Abstract
The persistence of human immunodeficiency virus (HIV) in optimally treated infected individuals poses a major therapeutic problem. In latently infected cells, one of the observed phenotypes is absent elongation of viral transcription. Thus, the positive elongation factor b (P-TEFb), which is usually recruited by NF-κB or Tat, is not present on the HIV long terminal repeat (LTR). Although most attempts to activate these proviruses centered on NF-κB, we investigated effects of Tat. To this end, we generated transgenic mice, which secreted a chimera between Tat and the green fluorescent protein from β cells of the pancreas. This extracellular Tat distributed widely, entered nuclei of resting cells, and specifically transactivated the HIV LTR. No deleterious side effects of Tat were found. Next, we determined that Tat can activate latent proviruses in optimally treated infected individuals. In their cells, T-cell activation or exogenous Tat could induce viral replication equivalently. Thus, P-TEFb could activate the majority of the latent HIV, in this case by Tat.
Human immunodeficiency virus type 1 (HIV-1) is the cause of AIDS. The development and clinical introduction of highly active antiretroviral therapy (HAART) has prolonged the survival of infected individuals (28, 29). HAART reduces HIV in plasma to levels below detection. However, the virus cannot be eradicated by current drug regimens. Instead, HIV persists in the body despite this optimal therapy and survives in a latent state (13, 17, 58). Thus, the need for new therapeutic strategies that target viral latency is evident.
The most important reservoir in infected individuals is composed of resting CD4+ T lymphocytes carrying integrated proviruses (12). This latency is established and maintained at the level of transcription. Inefficient and/or no initiation (34) and/or elongation of transcription (1) represent the two most important mechanisms whereby integrated proviruses are not expressed. In the first scenario, the provirus integrates into inactive heterochromatin, possibly near centromeres. No recruitment of RNA polymerase II (RNAPII) results in no transcription. Possibly, during cellular activation, growth, and proliferation, this chromatin is remodeled and HIV replication ensues. Nuclear factor kappa B (NF-κB), nuclear factor of activated T cells, and Ets family members that bind the HIV enhancer play important roles in this process (33, 44, 52). Absent elongation of viral transcription represents the second scenario (1, 36). RNAPII has cleared the promoter but is arrested at or slightly downstream from the transactivation response (TAR) RNA stem-loop (36, 43). The viral transactivator Tat and its cellular coactivator, the positive transcription elongation factor b (P-TEFb) then bind TAR and cause RNAPII to elongate (47, 56). Since the vast majority of HIV integrates into active chromatin and these cells harbor more than one provirus (35, 50), these findings favor the second scenario to account for the majority of latent proviruses. Indeed, this picture could be documented in both transformed cell lines (18, 19) and peripheral blood mononuclear cells (PBMC) of infected individuals (1).
The phosphorylation of the carboxy-terminal domain of RNAPII is critical for transcriptional elongation (14, 47). Whereas the unphosphorylated RNAPIIa assembles into preinitiation complexes, it is the phosphorylated RNAPIIo that copies the genome. After promoter clearance, negative transcription factors (47), which include the DRB sensitivity-inducing (54) and the negative transcription elongation (60) factors, cause a transcriptional arrest during which only short transcripts are synthesized and RNAPIIa can fall off. To release this block, the carboxy-terminal domain must be phosphorylated by P-TEFb, which contains a kinase (Cdk9) and a C-type cyclin (T1, T2, or K: CycT1, CycT2, or CycK) (47). The HIV long terminal repeat (LTR) recruits P-TEFb by two mechanisms. The first is via NF-κB, whose RelA subunit binds CycT1 (5). This activation leads to the synthesis of Tat, which also recruits P-TEFb via TAR (56). The tripartite complex between P-TEFb, Tat, and TAR leads to the efficient replication of HIV. When Tat and/or P-TEFb is in low abundance and/or inactive, the elongation of viral transcription is not observed.
Although only the human, but not the murine, P-TEFb supports HIV replication, Tat binds both CycT1 proteins well. This statement is supported by direct binding data and heterologous RNA tethering of Tat, e.g., via the regulator of virion gene expression Rev and its Rev response element, where the Rev.Tat chimera functioned equally well on the modified HIV LTR in cells from both species (6, 24, 42). Rather, the cysteine at position 261 in the human CycT1, which is a tyrosine in the murine CycT1, coordinates the Zn2+-dependent interaction between the two proteins and brings arginine-rich motifs (ARM) in CycT1 and Tat into close proximity so that they bind the 5′ bulge and central loop in TAR (26). All other functions of Tat that do not depend on TAR should be indistinguishable between humans and mice. These other effects of Tat remain controversial. They include T-cell activation and proliferation (46), apoptosis (41, 57), the expression of lymphokines and cytokines (49), central nervous system damage (30), and growth of Kaposi cells (16). They could result from the engagement of specific receptors by extracellular Tat or, because the ARM in Tat causes rapid uptake and nuclear targeting of Tat and other attached proteins in cells (20, 27) from its direct transcriptional effects. Alternatively, these effects could be caused by denatured, aggregated Tat, which contains seven cysteines. There are also reports where transgenic mice, which expressed intracellular Tat from a variety of promoters, did not develop any phenotype other than reduced levels of glutathione (9) and atypical skin lesions after 2 years of age (53).
To determine effects of extracellular Tat on the host and HIV, we created mice in which Tat was secreted from β cells of the pancreas. This Tat distributed widely and entered resting cells without causing any side effects after 2 years of observation. Its levels in serum were equivalent to those that activated latent proviruses in peripheral blood lymphocytes (PBL) of infected individuals. Indeed, transcriptional profiles from optimally treated patients revealed that lack of Tat and/or P-TEFb contributes significantly to the latent reservoir in the host.
MATERIALS AND METHODS
Generation of sTat.GFP transgenic mice.
A tissue plasminogen activator (tPA) leader peptide sequence (37) was fused in frame with Tat at the N terminus, and the enhanced green fluorescent protein (GFP) is tagged at the C terminus of the Tat protein. A 9.7-kb rat insulin promoter sequence (RIP7), including the first intron of the rat insulin II gene, was used to direct pancreatic β-cell-specific expression of the fusion Tat.GFP protein. Three transgenic founder mice were generated by microinjection of this transgenic sTat.GFP cDNA at a concentration of ∼2 ng/ml into B6D2F1 mouse embryos.
Histology.
Frozen unfixed or 4% paraformaldehyde-fixed paraffin sections of different tissues of sTat.GFP transgenic mice or wild-type mice were cut into 10-nm-thick sections. Paraffin sections were stained with hematoxylin and eosin (H&E) for bright field imaging, and the frozen unstained sections were used for dark-field fluorescent microscopy. Different leukocytes were collected by cytospin preparations onto slides as duplicates from one blood sample. One of the duplicates is unstained for dark-field fluorescent microscopy, and the other one is stained with H&E for bright field imaging.
Cell proliferation and apoptosis.
CD4+ T cells isolated from the thymi of wild-type or transgenic littermates were cultured with phorbol myristate acetate and ionomycin in RPMI supplemented with 10% fetal bovine serum (FBS). After 24 h of incubation, we harvested the activated CD4+ T cells and added 1 ml of ice-cold 70% ethanol to cells with continuous vortexing. We then spun down the cells and left 100 μl of the supernatant to which we added 400 μl of propidium iodide (PI) staining solution (50 μg of PI/ml, 100 U of RNaseA/ml, and 1 g of glucose/liter in phosphate-buffered saline) into the cell suspension. After PI staining, data were acquired and analyzed on a FACScalibur with CellQuest software (Becton Dickinson, Franklin Lakes, N.J.). The cell proliferation and apoptosis profile of the activated T cells was illustrated by using Modifit software.
Immunoprecipitation and Western blotting.
The living colors full-length A.v. polyclonal antibody (Clontech, Palo Alto, Calif.) was used for immunoprecipitation and the living colors A.v. (JL-8) monoclonal antibody (Clontech) was used to detect the secreted Tat.GFP fusion protein in the Western blot. For serial dilutions of GST.Tat chimera and subsequent Western blotting, polyclonal anti-glutathione S-transferase (GST) antibody (Amersham Pharmacia Biotech, Piscataway, N.J.) was used.
Patient samples.
Samples from 30 HIV-positive patients were provided by the Positive Health Program's Center for AIDS Research at San Francisco General Hospital. Twenty subjects had been receiving HAART for various lengths of time (HAART responders), and 10 patients were either not receiving HAART or not responding to HAART at the time of the study. The criterion for responder status is a viral load below 50 copies of HIV-1 RNA/ml. The non-HAART and nonresponder patients all had detectable viral loads. The CD4 range was 330 to 1,100 cells/μl for the HAART responders and 53 to 400 cells/μl for the non-HAART and nonresponder patients. Procurement of samples for this study had the approval of the human subject committees from University of California—San Francisco and San Francisco State University. Negative controls for the study are from an HIV-negative leukocyte concentrate (buffy coat) purchased from Blood Centers of the Pacific (San Francisco, Calif.).
Isolation, coculture, and stimulation of patient PBMC.
Ficoll-Paque (Amersham Pharmacia Biotech) was used to separate the PBMC from EDTA anticoagulated whole blood (20 ml) from HIV-positive samples within 4 h of collection. The yield from 20 ml of whole blood was approximately 6 × 107 cells. The number of PBMC from each sample varied, but each aliquot contained approximately 6 × 106 cells. An aliquot of patient cells was cultured in minimal T cell growth medium with 20% FBS and 5% interleukin 2 (IL-2) (10 U/ml) and antibiotics, and the cells were then cultured with an equal number of phytohemagglutinin-stimulated, HIV-negative feeder cells.
RT-PCR.
RNA was extracted from the PBMC by using the Trizol (Invitrogen, Carlsbad, Calif.) protocol. The total cellular RNA concentration was measured by spectrophotometry. All reverse transcription (RT) procedures performed included a positive HIV RNA control. Positive-control RNA was synthesized in vitro with a T7 polymerase kit (Ambion, Inc., Austin, Tex.) from plasmid pTZ-LTR*. This plasmid contains 31 bp of heterologous DNA inserted between the binding sites for primers 1 and 2. RNA samples from the HIV-negative buffy coat served as a negative control. An RT reaction was performed on 106 copies of the RNA control for each setup. All specimens and controls were incubated as follows: 10 min at room temperature, 1 h at 37°C, and then 5 min at 95°C to terminate the reaction. The primer design for PCR is shown below (1). Sequences of primers were derived from HIV-1HBX2 and are as follows: primer 1, 5′-GGGCTCTCTGGTTAGA-3′ (positions 454 to 470); primer 2, 5′-GGGTTCCCTAGTTAGCC-3′ (positions 496 to 512); primer 3, 5′-GGGCGCCACTGCTAGAGATTT-3′ (positions 623 to 643).
All samples were initially analyzed by RT-PCR (Table 1). Following RT, cDNAs were amplified separately by primer pairs 1 and 2 and 1 and 3 (Operon Technologies, Inc., Alameda, Calif.). Primer pair 1 and 2 amplifies the first 59 nucleotides of all HIV species, and primer pair 1 and 3 amplifies the first 190 nucleotides of HIV. While primer pair 1 and 2 amplifies both short and long transcripts, primer pair 1 and 3 amplifies only the processive, long transcripts. Control samples without reverse transcriptase were run in parallel. The thermal profile optimized in this laboratory for amplification with these primers includes 40 cycles of 30 s at 95°C, 30 s at 61°C, and 30 s at 72°C. The PCR for the experiment was a 15-μl reaction mixture and included 1.5 μl of cDNA template, 1.5 μl of 10× Klentaq PCR buffer (Clontech), 1.5 μl of a 200 mM concentration of each deoxynucleoside triphosphate, 0.3 μl of 0.5 μM primer mixture, and 0.63 U of Klentaq polymerase.
TABLE 1.
Transcriptional analysis of 20 HAART responders (1 to 20) and 10 non-HAART patients (21 to 30) by RT-PCR
| Patient no. | No. of copies of HIV RNA/ml of plasma | Presence ofa
|
||||
|---|---|---|---|---|---|---|
| ST | LT | Initiation | Elongation | Infectivity | ||
| 1 | <50 | − | − | |||
| 2 | <50 | + | − | + | ||
| 3 | <50 | + | + | + | + | |
| 4 | <50 | + | + | + | + | |
| 5 | <50 | + | − | + | ||
| 6 | <50 | + | − | + | ||
| 7 | <50 | + | + | + | + | |
| 8 | <50 | + | − | + | ||
| 9 | <50 | + | + | + | + | |
| 10 | <50 | + | + | + | + | |
| 11 | <50 | + | − | + | + | |
| 12 | <50 | + | − | + | ||
| 13 | <50 | + | + | + | + | |
| 14 | <50 | + | + | + | + | |
| 15 | <50 | + | − | + | + | |
| 16 | <50 | + | + | + | + | |
| 17 | <50 | + | + | + | + | |
| 18 | <50 | + | + | + | + | |
| 19 | <50 | − | − | + | ||
| 20 | <50 | + | − | + | ||
| 21 | 71,160 | + | + | + | + | |
| 22 | 9,415 | + | + | + | + | |
| 23 | 3,000 | + | + | + | + | |
| 24 | 160,000 | + | + | + | + | |
| 25 | 115,000 | + | + | + | + | |
| 26 | 3,000 | + | + | + | + | |
| 27 | 44,000 | + | + | + | + | |
| 28 | 30,000 | + | + | + | + | |
| 29 | 6,000 | + | + | + | + | |
| 30 | 4,800 | + | + | + | + | |
ST indicates the presence of short, promoter-proximal transcripts, and LT indicates the presence of long, elongated transcripts. Where there are no short or long transcripts, the sample is classified as initiation negative; where there are only short transcripts, the sample is classified as initiation positive; and where there are both long and short transcripts, the sample is classified as elongation positive. Pluses in the infectivity column indicate that the fully infectious virus was recovered by coculture with MAGI cells from these samples. No pluses or minuses indicate that these samples were not tested.
Tat functional assays.
GST.Tat protein was purified from the BL21 strain of Escherichia coli (Novagen, Madison, Mich.) transformed with plasmid pGEX4T1. Plasmid pGEX4T1 contains amino acid sequences 1 to 101 of Tat derived from HXB2 wild-type virus. The purified protein was quantified by using Bradford reagent (Bio-Rad, Hercules, Calif.); protein size was determined by Coomassie blue staining. The functional capacities of the recombinant Tat protein and Tat protein secreted to the mouse serum were assessed by a transactivational assay with HL3T1 cells. HL3T1 cells are HeLa cells stably expressing a chloramphenicol acetyltransferase (CAT) gene under the control of the HIV LTR (59). For the assay, 105 cells were aliquoted in 60-mm-diameter tissue culture plates. Various amounts (0.3 to 2.4 μg) of the purified GST.Tat or mouse serum from wild-type or sTat.GFP transgenic mice was added to the culture medium. As a negative transactivation control, 2.4 μg of the mutant GST.Tat (C30G) chimera was also tested. Chloroquine (Sigma, St. Louis, Mo.) at a concentration of 100 μM was also added to prevent lysosomal degradation of the protein. The cells were cultured for 48 h before the CAT activities were analyzed. The CAT assay is performed as described previously (22).
Culture of HAART responder PBMC with recombinant Tat.
PBMC (5 × 106 cells) were cultured in RPMI 1640 containing 20% FBS and 5% IL-2 (10 U/ml) for 21 days. Recombinant GST.Tat or mutant GST.Tat (C30G) chimeras were added to culture media at a concentration of 1.0 μg/ml. Protamine sulfate was added to culture media at a concentration of 100 μg/ml, and fresh Tat was added every 48 h. A negative control containing no exogenous protein and a coculture control as a positive control were also included. An aliquot of cells was removed from culture at 4, 8, 12, 16, and 21 days followed by RNA extraction and RT-PCR as described previously.
RESULTS
Tat is secreted and distributes widely with no deleterious effects in sTat.GFP transgenic mice.
In vitro, extracellular Tat can activate viral transcription inside cells. To determine if these concentrations of Tat have deleterious side effects on the host, we developed a transgenic mouse where the tPA leader peptide was fused in frame with the N terminus of Tat (37) and the C terminus of the chimera was linked to the GFP. Of note, Tat is fully active when both its N and C termini are fused to heterologous proteins (23). The expression of this transgene, the sTat.GFP chimera, was driven by the rat insulin promoter (RIP7) and targeted to β cells of the pancreas (15, 55) (Fig. 1a). Three founder sTat.GFP transgenic mice were generated. Breeding the founder animals with wild-type B6D2F mice generated heterozygous sTat.GFP transgenic mice. Crossing these heterozygotes resulted in homozygous transgenic mice.
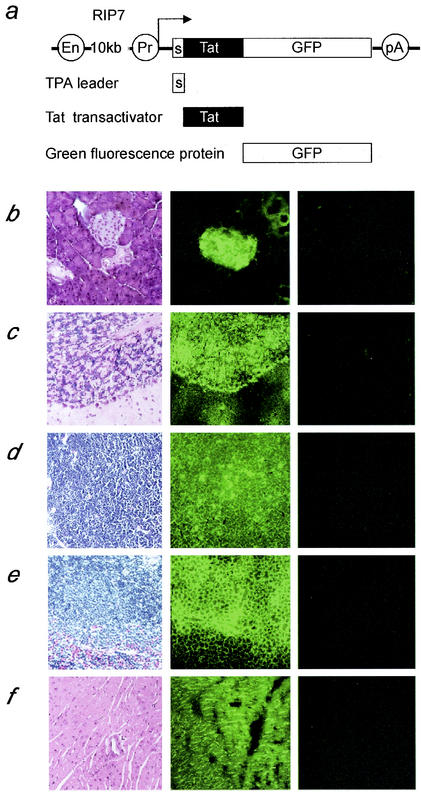
FIG. 1.
The sTat.GFP fusion protein is expressed abundantly in the pancreas and distributes widely in different organs and tissues. (a) Strategy for a secreted Tat transgenic mouse. The grey box represents the tPA leader sequence, the black box represents the 101-amino-acid Tat cDNA sequence, and the white box represents the GFP cDNA sequence. The expression of this chimeric protein was driven by the RIP7 promoter. The RIP7 promoter is 10 kb in length and contains enhancer (En) and promoter (Pr) sequences. (b to f) Histological analyses of different tissues from our transgenic mouse. The left column presents H&E stainings of paraffin sections from different tissues of the transgenic mouse; the middle column presents the green fluorescence with dark-field micrographs of frozen sections from different tissues of our transgenic mouse; and the right column presents the green fluorescence with dark-field micrographs of frozen sections from different tissues of a wild-type littermate. Tissues presented are from the pancreas (b), cerebellum (c), thymus (d), spleen (e), and heart (f). Note the strong green fluorescent staining of the pancreatic islet in the frozen section from our transgenic mouse (b).
We analyzed the histology of different tissues in our mice. High levels of the sTat.GFP chimera were detected in the pancreatic islets (Fig. 1b, middle panel). These pancreases remained morphologically normal after 2 years of age. No green fluorescence was detected in wild-type littermates (Fig. 1b, right panel). Despite the copious expression from the pancreas, blood sugar levels have also remained normal (data not shown). We also examined other organs including the brain, thymus, spleen, heart (Fig. 1c to f), lung, kidney, and liver (data not shown). We detected the presence of the sTat.GFP chimera in all of these organs from our transgenic mice (Fig. 1c to f, middle panels) but not from their wild-type littermates (Fig. 1c to f, right panels). The staining intensity correlated with the density of cells and nuclei (Fig. 1c to f, middle panels). This result indicates that the secreted Tat protein circulates in the body and penetrates all organs. High levels of Tat in the brain (Fig. 1c, middle panel) indicate that the fusion protein also crosses the blood-brain barrier. This finding is consistent with studies where the transduction domain of Tat could deliver biologically active fusion protein to all tissues, including the brain, in mice (51). More importantly, all tissues were and remain normal.
Secreted Tat protein has no effect on the immune system.
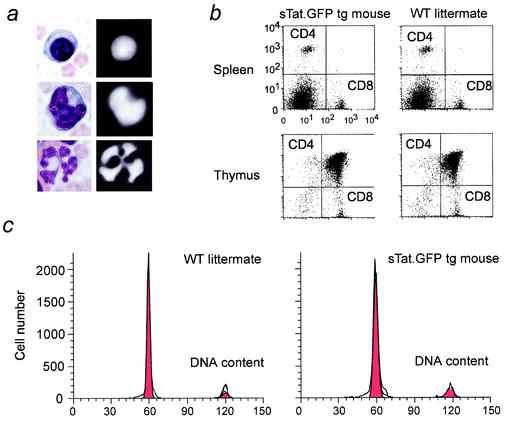
The immune system is the main target of HIV infection. Moreover, extracellular Tat protein could induce proliferative and apoptotic effects in PBL (41, 46, 57). To address this issue directly, we collected PBMC from our transgenic mice. First, Tat was detected in the nuclei of lymphocytes, monocytes, and neutrophils (Fig. 2a). Second, we isolated CD3+ T cells from the thymus and spleen, stained them with fluorochrome-tagged anti-CD4 and anti-CD8 antibodies, and analyzed ratios of CD4+ and CD8+ T cells by fluorescence activated cell sorting (FACS). No differences were observed in the thymus or spleen between these animals (Fig. 2b). CD4+ T cells were also collected from the thymus and cultured in minimal T cell growth medium with and without ionomycin and phorbol myristate acetate for 24 h. Cells from transgenic mice and their wild-type littermates displayed equivalent apoptotic and proliferative profiles (Fig. 2c). The founder animals are now more than 2 years old and remain phenotypically normal. Thus, at these concentrations, the sTat.GFP chimera caused no discernible pathology in the mouse.
FIG. 2.
The secreted Tat protein accumulates in the nucleus of different PBMC, and our transgenic mouse is phenotypically normal. (a) H&E staining of a lymphocyte (top left), monocyte (middle left), and neutrophil (bottom left). The fluorescent images of the secreted Tat protein accumulation in the nuclei of these leukocytes are presented to the right. (b) CD3+ T cells were isolated from the thymi (top panels) and the spleens (bottom panels) of transgenic (left panels) and wild-type (WT) (right panels) mice. The ratios of CD4+ and CD8+ T cells were analyzed by FACS. (c) CD4+ T cells were isolated from the thymi of wild-type (left) and transgenic (right) mice, and the proliferative and apoptotic profiles were analyzed by FACS.
Secreted sTat.GFP chimera activates transcription from the HIV LTR.
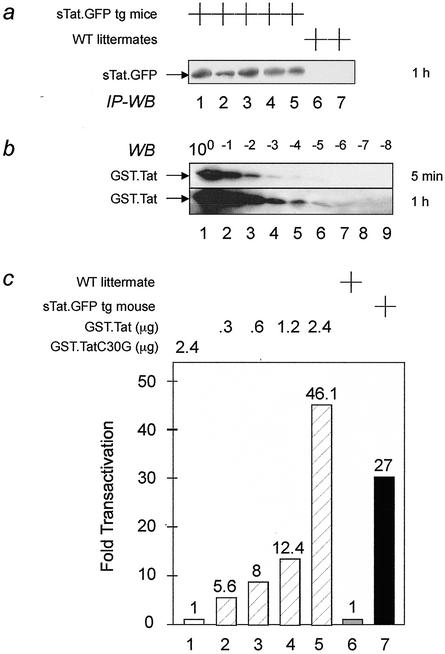
To determine the biological activity of our secreted Tat protein, we compared the ability of the mouse serum and GST.Tat fusion protein to activate the HIV LTR in cells. As presented in Fig. 3a, the expression of the hybrid sTat.GFP protein could be detected only in the blood of our transgenic mice but not in their wild-type littermates. When compared to serial dilutions of the GST.Tat fusion protein in Fig. 3b, the concentration of sTat.GFP chimera was between 20 and 200 pM (compare Fig. 3a, lanes 1 to 5, to Fig. 3b, bottom panel, lanes 4 and 5). This concentration compares favorably to fasting insulin levels in humans (50 to 200 pM) (8) and mice (250 pM) (55). This sTat.GFP chimera was biologically active and activated the HIV LTR in HL3T1 cells (Fig. 3c). Indeed, diluted to 2 ml of culture medium, 500 μl of serum from the transgenic mouse increased CAT activity to levels that were between those observed with 1.2 and 2.4 μg of the purified GST.Tat chimera (Fig. 3c, compare lanes 4, 5, and 7). Importantly, 2.4 μg of the mutant GST.Tat (C30G) had no effect (Fig. 3c, lane 1). Thus, the biological activity of the sTat.GFP chimera was similar to that of 0.8 μg of the hybrid GST.Tat protein/ml. However, when amounts of protein are compared between Fig. 3a and b, most of the GST.Tat chimera was inactive. Indeed, only 0.01 to 0.1% of this recombinant Tat, which was expressed from E. coli, was biologically active. We conclude that our transgenic mice secrete adequate levels of biologically active Tat. This sTat.GFP chimera distributes widely, enters resting cells, and activates the HIV LTR. Since no pathology was detected, effects of Tat are also very specific for the HIV LTR.
FIG. 3.
The sTat.GFP fusion protein is secreted from pancreatic β cells into the blood of our transgenic mouse. The secreted Tat protein is biologically active. (a) The sTat.GFP fusion protein was detected in different sera (lanes 1 to 5) by immunoprecipitation followed by Western blotting (IP-WB). These sera were derived from five transgenic mice but not detected in lanes 6 and 7, which represent sera from two wild-type (WT) littermates. (b) Western blotting (WB) of serial dilutions of the GST.Tat chimera used in panel c. Five micrograms of the GST.Tat fusion protein was diluted 10-fold 8 times (above the lanes) and detected by an anti-GST antiserum. Immunoblots were exposed for 5 min and 1 h, respectively. (c) The biological activity of the sTat.GFP chimera was measured by the CAT assay. To a total of 2 ml of culture medium, 500 μl of wild-type (lane 6) or transgenic (lane 7) mouse sera or different amounts of GST.Tat fusion protein (lanes 2 to 5) were added.
Transcriptional profiles of PBL from optimally treated infected individuals reveal a lack of Tat phenotype.
Since levels of Tat were achieved in vivo that transactivated the integrated HIV LTR in cells and since this sTat.GFP chimera had no deleterious effects on the host or growth and proliferation of HL3T1 cells (data not shown), we wanted to determine if exogenous Tat could also activate latent proviruses in humans. Previously, a model of proviral latency was studied in monocytic U937 cells (U1 cells) and in infected untreated individuals, most at seroconversion (1). In these cells, only promoter-proximal but not promoter-distal viral transcripts could be detected and Tat transduced by retroviral infection induced the elongation of viral transcription and replication of HIV (1). A similar state should predominate in infected individuals on HAART who have low levels of plasma viremia. Therefore, using RT-PCR approaches, we looked for promoter-proximal short transcripts and elongated long transcripts in RNA samples extracted from PBMC of infected individuals on HAART (Fig. 4a and b).
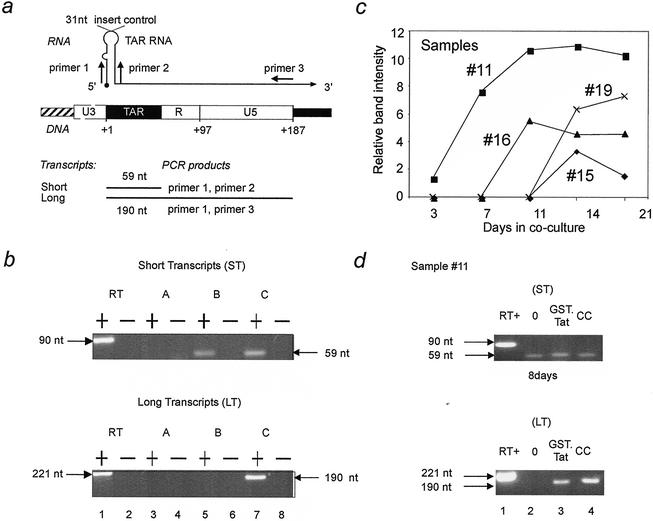
FIG. 4.
Detection of transcripts initiated from the HIV LTR in PBMC from optimally treated individuals. (a) Primer combinations for the amplification of the HIV LTR. Primers 1 and 2 amplify TAR and thus all HIV-1 transcripts. Primers 1 and 3 amplify only the long transcripts. The RT positive control was obtained with RNA generated from plasmid pTZ-LTR*. (b) Representative RT-PCR results from HAART patients. RT-PCR was performed with primer sets 1 and 2 (top panel) and 1 and 3 (bottom panel) from optimally treated infected individuals. In the images shown above, sample A (lane 3) contains neither short nor long transcripts and was classified as initiation negative, sample B (lane 5) contains short transcripts only and was classified as initiation positive and elongation negative, and sample C (lane 7) contains both short and long transcripts and was classified as elongation positive. Lane 1 is an RT positive control containing 106 copies of in vitro-generated RNA from plasmid pTZ-LTR*. Lanes 2, 4, 6, and 8 are RT-PCR controls which used total RNA without the RT reaction mixture as a template for the PCR. Results of this analysis are summarized in Table 1. +, RT positive; −, RT negative; nt, nucleotides. (c) Processive band intensities from coculture. Shown are the relative band intensities obtained from digital image analysis of elongated transcripts (primers 1 and 3) versus days in coculture for samples 11, 15, 16, and 19. The relative band intensity is the area continued within a peak representing the number of pixels following digital image capture and analysis. (d) Exogenous Tat protein activated latent proviruses equivalently to coculture in optimally treated infected individuals. Presented are the RT-PCR results from day 8. The top and bottom panels present the amplification with primers 1 and 2 and primers 1 and 3, respectively. Lanes: 1, positive RT control; 2, negative Tat control containing no exogenous protein; 3, sample containing 1.0 μg of recombinant Tat/ml; 4, coculture control. RΤ-PCR on β-actin was performed on all samples from panels b, c, and d and represents an internal RT-PCR control (data not shown). Results shown in panels b, c, and d are representative of 3 independent experiments.
Figure 4a diagrams the RT-PCR approach used to detect short and long transcripts. Primer sequences were derived from HIV-1HXB2. It and other HIV subtype B sequences are identical in the regions that serve as binding sites for primers 1 and 3. In sequences of the B subtype there is one polymorphism in the binding site for primer 2. Nevertheless, for the design of primer 2, the HIV-1HXB2 sequence allows for perfect binding to 35% of all viral isolates. With other templates, one G-T base pair is formed that still allows adequate binding. Representative data for the RT-PCR are presented in Fig. 4b. Using limiting dilutions of the positive-control RNA in 1 μg of total cellular RNA from HIV-negative buffy coats, the sensitivity of our assay for both primer sets was determined to be approximately 50 copies per PCR (data not shown).
Table 1 summarizes the results from 30 samples. Ten of these samples were from individuals who were not treated with HAART, and we detected both short and long transcripts in these individuals. The other 20 samples are from individuals on HAART who had viral loads of less than 50 RNA copies per ml and CD4 counts between 330 and 1,100. These results can be divided into three groups (Table 1; Fig. 4b). Neither short nor long transcripts were detected in samples 1 and 19. These samples might harbor transcriptionally silent proviruses, or viral transcripts in these samples may be below detectable levels. Both short and long transcripts were detected in samples 3, 4, 7, 9, 10, 13, 14, 17, and 18, which is similar to results for the 10 patients who were not on HAART (Table 1; Fig. 4b). This result indicates that there is continued productive viral replication in these nine patients despite undetectable plasma virus. Short transcripts only were detected in samples 2, 5, 6, 8, 11, 12, 15, 16, and 20. These PBMC harbor proviruses that had initiated but not elongated viral transcription (Table 1; Fig. 3b). This picture is similar to what we observed previously in U1 cells and PBMC and represents the lack of the Tat phenotype (1). Thus, the failure to recruit P-TEFb to the HIV LTR by NF-κB or Tat might be responsible for the majority of proviral latency also in optimally treated individuals.
T-cell activation and/or addition of exogenous Tat protein activates latent proviruses equivalently in PBL.
Given these patient data, we hypothesized that the introduction of exogenous Tat to these cells should activate the replication of latent HIV. First, we cocultured representative infected cells with mitogen-stimulated uninfected PBMC and induced the synthesis of long transcripts within 2 weeks in all samples (Fig. 4c). Fully infectious virus was recovered from the culture supernatants of the four activated samples (Table 1). This finding agrees with the results of many studies where allogeneic cells could trigger cellular activation and induce replication of latent proviruses (32, 40). The various time points for the appearance of long transcripts could have been due to differences in proviral loads, the number of latently infected cells in the aliquots, or differences in the reaction of patient cells to allogeneic stimuli, among others.
Next, we examined the ability of exogenous Tat to duplicate the induction effect of coculture (Fig. 4d). Because murine serum did not promote the growth and proliferation of human PMBC, we could not use the sTat.GFP chimera. Nevertheless, data from Fig. 3 allowed us to choose the optimal concentration of the GST.Tat hybrid protein (1.0 μg/ml) with the same biological activity. Indeed, when this GST.Tat chimera was added, long transcripts were detected in all samples examined. Our control mutant GST.Tat (C30G) had no effect (Fig. 3 and data not shown). Representative results of sample 11 are presented in Fig. 4d. These results suggest that cellular activation and/or the addition of exogenous Tat can activate the transcription of latent proviruses in optimally treated infected individuals.
DISCUSSION
Our study demonstrates that viral persistence in optimally treated infected individuals can represent absent elongation of viral transcription. T-cell activation or exogenous Tat protein activated latent proviruses in their PBMC. In another study, Tat could also equivalently reactivate viral replication to cellular activation in transformed Jurkat T and THP-1 monocytic cells (39). The secreted Tat protein expressed from β cells of the pancreas of sTat.GFP transgenic mice circulated in the body and penetrated all tissues and organs. It was well tolerated with no obvious side effects. These results point to the possibility of systemic administration of Tat or other recruitment of P-TEFb as an adjunct treatment with HAART to eliminate the latent reservoir of HIV in humans.
Although it was expected that Tat would cause deleterious effects, such as apoptosis and/or proliferation of T cells and somatic cells (41, 46, 57), central nervous system and vascular abnormalities (2, 30), anergy or immunodeficiency (62), none were observed in our transgenic mouse. Importantly, although the abundant expression of many proteins in β cells of the pancreas from the insulin promoter leads to diabetes mellitus (4, 48), the sTat.GFP chimera was innocuous. Because Tat can interact productively with the murine P-TEFb for all other purposes other than HIV transcription (6, 24, 42) and can enter murine and human cells equivalently (20, 27, 51), we conclude that, at these levels, Tat is highly specific for the HIV LTR.
The transcriptional profiles of PBMC from 20 optimally treated infected individuals were highly informative. In our study, only two infected individuals might have harbored completely silent proviruses. Since using additional real-time PCR approaches, no differences in any of these profiles were observed, their proviruses were also below the level of detection. Otherwise, in these two individuals, HIV could have integrated into silent heterochromatin, where no initiation or elongation of transcription from the HIV LTR occurs. In a recent study, this represented a very small number of all integration events (50). In 9 of 20 samples, we could detect only short viral transcripts, where viral transcription was initiated but not elongated past TAR. This group represents the lack of Tat or P-TEFb phenotype. We detected both short and long transcripts in 9 of 20 samples. These levels of viral replication resemble those in infected individuals not on HAART. They could also mark the involution of the lymphoid organs, which would allow activated cells to circulate.
Our study argues that inadequate levels and/or activity of P-TEFb at the initiating RNAPIIa on the HIV LTR is one of the mechanisms that contributes to proviral latency in optimally treated infected individuals. A previous study revealed that the same situation pertains to infected individuals that are not treated with HAART (1). These observations can be reconciled because most circulating PBMC are not activated and thus do not contain active NF-κB and do not synthesize Tat.
How does this form of proviral latency develop? NF-κB, Tat, and P-TEFb are all involved. First, NF-κB is activated only following cellular activation and/or proliferation (44, 52). Second, without transcription, no Tat is made. Third, although levels of Cdk9 are constant during the cell cycle and during cellular activation, CycT1 is limiting in resting cells (31). Autophosphorylation of Cdk9 is another important regulatory step for P-TEFb activity on TAR, i.e., unphosphorylated P-TEFb is unable to form the tripartite complex with Tat and TAR (25). In cells, P-TEFb can also form a stable and inactive 500-mDa complex with the small nuclear 7SK RNA in a transcription-dependent manner (45, 61). It is possible that in resting CD4+ T cells, most of P-TEFb is associated with 7SK RNA, which contains an inactive Cdk9 that is unable to interact with Tat and TAR. Thus, the complex regulation of P-TEFb could contribute to proviral latency that is established in the host.
Current approaches for clearing the reservoir use cytokines to activate latently infected cells. A recent study demonstrated that a combination of IL-2, IL-6, and tumor necrosis factor alpha could activate viral replication in these cells in vitro (10). While this strategy is promising, its application in vivo is limited because most cytokines cause nonspecific activation of large numbers of T cells and many infected cells do not display the appropriate receptors (7). To this end, limited clinical studies with these agonists have been unsuccessful (11, 21, 38). However, Tat is a viral protein that is already present in infected individuals. According to our study, at these concentrations, it is also rather harmless. Additionally, little to no sustained immune response to Tat has been observed in infected individuals (3). From our data, Tat can activate latent proviruses similarly to cellular activation. Nevertheless, it is possible that P-TEFb is inactive in many other resting infected cells. To this end, we created tripartite mutant chimeras between Tat, CycT1, and Cdk9 that are fully active on the HIV LTR (23). Moreover, they contain glutamic acid residues in place of C-terminal serines and threonines in Cdk9 (23). These findings increase the attractiveness of moving our studies to the SCID/hu mouse or monkey models of AIDS. In adjunction with HAART, these approaches might point to a new direction for the eradiation of the latent reservoir.
Acknowledgments
We thank members of our laboratories for helpful discussions. Morphological analyses were performed at the San Francisco Veterans Administration Microscopy and Advanced Imaging Core Facility, with support from the University of California—San Francisco Liver Center.
This work was supported by the Rosalind Russell Medical Research Foundation, the NIH (RO1 AI49104-01) (to B.M.P), and the Campbell Foundation (to X.L.).
REFERENCES
- 1.Adams, M., L. Sharmeen, J. Kimpton, J. M. Romeo, J. V. Garcia, B. M. Peterlin, M. Groudine, and M. Emerman. 1994. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc. Natl. Acad. Sci. USA 91:3862-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albini, A., R. Soldi, D. Giunciuglio, E. Giraudo, R. Benelli, L. Primo, D. Noonan, M. Salio, G. Camussi, W. Rockl, and F. Bussolino. 1996. The angiogenesis induced by HIV-1 tat protein is mediated by the Flk-1/KDR receptor on vascular endothelial cells. Nat. Med. 2:1371-1375. [DOI] [PubMed] [Google Scholar]
- 3.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 4.Allison, J., I. L. Campbell, G. Morahan, T. E. Mandel, L. C. Harrison, and J. F. Miller. 1988. Diabetes in transgenic mice resulting from over-expression of class I histocompatibility molecules in pancreatic beta cells. Nature 333:529-533. [DOI] [PubMed] [Google Scholar]
- 5.Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327-337. [DOI] [PubMed] [Google Scholar]
- 6.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1998. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 17:7056-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blankson, J. N., D. Persaud, and R. F. Siliciano. 2002. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 53:557-593. [DOI] [PubMed] [Google Scholar]
- 8.Cersosimo, E., P. Garlick, and J. Ferretti. 2001. Abnormal glucose handling by the kidney in response to hypoglycemia in type 1 diabetes. Diabetes 50:2087-2093. [DOI] [PubMed] [Google Scholar]
- 9.Choi, J., R. M. Liu, R. K. Kundu, F. Sangiorgi, W. Wu, R. Maxson, and H. J. Forman. 2000. Molecular mechanism of decreased glutathione content in human immunodeficiency virus type 1 Tat-transgenic mice. J. Biol. Chem. 275:3693-3698. [DOI] [PubMed] [Google Scholar]
- 10.Chun, T. W., D. Engel, S. B. Mizell, L. A. Ehler, and A. S. Fauci. 1998. Induction of HIV-1 replication in latently infected CD4+ T cells using a combination of cytokines. J. Exp. Med. 188:83-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun, T. W., D. Engel, S. B. Mizell, C. W. Hallahan, M. Fischette, S. Park, R. T. Davey, Jr., M. Dybul, J. A. Kovacs, J. A. Metcalf, J. M. Mican, M. M. Berrey, L. Corey, H. C. Lane, and A. S. Fauci. 1999. Effect of interleukin-2 on the pool of latently infected, resting CD4+ T cells in HIV-1-infected patients receiving highly active anti-retroviral therapy. Nat. Med. 5:651-655. [DOI] [PubMed] [Google Scholar]
- 12.Chun, T. W., D. Finzi, J. Margolick, K. Chadwick, D. Schwartz, and R. F. Siliciano. 1995. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat. Med. 1:1284-1290. [DOI] [PubMed] [Google Scholar]
- 13.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahmus, M. E. 1996. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 271:19009-19012. [DOI] [PubMed] [Google Scholar]
- 15.Efrat, S., and D. Hanahan. 1987. Bidirectional activity of the rat insulin II 5′-flanking region in transgenic mice. Mol. Cell. Biol. 7:192-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ensoli, B., G. Barillari, S. Z. Salahuddin, R. C. Gallo, and F. Wong-Staal. 1990. Tat protein of HIV-1 stimulates growth of cells derived from Kaposi's sarcoma lesions of AIDS patients. Nature 345:84-86. [DOI] [PubMed] [Google Scholar]
- 17.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 18.Folks, T., D. M. Powell, M. M. Lightfoote, S. Benn, M. A. Martin, and A. S. Fauci. 1986. Induction of HTLV-III/LAV from a nonvirus-producing T-cell line: implications for latency. Science 231:600-602. [DOI] [PubMed] [Google Scholar]
- 19.Folks, T. M., J. Justement, A. Kinter, C. A. Dinarello, and A. S. Fauci. 1987. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science 238:800-802. [DOI] [PubMed] [Google Scholar]
- 20.Frankel, A. D., and C. O. Pabo. 1988. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 55:1189-1193. [DOI] [PubMed] [Google Scholar]
- 21.Fraser, C., N. M. Ferguson, A. C. Ghani, J. M. Prins, J. M. Lange, J. Goudsmit, R. M. Anderson, and F. de Wolf. 2000. Reduction of the HIV-1-infected T-cell reservoir by immune activation treatment is dose-dependent and restricted by the potency of antiretroviral drugs. AIDS 14:659-669. [DOI] [PubMed] [Google Scholar]
- 22.Fujinaga, K., T. P. Cujec, J. Peng, J. Garriga, D. H. Price, X. Grana, and B. M. Peterlin. 1998. The ability of positive transcription elongation factor B to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J. Virol. 72:7154-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujinaga, K., D. Irwin, M. Geyer, and B. M. Peterlin. 2002. Optimized chimeras between kinase-inactive mutant Cdk9 and truncated cyclin T1 proteins efficiently inhibit Tat transactivation and human immunodeficiency virus gene expression. J. Virol. 76:10873-10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujinaga, K., D. Irwin, R. Taube, F. Zhang, M. Geyer, and B. M. Peterlin. 2002. A minimal chimera of human cyclin T1 and Tat binds TAR and activates human immunodeficiency virus transcription in murine cells. J. Virol. 76:12934-12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garber, M. E., T. P. Mayall, E. M. Suess, J. Meisenhelder, N. E. Thompson, and K. A. Jones. 2000. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 tat-P-TEFb complex to TAR RNA. Mol. Cell. Biol. 20:6958-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garber, M. E., P. Wei, V. N. KewalRamani, T. P. Mayall, C. H. Herrmann, A. P. Rice, D. R. Littman, and K. A. Jones. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12:3512-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green, M., and P. M. Loewenstein. 1988. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell 55:1179-1188. [DOI] [PubMed] [Google Scholar]
- 28.Gulick, R. M., J. W. Mellors, D. Havlir, J. J. Eron, C. Gonzalez, D. McMahon, D. D. Richman, F. T. Valentine, L. Jonas, A. Meibohm, E. A. Emini, and J. A. Chodakewitz. 1997. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N. Engl. J. Med. 337:734-739. [DOI] [PubMed] [Google Scholar]
- 29.Hammer, S. M., K. E. Squires, M. D. Hughes, J. M. Grimes, L. M. Demeter, J. S. Currier, J. J. Eron, Jr., J. E. Feinberg, H. H. Balfour, Jr., L. R. Deyton, J. A. Chodakewitz, and M. A. Fischl. 1997. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N. Engl. J. Med. 337:725-733. [DOI] [PubMed] [Google Scholar]
- 30.Hayman, M., G. Arbuthnott, G. Harkiss, H. Brace, P. Filippi, V. Philippon, D. Thomson, R. Vigne, and A. Wright. 1993. Neurotoxicity of peptide analogues of the transactivating protein tat from Maedi-Visna virus and human immunodeficiency virus. Neuroscience 53:1-6. [DOI] [PubMed] [Google Scholar]
- 31.Herrmann, C. H., R. G. Carroll, P. Wei, K. A. Jones, and A. P. Rice. 1998. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J. Virol. 72:9881-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hollinger, F. B., J. W. Bremer, L. E. Myers, J. W. M. Gold, L. McQuay, and The NIH/DAIDS/ACTG Virology Laboratories. 1992. Standardization of sensitive human immunodeficiency virus coculture procedures and establishment of a multicenter quality assurance program for the AIDS Clinical Trials Group. J. Clin. Microbiol. 30:1787-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones, K. A., and B. M. Peterlin. 1994. Control of RNA initiation and elongation at the HIV-1 promoter. Annu. Rev. Biochem. 63:717-743. [DOI] [PubMed] [Google Scholar]
- 34.Jordan, A., P. Defechereux, and E. Verdin. 2001. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20:1726-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung, A., R. Maier, J. P. Vartanian, G. Bocharov, V. Jung, U. Fischer, E. Meese, S. Wain-Hobson, and A. Meyerhans. 2002. Multiply infected spleen cells in HIV patients. Nature 418:144.. [DOI] [PubMed] [Google Scholar]
- 36.Kao, S. Y., A. F. Calman, P. A. Luciw, and B. M. Peterlin. 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330:489-493. [DOI] [PubMed] [Google Scholar]
- 37.Kohne, C., A. Johnson, S. Tom, D. H. Peers, R. L. Gehant, T. A. Hotaling, D. Brousseau, T. Ryll, J. A. Fox, S. M. Chamow, and P. W. Berman. 1999. Secretion of glycosylation site mutants can be rescued by the signal/pro sequence of tissue plasminogen activator. J. Cell. Biochem. 75:446-461. [PubMed] [Google Scholar]
- 38.Kulkosky, J., G. Nunnari, M. Otero, S. Calarota, G. Dornadula, H. Zhang, A. Malin, J. Sullivan, Y. Xu, J. DeSimone, T. Babinchak, J. Stern, W. Cavert, A. Haase, and R. J. Pomerantz. 2002. Intensification and stimulation therapy for human immunodeficiency virus type 1 reservoirs in infected persons receiving virally suppressive highly active antiretroviral therapy. J. Infect. Dis. 186:1403-1411. [DOI] [PubMed] [Google Scholar]
- 39.Kutsch, O., E. N. Benveniste, G. M. Shaw, and D. N. Levy. 2002. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J. Virol. 76:8776-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levy, J. A., and J. Shimabukuro. 1985. Recovery of AIDS-associated retroviruses from patients with AIDS or AIDS-related conditions and from clinically healthy individuals. J. Infect. Dis. 152:734-738. [DOI] [PubMed] [Google Scholar]
- 41.Li, C. J., D. J. Friedman, C. Wang, V. Metelev, and A. B. Pardee. 1995. Induction of apoptosis in uninfected lymphocytes by HIV-1 Tat protein. Science 268:429-431. [DOI] [PubMed] [Google Scholar]
- 42.Madore, S. J., and B. R. Cullen. 1993. Genetic analysis of the cofactor requirement for human immunodeficiency virus type 1 Tat function. J. Virol. 67:3703-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marciniak, R. A., B. J. Calnan, A. D. Frankel, and P. A. Sharp. 1990. HIV-1 Tat protein trans-activates transcription in vitro. Cell 63:791-802. [DOI] [PubMed] [Google Scholar]
- 44.Nabel, G., and D. Baltimore. 1987. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326:711-713. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322-325. [DOI] [PubMed] [Google Scholar]
- 46.Ott, M., S. Emiliani, C. Van Lint, G. Herbein, J. Lovett, N. Chirmule, T. McCloskey, S. Pahwa, and E. Verdin. 1997. Immune hyperactivation of HIV-1-infected T cells mediated by Tat and the CD28 pathway. Science 275:1481-1485. [DOI] [PubMed] [Google Scholar]
- 47.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarvetnick, N., D. Liggitt, S. L. Pitts, S. E. Hansen, and T. A. Stewart. 1988. Insulin-dependent diabetes mellitus induced in transgenic mice by ectopic expression of class II MHC and interferon-gamma. Cell 52:773-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scala, G., M. R. Ruocco, C. Ambrosino, M. Mallardo, V. Giordano, F. Baldassarre, E. Dragonetti, I. Quinto, and S. Venuta. 1994. The expression of the interleukin 6 gene is induced by the human immunodeficiency virus 1 TAT protein. J. Exp. Med. 179:961-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schroder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 51.Schwarze, S. R., A. Ho, A. Vocero-Akbani, and S. F. Dowdy. 1999. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science 285:1569-1572. [DOI] [PubMed] [Google Scholar]
- 52.Tong-Starksen, S. E., P. A. Luciw, and B. M. Peterlin. 1987. Human immunodeficiency virus long terminal repeat responds to T-cell activation signals. Proc. Natl. Acad. Sci. USA 84:6845-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vogel, J., S. H. Hinrichs, R. K. Reynolds, P. A. Luciw, and G. Jay. 1988. The HIV tat gene induces dermal lesions resembling Kaposi's sarcoma in transgenic mice. Nature 335:606-611. [DOI] [PubMed] [Google Scholar]
- 54.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walker, M. D., T. Edlund, A. M. Boulet, and W. J. Rutter. 1983. Cell-specific expression controlled by the 5′-flanking region of insulin and chymotrypsin genes. Nature 306:557-561. [DOI] [PubMed] [Google Scholar]
- 56.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 57.Westendorp, M. O., R. Frank, C. Ochsenbauer, K. Stricker, J. Dhein, H. Walczak, K. M. Debatin, and P. H. Krammer. 1995. Sensitization of T cells to CD95-mediated apoptosis by HIV-1 Tat and gp120. Nature 375:497-500. [DOI] [PubMed] [Google Scholar]
- 58.Wong, J. K., M. Hezareh, H. F. Gunthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
- 59.Wright, C. M., B. K. Felber, H. Paskalis, and G. N. Pavlakis. 1986. Expression and characterization of the trans-activator of HTLV-III/LAV virus. Science 234:988-992. [DOI] [PubMed] [Google Scholar]
- 60.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 61.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414:317-322. [DOI] [PubMed] [Google Scholar]
- 62.Zagury, D., A. Lachgar, V. Chams, L. S. Fall, J. Bernard, J. F. Zagury, B. Bizzini, A. Gringeri, E. Santagostino, J. Rappaport, M. Feldman, A. Burny, and R. C. Gallo. 1998. Interferon alpha and Tat involvement in the immunosuppression of uninfected T cells and C-C chemokine decline in AIDS. Proc. Natl. Acad. Sci. USA 95:3851-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]