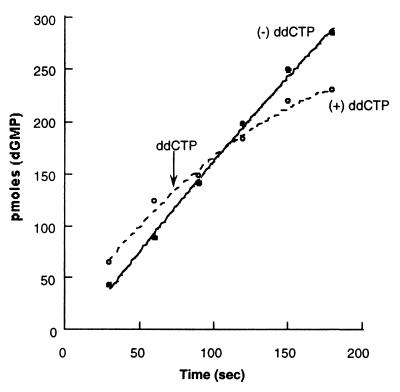
Additional protein-protein interactions are of course involved in the assembly and coordination of the multiprotein complex that constitutes the T4 DNA replication fork (29). The unique base composition of the minicircle primer-template allows us to explore coupling of leading- and lagging-strand synthesis in a direct fashion. Because dCMP is exclusively incorporated into the lagging strand, it is possible to specifically inhibit lagging-strand synthesis by incorporation of the chain terminating dideoxynucleotide ddCMP. It is reasonable to presume that suppression of synthesis on the lagging strand will inhibit leading-strand synthesis if events occurring on one strand are physically coupled to the other via protein-protein interactions. This has previously been demonstrated using the bacteriophage T7 system (18). As is shown in Fig. 5, synthesis on the leading strand is strongly inhibited when ddCTP is added to inhibit lagging-strand synthesis 75 sec into a coupled reaction. Moreover, in the absence of lagging-strand synthesis (by omission of rNTPs), leading-strand synthesis is not effected by the addition of ddCTP (data not shown). This is strong evidence that, on addition of ddCTP to stop lagging-strand synthesis, we have stalled a replisome complex, which includes two associated holoenzyme complexes.
Figure 5.
Inhibition of lagging- strand synthesis results in the concomitant inhibition of leading-strand synthesis. Leading-strand synthesis in the complete replication reaction was again determined by monitoring the incorporation of [α-32P]dGMP. The nucleotide analog ddCTP was added 75 sec after the initiation of the reaction, as described in Materials and Methods, to inhibit lagging-strand synthesis.
A Two-Hybrid System Used to Characterize a Putative 43–43 Protein-Protein Interaction.
Alberts was the first to propose that a physical coupling of the two holoenzyme complexes would ensure the rapid and efficient recycling of the lagging-strand polymerase to the next primer after completion of an Okazaki fragment. Many of the specific protein-protein interactions involved in the assembly and coordination of the large machine that constitutes the T4 replication fork are relatively weak and have been difficult to detect by conventional means. The technique of affinity chromatography proved useful because even relatively weak protein-protein interactions were detected when the protein immobilized on the column matrix was high (≈1 mg/ml). Results obtained by using a gp43 affinity column were most notable with regard to modeling a replication fork with coupled holoenzyme complexes. Alberts and coworkers showed that a gp43 affinity column retained gp43 when exposed to a cleared lysate of T4-infected E. coli that contained radiolabeled T4 early proteins (15). Self-aggregations of the gp43 proteins were disrupted by prolonged washing with low salt buffer and were therefore unstable. However, the lysate that was passed through the gp43 column was completely cleared of any gp43 protein.
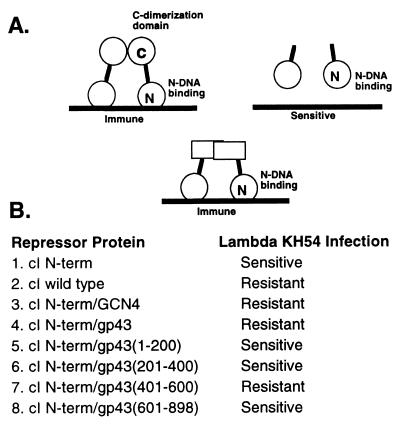
To further characterize this putative 43 homodimer interaction, we used a two-hybrid-like system in E. coli that utilizes gene fusions to the lambda cI repressor protein (Fig. 6A). The lambda phage cI repressor has been used extensively as a genetic tool to characterize homodimer interactions in E. coli (20). The lambda cI repressor consists of an N-terminal helix-turn-helix DNA binding domain (amino acids 1–92) and a globular C-terminal dimerization domain connected by a flexible hinge region of about 40 amino acids. The cI repressor binds a dyad symmetric DNA element (lambda operator site) in a dimerization-dependent manner and represses transcription of the genes in the lytic pathway of the phage life cycle by blocking the binding of RNA polymerase to its promoter site. The system used takes advantage of the observation that a functional transcriptional repressor can be created by fusing a heterologous dimerization domain to the DNA binding domain of the lambda cI repressor protein (20). Intracellular expression of such a repressor fusion will confer a level of phage immunity that will depend on the homodimerization tendency of the particular peptide fused to the repressor.
Figure 6.
(A) Schematic overview of the lambda cI repressor-based two-hybrid assay used to screen for a 43–43 homodimer interaction. (B) Fusion protein activity was assayed after transformation into E. coli strain AG1688. Activity was evaluated by scoring sensitivity to phage (KH54) superinfection as described in Materials and Methods.
We determined that the gp43 repressor fusion (gene-43/cI N-domain) supported immunity to KH54 phage infection, providing a phenotype that can be exploited in genetic studies of gp43 homodimer formation (Fig. 6B). In an attempt to identify the regions of the gene 43 polymerase that support dimer formation, various regions of the polymerase were fused to the cI DNA binding domain followed by analysis using the same two-hybrid assay. The gene 43 protein was split into various fragments, including amino acid regions 1–200, 301–400, 401–600, and finally 601–898 (Fig. 6B). It was found that only the region corresponding to amino acids 401–600 of the 43 polymerase resulted in an immunity to lambda phage KH54 infection, suggesting that this is the primary region responsible for mediating a 43–43 protein-protein interaction. As a control infection, lambda imm21c was used. This phage contains a different immunity region that is not recognized by the cI N-terminal DNA binding domain, and, therefore, its infection should not be repressed. As expected, all of the above repressor fusions were sensitive to lambda imm21c.
The bacteriophage RB69 and T4 gp43 proteins are either identical or chemically similar at 74% of all amino acid sites (30). The bacteriophage RB69 DNA polymerase structure has been solved and includes two striking protrusions (31). One is a hydrophobic stretch of about 12 amino acids at the C terminus that extends outward. Removal of these residues from the T4 polymerase abolishes interaction with the gene product 45 sliding clamp and eliminates processive DNA synthesis (12). The other striking protrusion is an antiparallel coiled-coil extension of the finger domain located next to the central core. On alignment of the T4 gene 43 with that of the RB69 polymerase, the amino acid region 401–600 includes this long protrusion of the finger domain. It has been suggested that this structure might play some role in the interaction of two polymerases during coupled leading- and lagging-strand DNA synthesis (30, 31).
To identify possible mutations that would result in the loss of homodimer formation, mutagenic PCR was used to generate random mutations within the 401–600 amino acid region of the 43 gene followed by fusion to the cI N-terminal DNA binding domain and transformation into the E. coli strain JH372. The protocol for mutagenic PCR was derived from standard PCR conditions with changes made as described in Materials and Methods. The strain JH372 has the same phenotype as AG1688 except for a chromosomal copy of the LacZ gene driven by the lambda operator site. The intracellular expression in JH372 of a repressor fusion with homodimerization tendency will result in repression of lacZ expression and the formation of a white colony. As expected, the transformation of JH372 with the wild-type 401–600/cI N terminus repressor fusion construct resulted in the expected repression of lacZ expression and the growth of white colonies, thus providing a screen from which to isolate nondimerizing mutants. Mutations in the region 401–600 resulting in the loss of dimer formation included two double mutants, Leu538Pro/Lys480Arg and Glu526Val/Lys497Arg. We are currently in the process of characterizing these mutations. However, when compared with the structure of the Rb69 polymerase, the proline mutation lies at the base of the second helix of the anti-parallel coiled-coil extension of the finger domain and was chosen as the first mutation to be characterized.
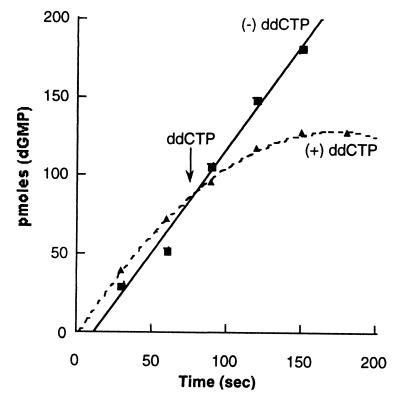
To determine the effect the leu538pro mutation might have on the formation of a replisome complex encompassing two holoenzyme complexes, we again used ddCTP to stop lagging-strand synthesis and followed the effect on leading-strand synthesis. As is shown in Fig. 7, inhibition of lagging-strand synthesis with the incorporation of ddCTP has a diminished effect on the resultant leading-strand synthesis. We interpret these results to mean that this particular contact point between polymerases does sensitize one holoenzyme to events occurring on the other strand. However, failure to completely bypass inhibition of leading-strand synthesis after repression of lagging-strand synthesis does suggest that additional contacts are involved during the coupled replication of both strands.
Figure 7.
Effect of Leu538Pro mutation in gp43 on inhibition of leading-strand synthesis with the addition of ddCTP to inhibit lagging-strand synthesis. Reactions were performed as in Fig. 5.
Furthermore, the sizes of both the leading- and lagging-strand products using the mutant (Leu538Pro) polymerase compare favorably against the wild-type enzyme (data not shown). The absence of a noticeable change in the length of the Okazaki fragments from a system reconstituted from either the wild type or mutant polymerase would suggest that another factor predominates in controlling their length. The simple presence of the required template triplet sequence appears too frequently in the minicircle (twice every 70 bp) to be used as the sole signal for initiation because the average size of the Okazaki fragments observed both in vivo and with the minicircle system is 1–2 kb (32). One possibility is a special temporal control mechanism such as the accumulation of a gp32-coated single-stranded structure that mediates a priming event. The effect of other replication proteins on primer synthesis has suggested that the gp32 protein regulates primase activity in coordination with the polymerase accessory proteins. Richardson and Nossal have shown that primer synthesis by gp61 is strongly inhibited by the presence of 32 protein on DNA and that primase activity is restored by the addition of the 45 and 44/62 proteins (33). A model was proposed in which the primase/helicase complex is not able to make primers on templates covered with 32 protein whereas the fragment initiated by the previous primer is being elongated. When the holoenzyme has completed synthesis of this fragment, the accessory factors are released and free to move relative to the 61 primase to displace the 32 protein, which was inhibiting primer synthesis, and facilitate holoenzyme complex formation at the start of the next Okazaki fragment. The primase/helicase should continue to expose single-stranded DNA, which is again coated by the 32 protein, inhibiting primer synthesis until completion of the next Okazaki fragment.
Results with the E. coli system have shown that the E. coli primase tightly interacts with its RNA primer and must be displaced before the primer can be extended during Okazaki fragment synthesis (34). The clamp loader (γ complex) is responsible for the displacement of the primase, followed by the loading of the β clamp onto the primer. The underlying mechanism was reported to be a competition between primase and the clamp loader for the single-stranded binding protein. Moreover, work with the bacteriophage T7 system has shown that the gene 2.5 single-stranded binding protein is essential for the coordination of leading- and lagging-strand synthesis (18).
Although there is functional evidence in bacteriophage T4 for an accessory protein-primase/helicase interaction, direct physical interaction has not yet been demonstrated. It is also not clear that during T4 replication the holoenzyme accessory factors stimulating primer synthesis are those released on completion of the previous fragment, or bind free from solution. At any rate, this model is consistent with the requirement for full-length gp32 during coordinated leading- and lagging-strand synthesis and for the high efficiency with which primers are elongated by the polymerase (19).
We are currently pursuing the characterization of the region corresponding to amino acids 401–600 of the 43 polymerase with regard to its participation in the assembly and continued stability of a replisome complex encompassing two holoenzyme complexes. We have not ruled out the possibility that processive synthesis by the T4 gp43 polymerase during multiple rounds of Okazaki fragment synthesis is regulated by additional protein-protein interactions. For example, in bacteriophage T7, the gene 4 primase/helicase, as well as the gene 2.5 single-stranded binding protein, appears to be the central elements mediating the physically coupling between the two polymerase, and, in E. coli, the tau subunit serves to bridge the two holoenzyme complexes (17, 18).
Acknowledgments
The authors thank Ann Valentine, Faoud Ishmael, and Uljana Mayer for help in the purification of the proteins used in this study. This work was supported by National Institutes of Health Grants 3 R01 GM13306-35 to SJB and 1 F32 GM19317-01A1 to F.S.
Abbreviation
- gp
gene product
References
- 1.Alberts B M, Frey L. Nature (London) 1970;227:1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- 2.Nossal N G, Peterlin B M. J Biol Chem. 1979;254:6032–6040. [PubMed] [Google Scholar]
- 3.Huang C C, Hearst J E, Alberts B M. J Biol Chem. 1981;256:4087–4094. [PubMed] [Google Scholar]
- 4.Moarefi I, Jeruzalmi D, Turner J, O'Donnell M, Kuriyan J. J Mol Biol. 2000;296:1215–1223. doi: 10.1006/jmbi.1999.3511. [DOI] [PubMed] [Google Scholar]
- 5.Mace D C, Alberts B M. J Mol Biol. 1984;177:279–293. doi: 10.1016/0022-2836(84)90457-1. [DOI] [PubMed] [Google Scholar]
- 6.Nossal N G. J Biol Chem. 1979;254:6026–6031. [PubMed] [Google Scholar]
- 7.Cha T A, Alberts B M. J Biol Chem. 1989;264:12220–12225. [PubMed] [Google Scholar]
- 8.Morrical S W, Hempstead K, Morrical M D. J Biol Chem. 1994;256:4087–4094. [PubMed] [Google Scholar]
- 9.Liu C C, Alberts B M. Proc Natl Acad Sci USA. 1980;77:5698–5703. doi: 10.1073/pnas.77.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sexton D J, Berdis A J, Benkovic S J. Curr Opin Chem Biol. 1997;1:316–322. doi: 10.1016/s1367-5931(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 11.Berdis A J, Benkovic S J. Biochemistry. 1996;35:9253–9265. doi: 10.1021/bi952569w. [DOI] [PubMed] [Google Scholar]
- 12.Berdis A J, Soumillion P J, Benkovic S J. Proc Natl Acad Sci USA. 1996;93:12822–12827. doi: 10.1073/pnas.93.23.12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogawa T, Okazaki T. Annu Rev Biochem. 1980;49:421–457. doi: 10.1146/annurev.bi.49.070180.002225. [DOI] [PubMed] [Google Scholar]
- 14.Carver T E, Sexton D J, Benkovic S J. Biochemistry. 1997;31:10984–10994. [Google Scholar]
- 15.Alberts B M, Barry J, Bedinger P, Formosa T, Jongeneel C V, Kreuzer K N. Cold Spring Harbor Symp Quant Biol. 1983;47:655–658. doi: 10.1101/sqb.1983.047.01.077. [DOI] [PubMed] [Google Scholar]
- 16.Sungsub K, Dallmann H G, McHenry C S, Marians K J. J Biol Chem. 1996;271:21406–21412. doi: 10.1074/jbc.271.35.21406. [DOI] [PubMed] [Google Scholar]
- 17.Debyser Z, Tabor S, Richardson C C. Cell. 1994;77:157–166. doi: 10.1016/0092-8674(94)90243-7. [DOI] [PubMed] [Google Scholar]
- 18.Lee J, Chastain P D, Kusakabe T, Griffith T, Richardson C C. Mol Cell. 1998;1:1001–1010. doi: 10.1016/s1097-2765(00)80100-8. [DOI] [PubMed] [Google Scholar]
- 19.Burke R L, Alberts B M, Hosoda J. J Biol Chem. 1980;255:11484–11493. [PubMed] [Google Scholar]
- 20.Hu J C, O'Shea E K, Jim P S, Sauer R T. Science. 1990;250:1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- 21.Frey M W, Nossal N G, Capson T L, Benkovic S J. Proc Natl Acad Sci USA. 1993;90:2579–2583. doi: 10.1073/pnas.90.7.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nossal N G. J Biol Chem. 1979;254:6026–6031. [PubMed] [Google Scholar]
- 23.Shamoo Y, Adari H, Konigsberg W H, Williams K R, Chase J W. Proc Natl Acad Sci USA. 1986;83:8844–8848. doi: 10.1073/pnas.83.23.8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spacciapoli P, Nossal N G. J Biol Chem. 1994;269:447–455. [PubMed] [Google Scholar]
- 25.Hinton D M, Nossal N G. J Biol Chem. 1985;260:12858–12865. [PubMed] [Google Scholar]
- 26.Christopher A, Kingston R E. Nucleic Acids Res. 1995;23:269–276. doi: 10.1093/nar/23.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krassa K B, Green L S, Gold L. Proc Natl Acad Sci USA. 1991;88:4010–4014. doi: 10.1073/pnas.88.9.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spicer E, Williams K R, Konigsberg W. J Biol Chem. 1979;254:6433–6436. [PubMed] [Google Scholar]
- 29.Nossal N. FASEB J. 1992;6:871–878. doi: 10.1096/fasebj.6.3.1310946. [DOI] [PubMed] [Google Scholar]
- 30.Wang C-C, Yeh L-S, Karam J D. J Biol Chem. 1995;270:26558–26564. doi: 10.1074/jbc.270.44.26558. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Sattar A K, Wang C C, Karam J D, Konigsberg W H, Steitz T A. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 32.Kurosawa Y, Okazaki T. J Mol Biol. 1979;135:841–861. doi: 10.1016/0022-2836(79)90515-1. [DOI] [PubMed] [Google Scholar]
- 33.Richardson R W, Nossal N G. J Biol Chem. 1989;254:4732–4739. [PubMed] [Google Scholar]
- 34.Yuzhakov A, Kelman Z, O'Donnell M. Cell. 1999;96:153–163. doi: 10.1016/s0092-8674(00)80968-x. [DOI] [PubMed] [Google Scholar]