Abstract
Replication-deficient human adenovirus type 5 (Ad5) can be produced to high titers in complementing cell lines, such as PER.C6, and is widely used as a vaccine and gene therapy vector. However, preexisting immunity against Ad5 hampers consistency of gene transfer, immunological responses, and vector-mediated toxicities. We report the identification of human Ad35 as a virus with low global prevalence and the generation of an Ad35 vector plasmid system for easy insertion of heterologous genes. In addition, we have identified the minimal sequence of the Ad35-E1B region (molecular weight, 55,000 [55K]), pivotal for complementation of fully E1-lacking Ad35 vector on PER.C6 cells. After stable insertion of the 55K sequence into PER.C6 cells a cell line was obtained (PER.C6/55K) that efficiently transcomplements both Ad5 and Ad35 vectors. We further demonstrate that transduction with Ad35 is not hampered by preexisting Ad5 immunity and that Ad35 efficiently infects dendritic cells, smooth muscle cells, and synoviocytes, in contrast to Ad5.
It has been shown in diverse in vivo models that recombinant adenovirus type 5 (Ad5) has potential as a vehicle to transfer genes for treatment or prevention of disease (49, 52). Although encouraging, the extrapolation from animal models to humans faces at least one extra hurdle, i.e., the presence of anti-Ad5 neutralizing activity (NA) in sera from human individuals. The humoral response to Ad5 is strong and has been found to impede, depending on the administration route, the infection efficiency in animal models as well as in humans (7, 9, 18, 29, 30, 35, 37, 42, 45). Concomitant with the decrease in transduction, high NA against the vector also abolishes Ad5-mediated toxicity (8). Importantly, when very high vector doses were used in preimmunized nonhuman primates, new toxic effects were found that were not observed in naive animals (54). These findings show that preexisting immunity severely hampers accurate dose control, since human individuals differ in their NA against Ad5-based vectors. Strategies to bypass NA to Ad5 viruses include switching of adenovirus type (28, 32, 36) and use of animal adenoviruses (13, 25, 34). Animal adenoviruses have the advantage that NA is predicted to be absent in humans. Disadvantages of this strategy include the lack of knowledge regarding the biology of these viruses including tropism on human cells, potential difficulties in manufacturing, and the possibility of in vivo recombination with human types leading to unknown disease. Human adenoviruses on the other hand are better characterized and their subclinical disease association in humans is known (10, 17, 55). However, recent knowledge on the prevalence of NA towards human adenoviruses worldwide is not available and therefore it is difficult to predict which type would be the best alternative for Ad5. To identify human adenovirus types with low seroprevalence, an extensive screen was performed using most human adenovirus types and serum samples derived from healthy blood donors from 6 different geographical locations. Of the 47 types tested, subgroup B viruses Ad35 and Ad11 proved rarely neutralized by human sera. Next, we generated E1-lacking Ad35 viruses carrying marker genes using newly developed PER.C6/55K, Ad35 packaging cells. High-titer, purified E1-lacking Ad35 virus was subsequently tested in vitro and in vivo, showing that a recombinant Ad35 vector successfully circumvents anti-Ad5 NA and that the tropism of Ad35 is favorable, compared to Ad5, on human dendritic cells, smooth muscle cells (SMC), and synoviocytes, considered important target cells for treatment or prevention of disease.
MATERIALS AND METHODS
Cells and wild-type viruses.
PER.C6 cells (12, 38) were maintained in Dulbecco's modified Eagle medium (Life Technologies, Inc. [LTI]) containing 10% fetal bovine serum (LTI) and 10 mM MgCl2 at 37°C in 5% CO2. All human adenovirus types, Ad1 through Ad51 (a kind gift from Jan de Jong, University of Rotterdam, Rotterdam, The Netherlands), were inoculated on PER.C6 cells. Batches of CsCl-purified viruses were generated according to standard procedures. Virus particles (vp) were quantified by high-performance liquid chromatography (46) and titrated on PER.C6 cells to determine the amount of virus necessary to obtain full cytopathic effect (CPE) in 5 days. To this end, 100 μl of medium was dispensed into 96-well plates, and 25 μl of a prediluted adenovirus stock (10−4, 10−5, 10−6, or 10−7) was added to eight wells of column 2 and then further diluted (fivefold steps) until column 11. Next, 3 × 104 PER.C6 cells were added in a 100-μl volume, and the plates were incubated for 5 days. CPE was monitored microscopically to determine the 50% cell culture infectious dose and was monitored with an dimethyl-thiazol-diphenyl-tetrazolium bromide (MTT) cell viability assay (Promega).
Human sera and neutralization assays.
Serum was collected in Europe (Belgium, United Kingdom, and The Netherlands), the United States (Stanford, Calif., and Great Neck, N.Y.), and Japan (Tokyo) from healthy volunteers (age, 20 to 70 years; both female and male) after informed consent was obtained. In each location, except The Netherlands (n = 56), 100 serum samples were obtained. Sera were heat-inactivated at 55°C for 15 min. To determine the NA, sera were diluted (1/2, 1/4, 1/8, 1/16, or 1/32) and 100 μl was dispensed in duplicate in 96-well plates (eight serum samples per plate). Next, 50 μl of adenovirus stock, diluted to 200 50% cell culture infectious doses, was added to the wells containing serum. Plates were incubated for 1 h at 37°C in 5% CO2 before the addition of 50 μl (6 × 105/ml) of PER.C6 cell suspension, after which plates were further incubated overnight. As a positive control for virus replication, only virus was added to cells (column 11), and as a negative control, cells were cultured in the absence of virus (column 12). Supernatant was removed the next day, and 200 μl of fresh medium was added to all wells, followed by an incubation period for another 4 days. On day 5 to 6, plates were analyzed by the MTT assay (Promega) for inhibition of CPE. Positive-control plates were set up in parallel using type-specific sera generated in rabbits and having a known neutralization titer (kind gift of Jan de Jong, Erasmus University, Rotterdam, The Netherlands). Sera were scored positive for NA when the protection of CPE was >90%. The percentage of replication inhibition was calculated by dividing the optical density (OD) of the corresponding well minus the OD value of the virus control by the OD of the noninfected control minus the OD value of the virus control and multiplying the result by 100. In this formula the corresponding well is the well containing virus and serum and the virus control is the well with virus only.
Ad35 plasmids and recombinant viruses.
Many different cloning steps were executed to obtain an Ad35-based plasmid system for the generation of fully E1-lacking Ad35-based recombinant vectors. Here, plasmids and cosmids generated are described in general terms. An Ad35 prototype stock derived from the American Type Culture Collection strain was propagated on PER.C6 cells, and viral DNA was isolated. The viral genome was sequenced using the shotgun method (Qiagen GmbH, Hilden, Germany) and is available under GenBank accession no. AY271307. The following primers were used to amplify nucleotides (nt) 1 to 464, 35F1 (5′-CGGAATTCTTAATTAATCGACATCATCAATAATATACCTTATAG-3′) and 35R2 (5′-GGTGGTCCTAGGCTGACACCTACGTAAAAACAG-3′), and nt 3401 to 4669, 35F3 (5′-TGGTGGAGATCTGGTGAGTATTGGGAAAAC-3′) and 35R4 (5′-CGGAATTCTTAATTAAGGGAAATGCAAATCTGTGAGG-3′). Both PCR products were used to generate a pBr-based plasmid-coded pAdApt35IP1. This plasmid contains the left end of the Ad35 genome (nt 1 to 464) linked to an expression cassette followed by Ad35 sequences (nt 3401 to 4669). The expression cassette was derived from the pAdApt plasmid (21) and consists of a cytomegalovirus promoter (nt −735 to +95 [5]), a multiple cloning site, and the simian virus 40 pA.
The remaining part of the Ad35 genome was cloned into a cosmid. To this end, again two Ad35 genome sequences were amplified using oligonucleotides 35F5 (5′-CGGAATTCGCGGCCGCGGTGAGTATTGGGAAAAC-3′), 35R6 (5′-CGCCAGATCGTCTACAGAACAG-3′), 35F7 (5′-GAATGCTGGCTTCAGTTGTAATC-3′), and 35R8 (5′-CGGAATTCGCGGCCGCATTTAAATCATCATCAATAATATACC-3′). These oligonucleotide sets amplify Ad35 sequence from nt 3401 to 6771 with a NotI site added to the 5′ end and from nt 33097 to the end of the right inverted terminal repeat, with a NotI site at the 3′ end. Both fragments were digested with NotI and NdeI (NdeI sites present in amplified sequences) and cloned into cosmid vector pWE15 (Clontech). The resulting cosmid was next digested with NdeI and ligated with a 26.6-kb fragment resulting from NdeI digestion of wild-type Ad35 (wtAd35) DNA. The ligation mix was packaged using λ-phage packaging extracts (Stratagene), resulting in cosmid pWE.Ad35.pIX-rITR. To generate E1/E3-lacking viruses, a second cosmid clone, pWE.Ad35.pIX-rITRΔE3, was generated that has a deletion of nt 27648 to 30320 (numbers as in wtAd35) corresponding to most of the E3 region. The E3 coding regions (3, 4) are located between nt 27200 and 30622 in the Ad35 genome.
Ad35 viruses were generated using the appropriate adapter plasmid and cosmid clone. DNAs were digested to liberate plasmid vector sequences from adenovirus inserts and were transfected on PER.C6/55K cells. Homologous recombination between the shared sequences of the virus DNAs gives rise to full-length Ad35 genomes carrying the foreign gene (see Fig. 2). Viruses were plaque purified and propagated on adherent cultures in 10 to 24 triple-layer flasks. Purified stocks were obtained by standard two-step CsCl gradient banding and the isolated virus was dialyzed in three steps to a final formulation in phosphate-buffered saline (PBS)-5% sucrose.
FIG. 2.
Generation of recombinant Ad35 vectors. (A) Schematic of the genome organization of wtAd35. Indicated are the inverted terminal repeats (ITRs), packaging signal (Ψ), and early genes involved in replication (E1, E2A, E2B, and E4) and immune modulation (E3). Also, regions coding for the structural proteins (L1 to L5) are given. Total length is 34,794 bp. The NdeI sites are located at positions 6541 and 33166 in the wtAd35 sequence (GenBank accession no. AY271307). (B) Schematic overview of the two-plasmid vector system designed to allow generation of recombinant E1-lacking Ad35 virus (see Materials and Methods for details).
Generation of packaging cells for Ad35 vectors.
Construct pIG55Kneo contains the Ad35 E1B-55K sequence corresponding to nt 1916 (ATG) to 3400 (TAA) from the wtAd35 genome sequence, driven by the human phosphoglycerate kinase promoter and terminated by the hepatitis B virus pA (HBVpA). The plasmid also contains an RSV-neor-HBVpA expression cassette. ScaI-linearized pIG55Kneo DNA was transfected with Lipofectamine (Invitrogen) using the protocol provided by the manufacturer. Twenty-four hours after transfection, medium was replaced with culture medium containing G418 (0.5 mg/ml; Gibco BRL). Selection medium was refreshed twice weekly. After 3 to 4 weeks G418-resistant clones were picked, expanded to 175-cm2 flasks, and tested for the ability to complement replication of E1-lacking Ad35 viruses. Based on the level of transcomplementation and growth characteristics, both adherent and in suspension, one clone was selected that was subsequently subcloned twice. Suspension growth of the PER.C6/55K cells was done in roller bottles in proprietary serum-free medium (AEM; Gibco BRL).
Ad35 vector tropism.
Monocytes were purified from peripheral blood mononuclear cells by positive selection with CD14 microbeads (Miltenyi BioTec) and were cultured in the presence of granulocyte-macrophage colony-stimulating factor (Leukomax [800 U/ml]; Novartis) and interleukin-4 (500 U/ml; PreproTech). After 6 days, cells developed into typical immature DC that were CD14−, CD1a+, CD11c+, CD80+, CD86+, HLA-DR+, HLA-class I+, and CD83− (data not shown). A549 cells (ATCC, CCL1185), human saphenous vein smooth muscle cells (21), and human synoviocytes derived from rheumatoid arthritis patients (19) were cultured as described previously.
Whole-body bioluminescent reporter imaging of mice that received Ad5- and Ad35-Luc injections was done as previously described (56).
Quantification of adenoviral genomes by quantitative PCR (Q-PCR).
Harvested cells were first treated by freeze-thawing followed by addition of deoxycholate (ICN Biomedicals, Inc.) up to 0.5% and further treated as previously described (31).
The primers and probe were as follows: primer 35PSF-1 (5′-GTT CAG GGC CAG GTA GAC TTT G-3′), primer 35PSR-3 (5′-CGC GGA AAT TCA GGT AAA AAA C-3′), and probe 5′-VIC CCC ATT ACG TGG AGG TTT CGA TTA CCG-3′ (3′ labeled with 6-carboxy-tetramethylrhodamine). The 25-μl reaction mixtures contained 200 μM deoxynucleoside triphosphate, 1× buffer A, 3 mM MgCl2, 120 nM concentrations of primers, 200 nM probe, 0.6 U of Taqman Gold DNA polymerase, and 5 μl of DNA. The initial denaturation step (10 min at 95°C) was followed by 45 cycles of 15 s at 95°C and 1 min at 60°C.
Immunological assays and mice experiments.
All animal experiments were approved by the Institutional Review Board and the National Ethical Committee for animal experiments. BALB/c mice (Harlan BV) were immunized intravenously (i.v.) at day 0 and day 14 with 1010 vp of Ad5 (n = 5). At day 28 mice were sacrificed followed by serum and spleen cell isolation. Antiadenovirus NA was determined using a sensitive assay based on inhibition of Ad-luciferase infection (Sprangers et al., submitted for publication). Briefly, serial dilutions of the serum were added to 5 × 106 vp of either Ad5 or Ad35 carrying luciferase and added to wells of 96-well plates, each well containing 104 human A549 cells. Twenty-four hours after virus addition, cells were harvested and luciferase activity determined using steady-glow substrate (Promega).
Spleen cells (105) were added to syngeneic NIH 3T3 cells (5 × 104) loaded for 48 h with Ad5 or Ad35 virus (105 vp/cell). Vector-specific T cells were detected using a mouse gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) assay. Briefly, 96-well plates (Millipore, Bedford, Mass.) were precoated with anti-mouse IFN-γ antibody (Pharmingen) and blocked with culture medium. Spleen cells and target cells were cocultured for 18 h at 37°C in 10% CO2. Plates were washed (PBS-0.05% Tween) and incubated (overnight, 4°C) with 100 μl (2.5 mg/ml) of biotin-conjugated anti-mouse IFN-γ antibody (Pharmingen). Alkaline phosphatase-conjugated streptavidin and substrate (5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium) were both obtained from Sigma. Spots obtained were quantified using an ELISPOT reader (AELVIS GmbH).
For injections for SCID mice, human serum samples with high titers of Ad5 NA (n = 20) were pooled. Adeno-specific activity was determined in an adenovirus-neutralization assay. One hundred microliters of the serum pool was administered i.v. into SCID mice. From mice (n = 5) serum was retrieved 6 and 48 h after administration to determine survival of human serum via the maintenance of Ad5 neutralizing capacity. To assess in vivo neutralization of human sera, SCID mice injected with 200 μl of either anti-Ad5-positive serum pool or an anti-Ad5-negative pool received 6 h later a single intramuscular (i.m.) (m. gastrocnemius) administration of either Ad5 or Ad35 carrying luciferase. Forty-eight hours after vector administration, mice were sacrificed, skeletal muscle was isolated, and a protein lysate was generated by mincing for 30 s in 750 μl of ice-cold PBS. Luciferase activity was determined in 25-μl samples as described above. Statistical analyses were done using the Mann-Whitney test.
RESULTS
Seroprevalence of human adenoviruses in healthy blood donors.
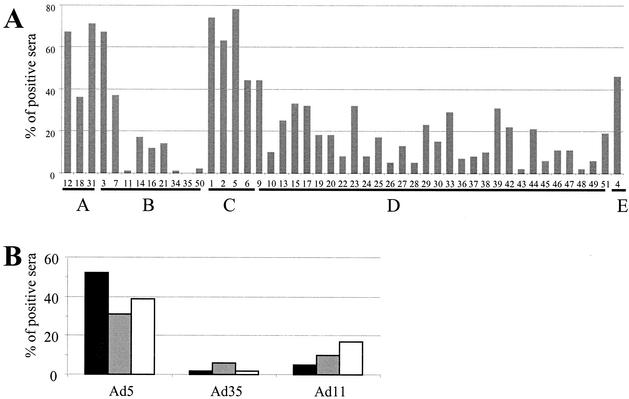
With the exception of Ad8, Ad32, Ad40, and Ad41, which replicated poorly on PER.C6 cells (12), all human adenovirus types were produced on PER.C6 cells, yielding between 8 × 1010 and 5 × 1012 virus particles (vp) per milliliter after cesium chloride (CsCl) purification. As an example of the neutralization frequency against most adenovirus types, NA is shown for 100 serum samples obtained from Belgium volunteers (Fig. 1A). These, and subsequent data from other locations in Europe, Asia, and the United States (Fig. 1B) showed that both Ad2 and Ad5, which are commonly used, are neutralized efficiently by the majority of sera tested and identified Ad35 and Ad11 as types with low prevalence in healthy blood donors.
FIG. 1.
Low seroprevalence toward Ad35 in healthy individuals. (A) One hundred serum samples from the Belgium population (average age, 30.8 years; range, 18 to 62 years; 50% female) were tested for NA against human adenovirus types. Shown on the y axis is the percentage of sera that inhibited virus replication >90% at the lowest dilution (end concentration in the well: 1/4). Types are ranked according to their classification in subgroups. (B) Approximately 560 serum samples from locations in Europe (black bar), the United States (gray bar), and Asia (white bar) were tested for NA against adenovirus types. Shown on the y axis is the percentage of sera that inhibited virus replication >90% at the lowest dilution for Ad5, Ad35, and Ad11 wild-type virus.
Generation of a manufacturing platform for Ad35-based vectors.
First the Ad35 virus genome was sequenced (GenBank accession no. AY271307). Based on the sequence homology with Ad5 (GenBank accession no. M72360) and Ad7 (partial sequence, GenBank accession no. X03000) and on the location of open reading frames, the genome organization of the Ad35 virus proves almost identical to other human adenoviruses, especially the subgroup B viruses to which Ad35 belongs. To obtain a manufacturing platform for production of replication-deficient Ad35 vectors, a two-plasmid system was designed that allows for virus generation following transfection and homologous recombination into transcomplementing cells (Fig. 2). However, Ad5-based complementing cell lines such as PER.C6 cells were unable to complement fully E1-lacking Ad35 vectors. To identify the protein(s) pivotal for an Ad35 vector to replicate on PER.C6, a series of vectors were constructed carrying different parts of the Ad35 E1 region, including a ΔE1A, ΔE1A/ΔΕ1β21k, and ΔE1A/ΔE1B55K vector. Subsequent transfections into PER.C6 demonstrated that the gene encoding for the E1B-derived 55,000-molecular-weight (55K) protein is minimally required to obtain a stable Ad35 virus that can be propagated on PER.C6 cells (data not shown). Using this observation, a packaging cell line for Ad35 was designed, based on PER.C6 cells, by transfection of a plasmid (pIG.55Kneo) carrying the Ad35 E1B-55K gene and a neomycin resistance gene to enable selection of stable transfectants. The ultimate clone selected was coded PER.C6/55K and transcomplemented with high efficiency, both E1-lacking Ad5 and E1-lacking Ad35 vectors. The PER.C6/55K cells were easily adapted to growth in suspension in the absence of serum components and maintained the ability to transcomplement recombinant Ad35 vector for >80 days in the absence of G418, indicative of stable 55K gene insertion in a transcriptionally active locus of PER.C6 cells (data not shown). Both cell-doubling time in suspension (30 h) and cell density (±5 × 106 cells/ml in a roller bottle) of PER.C6/55K proved equivalent to those of PER.C6. Vector yields of ΔE1-Ad35 vectors obtained prepurification (80,000 to 140,000 vp/cell) and postpurification (±18,000 vp/cell) on PER.C6/55K were determined by Q-PCR using oligonucleotides and a probe directed at the packaging signal located in the viral backbone. Since cells loaded with adenovirus represent the initial material for CsCl purification, the optimal harvest time for cells exposed to Ad35 was defined by the time at which the maximum number of virus particles could be detected in a cell pellet. Via Q-PCR analysis both on cell pellets and supernatant samples the optimal cell harvest time was set at 24 to 48 h after full CPE for Ad35 vectors (data not shown) that deviates from the typical Ad5 vector production for which cells are usually harvested at the day of full CPE.
Tropism of Ad35-based recombinant vectors.
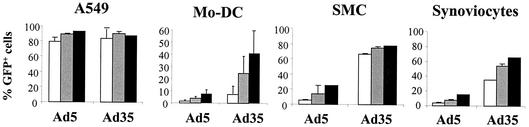
Human monocyte derived immature dendritic cells (mo-DC), SMC, and synoviocytes were exposed to Ad5 or Ad35 vectors carrying green fluorescent protein. Gene transfer efficacy mediated by Ad5 and Ad35 was assessed by marker gene expression. The results obtained (Fig. 3) show that gene transfer with an Ad35 vector is superior compared to Ad5 for these cell types and proved similar to an Ad5 vector carrying the fiber molecule of Ad35 (Ad5.Fib35) as reported earlier (19, 21, 22, 41).
FIG. 3.
Improved infection of (primary) human cells with Ad35-based viruses. Cells were exposed for 2 h, in triplicate, to Ad5 or Ad35 vector carrying green fluorescent protein (GFP) as a marker gene, after which the medium was replaced by culture medium without virus. A549 cells, SMC, and human synoviocytes were infected at a multiplicity of infection of 500 (white bar), 2,000 (grey bar), or 5,000 (black bar) vp/cell. Mo-DC (from three different donors; data were pooled) were infected with 100, 1,000, or 2,500 vp/cell. After 48 h cells were harvested and gene transfer efficacy was determined.
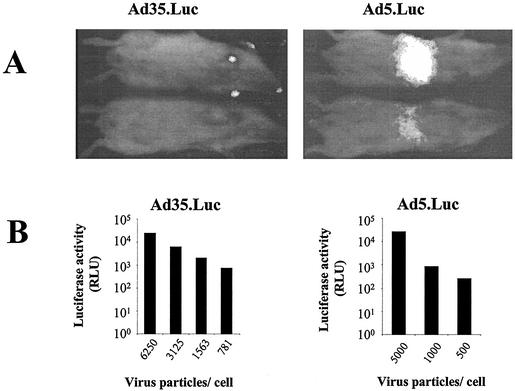
To investigate the biodistribution of Ad35 in a rodent, 1011 vp of Ad35.ΔE3.Luc was administered i.v. in BALB/c mice (n = 2), and after 72 h luciferase expression was determined using charge-coupled device detection technology (56). Luciferase activity was readily detectable in the liver of the mice injected with Ad5-based vectors but was not found following Ad35.Luc vector administration (Fig. 4A). This difference cannot be attributed to difference in batch quality since A549 cells were efficiently infected by both vectors (Fig. 4B). The signals on the footpads, ear, and nose of one of the Ad35-treated animals are currently unexplained.
FIG. 4.
Biodistribution of recombinant Ad35 vectors in mice. (A) Ad5.AdApt.Luc or Ad35ΔE3.AdApt.Luc (1011 vp) was injected i.v. into BALB/c mice, and luciferase expression was monitored 72 h later using charge-coupled device camera technology (56). (B) Quality of Ad5 and Ad35 vector batches. Human A549 cells were exposed for two hours to an increasing concentration of Ad5 or Ad35 carrying the luciferase reporter gene. Cells were harvested after 48 h and lysed, and luciferase activity was determined (expressed in relative light units [RLU]).
Recombinant Ad35 vectors circumvent in vitro and in vivo neutralization.
To evaluate the behavior of recombinant Ad35 vectors in the presence of Ad5 immunity Ad5.AdApt.Luc or Ad35.ΔE3.AdApt.Luc viruses were incubated with eight different human sera positive for Ad5 neutralization and analyzed for infectivity on A549 cells. The human sera efficiently neutralize Ad5-based vectors (90% inhibition titers, 1,024 to 4,096) whereas Ad35 viruses are not neutralized or, in some cases, at very low titers (Fig. 5A). The absence of cross-neutralization is confirmed in animal experiments. Mice that were immunized twice with Ad5.empty viruses on day 0 and 14 were sacrificed on day 28 and serum was analyzed for neutralization of Ad5- and Ad35-Luc viruses. Serum from Ad5-preimmunized mice neutralized Ad5 vector with high efficiency but not an Ad35 vector (Fig. 5B). Spleen cells from the same mice were incubated with 3T3 cells infected either with Ad5.empty or with Ad35.empty viruses to analyze virus specific T cells. IFN-γ ELISPOT analysis showed that Ad5 specific T cells do not cross-react with Ad35 (Fig. 5C). Confirmation of 3T3 cell loading in this assay was performed with Ad5- and Ad35-Luc viruses, demonstrating that 3T3 cells are capable of uptake of Ad5 and Ad35 viruses at high MOI (105 vp/cell, data not shown).
FIG. 5.
Ad35 vectors circumvent Ad5 immunity. (A) Human serum samples were tested for NA against Ad5- or Ad35-luciferase viruses. Black dots represent individual sera at the serum dilution that inhibited >90% of the luciferase activity in the absence of serum. The bars represent geometric mean titers. (B) Serum samples from mice twice immunized with Ad5.empty viruses (1010 vp) were tested for NA against Ad5- or Ad35-luciferase viruses. (C) Spleen cells from mice twice immunized with Ad5.empty viruses (1010 vp) were incubated with NIH 3T3 cells transduced with either Ad5 or Ad35 empty vectors and IFN-γ release was quantified.
In a second experiment, human serum was administered to SCID mice prior to adenovirus injection. We first showed that anti-Ad5 NA was still present after 6 and 48 h. in SCID mice (Fig. 6A). Next, SCID mice received i.v. injections with human serum (either with or without Ad5 NA), and after 6 h Ad5- or Ad35-Luc viruses were administered i.m. We chose the i.m. route because in contrast to i.v. administration where Ad35.Luc expression could not be detected, i.m. administration resulted in clearly detectable, albeit three- to fourfold reduced, luciferase expression (Fig. 6B). Two days later, analysis of luciferase reporter gene expression demonstrated a 90% decrease (P = 0.008) in Ad5-mediated luciferase activity in mice that received anti-Ad5 serum as opposed to mice receiving control serum. Luciferase gene transfer mediated by Ad35 was not significantly inhibited (P = 0.56) by the presence of anti-Ad5 serum (Fig. 6B).
FIG. 6.
(A) Human serum was administered i.v. to SCID mice, and serum was isolated at various time points. The undiluted human serum fraction was tested in vitro for the ability to inhibit Ad5-mediated luciferase gene transfer to human A549 cells (black circles). The serum fraction was subsequently diluted 10-fold in PBS (black squares) to mimic the dilution of the samples retrieved from the mice that received injections. Mouse serum was isolated at 6 h (black triangles) post-serum administration and after 48 h (black diamonds). Serum from mice that received Ad5-negative human serum served as negative controls (white circles). (B) Ad35 viruses transduce muscle in presence of human serum with NA against Ad5. Results are presented as luciferase activity in 25 μl of muscle lysates (left graph), showing that Ad5.Luc expression is largely reduced in the presence of human serum with Ad5 NA (+serum) compared to Ad5.Luc activity in mice that received human serum without Ad5 NA (−serum). This reduction is not seen in mice receiving Ad35.Luc viruses. In the graph at right the luciferase activity determined in muscle from mice that received human anti-Ad5 serum (+serum) is shown as a percentage of luciferase activity compared to that in mice that received anti-Ad5-negative serum (−serum).
DISCUSSION
The preexisting immunity data for Ad5 and Ad35 reported here correlate with reported virus isolation data, showing that Ad35 virus could not be isolated from adenovirus-positive clinical samples (n = 2,300) derived from immunocompetent patients whereas Ad5 was isolated with high (∼10%) frequency (10). Although at present this low seroprevalence of Ad35 is not completely understood, it has become evident that a potent humoral immune response can be elicited against Ad35 in rodents and nonhuman primates (unpublished data). However, most of these results were obtained with E3-lacking vectors. One striking difference between subgroup C and B viruses is that members of the latter subgroup have a larger E3 region containing homologues of the coding regions identified in Ad5 and two additional coding regions (4, 15, 23, 26, 50). The function of E3 encoded proteins in inhibition of peptide presentation by major histocompatibility complex class I molecules and in inhibition of cytolysis, has been amply described (reviewed in references 27 and 57). The function of the additional proteins encoded by the subgroup B adenovirus proteins is currently unknown. A possible causative relation with the low seroprevalence of Ad35 is questionable given the observation that Ad3, Ad7, and Ad16, to which humans frequently have NA, also contain these additional coding regions (26, 50). This suggests that Ad35 did not develop a new, or more effective, evasion mechanism to impair a host in boosting humoral responses against this virus. More detailed analysis of the expression levels and function of these E3 proteins may aid a better understanding of a possible involvement of the E3 proteins in the observed differences in seroprevalence. The fact that Ad35 and Ad11 are isolated from immune-compromised patients indicates that the virus is either circulating but rare or latently present (16, 24). It may be that the route of infection of Ad35 differs from the more common adenoviruses, necessitating a closer contact for transmission. This latter hypothesis is strengthened by a report covering an outbreak of Ad35 affecting healthy individuals residing in a chronic care psychiatric facility (43). Although speculative, the outbreak was attributed by the authors to an increased transmission caused by crowding and poor hygienic behaviors within the facility.
Towards the generation of a manufacturing platform for E1-lacking Ad35 vectors we found that the Ad5-E1 region present in PER.C6 cells is unable to transcomplement for the E1 deficiency in Ad35, and that in order to obtain virus from the PER.C6 cells, the E1B derived 55K protein of Ad35 is minimally required. These observations corroborate an earlier report (1) demonstrating that the subgroup B derived Ad7a vector could not be produced in the standard HEK293 packaging cell line. However, it was shown that an Ad7a virus that had reacquired the E1B region, due to an unexpected recombination, could be propagated, albeit efficiently only on 293 cells stably expressing the Ad5 E4-ORF6 region. This finding by Abrahamsen et al. differs from our results obtained with Ad35 since PER.C6 cells that do not express E4-ORF6 are well able to support the continuous production and plaque purification of an E1B+ Ad35 vector (not shown). The involvement of both E1B-55K and E4-Orf6 proteins strongly suggests that the inability of E1-lacking Ad35-based viruses to replicate on Ad5 packaging cell lines is caused by impairment of the complex formed by both proteins (44). The E1B-55K/E4-Orf6 complex has been described to increase the nuclear export of viral mRNAs and to inhibit nuclear export of cellular mRNAs (2, 6, 40). Both processes play an important role in virus formation and virus release from the cell (58).
Based on the above, PER.C6 cells were transfected with an Ad35 E1B-55K expression cassette and a new complementation cell line was established. To obtain purified research batches for Ad35, the production and purification protocols for Ad5 vectors were used and little attempts have been made so far to optimize these protocols for Ad35 vector production. Despite the latter, and the observation that virus harvest times may differ, Ad35 vector yields per cell on PER.C6/55K are comparable to Ad5 produced either on PER.C6 or PER.C6/55K, both in terms of virus particles per cell prepurification and postpurification. One major difference with Ad35-based viruses concerns the virus particle per infectious unit ratio of 68 ± 45 that is approximately sixfold higher than that for Ad5 and is attributed to the differences between Ad5 and Ad35 with respect to their ability to infect PER.C6/55K cells. So far, >25 virus batches of Ad35, carrying diverse transgenes, have been produced, purified, and tested. The first Ad35 virus batches generated were carrying marker genes like luciferase and green fluorescent protein to assess tropism of Ad35 and the ability to circumvent anti-Ad5 NA. The data obtained regarding the tropism of Ad35 show that Ad35, as was previously reported for Ad5Fib35 chimeric viruses, has a natural tropism for diverse primary human cell types of interest for those developing a treatment for cardiovascular disorders (21), rheumatoid arthritis (19), and for both ex vivo and in vivo vaccination strategies (22, 41). This increased infection is not seen on rodent cells and tissues where Ad5 vectors are more efficient than Ad35 vectors. Using the whole-body luminescence technique, we did not find luciferase expression in the liver following i.v. Ad35 vector administration, in contrast to the strong expression seen with the Ad5.Luc vector. These data correlate with biodistribution studies in mice (O. Ophorst and M. Havenga, unpublished results) where i.v. injection with a chimeric Ad5fib35.Luc vector resulted in a 2- to 3-log-decreased luciferase activity in liver compared to that obtained with an Ad5.Luc virus. In the latter study, luciferase activity was determined in minced tissues. The fact that no luciferase activity was found in the liver of Ad35 transduced mice not necessarily reflects a significant difference with the reduced luciferase levels found with the Ad5fib35 vector but may be attributable to differences in sensitivities of the used techniques. These results are also in line with other results obtained with an Ad5-fiber35 chimeric virus in a different mouse strain (CB17) that show a 10-fold-reduced infection in mouse liver compared to unmodified Ad5 vectors, both in copy number and expression level (47). Reduced infection of rodent cells with viruses carrying a subgroup B fiber, also observed previously with an Ad5.Fib16 vector (21, 22), is possibly caused by lack of, or inefficient binding to, the (unknown) cellular receptor for Ad35 and other subgroup B viruses in these species. In addition, it cannot be excluded that the reduced transgene expression is in part due to differences in intracellular trafficking. It has been recently reported that the fiber of subgroup B adenoviruses directs a different intracellular route for internalized virus particles compared to Ad5 fibers, leading to a reduction of the amount of genomes that reach the nucleus (33, 48). Direct injection into the muscles of mice results in a three- to fourfold reduced, but clearly measurable, expression using Ad35 vectors compared to Ad5 vectors. The reason that Ad35 viruses do transduce mouse muscle upon i.m. injection is currently unknown. It may be similar to previous findings regarding Ad5 muscle transduction in mice, which was suggested to be fiber-independent (11, 53).
Besides its superior in vitro tropism over Ad5, Ad35 was primarily selected to overcome preexisting immunity. We have shown that, at least after induced immunity in rodents, no cross-reaction, either by antibodies or T cells, occurs between recombinant Ad5 and Ad35 vectors resulting in a maintained gene transfer efficacy with Ad35 in a host positive for anti-Ad5 NA. The absence of cross-reactivity of T cells in mice may not be predictive for the human situation. Two injections of Ad5 viruses in otherwise naive mice most likely does not give the complex immune response expected in humans which are frequently infected with different types. Sequential infections with antigenic variant strains is thought to select for cross-reactive T cells in vivo, as has been found for influenza A virus (20). In this respect, both CD4+ and CD8+ cross-reactive T cells against Ad types from different subgroups were found in human peripheral blood mononuclear cells (14, 51). Also, a dominant CD4+ T-cell epitope located in a C-terminal conserved region in hexon (39) is also present in the Ad35 hexon protein. In this respect, it should be noted that CD4+ cytotoxic T cells are not likely detected in our assay because 3T3 cells do not express major histocompatibility complex class II antigens.
Naturally, the studies described here need detailed follow-up to determine in diverse animal models the safety, toxicity, and efficacy profile of Ad35-based vectors, but based on the ability to produce the replication-deficient Ad35 virus and on the first data regarding tropism and lack of preexisting humoral immunity in humans, we are confident that this vector system provides a superior platform for gene transfer and vaccination strategies.
Acknowledgments
This work was supported by grants from the Dutch government (BTS 99-107 and S&O).
We thank Alexander Zakhartchouk for constructing the E3 deletion in the Ad35 cosmid clone. Furthermore, we thank J. C. de Jong (Erasmus University, Rotterdam, The Netherlands) for providing the wt adenoviruses and for his input during the neutralization study. We thank E. Schraa, P. Tjou Tam Sin, and C. Pont for excellent work in animal studies.
REFERENCES
- 1.Abrahamsen, K., H. L. Kong, A. Mastrangeli, D. Brough, A. Lizonova, R. G. Crystal, and E. Falck-Pedersen. 1997. Construction of an adenovirus type 7a E1A- vector. J. Virol. 71:8946-8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babiss, L. E., H. S. Ginsberg, and J. E. Darnell, Jr. 1985. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol. Cell. Biol. 5:2552-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basler, C. F., G. Droguett, and M. S. Horwitz. 1996. Sequence of the immunoregulatory early region 3 and flanking sequences of adenovirus type 35. Gene 170:249-254. [DOI] [PubMed] [Google Scholar]
- 4.Basler, C. F., and M. S. Horwitz. 1996. Subgroup B adenovirus type 35 early region 3 mRNAs differ from those of the subgroup C adenoviruses. Virology 215:165-177. [DOI] [PubMed] [Google Scholar]
- 5.Boshart, M., F. Weber, G. Jahn, K. Dorsch-Häler, B. Fleckenstein, and W. Scaffner. 1985. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell 41:521-530. [DOI] [PubMed] [Google Scholar]
- 6.Bridge, E., and G. Ketner. 1990. Interaction of adenoviral E4 and E1b products in late gene expression. Virology 174:345-353. [DOI] [PubMed] [Google Scholar]
- 7.Chen, P., I. Kovesdi, and J. T. Bruder. 2000. Effective repeat administration with adenovirus vectors to the muscle. Gene Ther. 7:587-595. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Y., D. C. Yu, D. Charlton, and D. R. Henderson. 2000. Pre-existent adenovirus antibody inhibits systemic toxicity and antitumor activity of CN706 in the nude mouse LNCaP xenograft model: implications and proposals for human therapy. Hum. Gene Ther. 11:1553-1567. [DOI] [PubMed] [Google Scholar]
- 9.Dai, Y., E. M. Schwarz, D. Gu, W. W. Zhang, N. Sarvetnick, and I. M. Verma. 1995. Cellular and humoral immune responses to adenoviral vectors containing factor IX gene: tolerization of factor IX and vector antigens allows for long-term expression. Proc. Natl. Acad. Sci. USA 92:1401-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Jong, J. C., A. G. Wermenbol, M. W. Verweij-Uijterwaal, K. W. Slaterus, P. Wertheim-Van Dillen, G. J. J. Van Doornum, S. H. Khoo, and J. C. Hierholzer. 1999. Adenoviruses from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotypes Ad50 and Ad51 of species B1 and D, respectively. J. Clin. Microbiol. 37:3940-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Einfeld, D. A., R. Schroeder, P. W. Roelvink, A. Lizonova, C. R. King, I. Kovesdi, and T. J. Wickham. 2001. Reducing the native tropism of adenovirus vectors requires removal of both CAR and integrin interactions. J. Virol. 75:11284-11291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fallaux, F. J., A. Bout, I. van der Velde, D. J. M. van den Wollenberg, K. Hehir, J. Keegan, C. Auger, S. J. Cramer, H. van Ormondt, A. J. van der Eb, D. Valerio, and R. C. Hoeben. 1998. New helper cells and matched early region 1-deleted adenovirus vectors prevent generation of replication-competent adenoviruses. Hum. Gene Ther. 9:1909-1917. [DOI] [PubMed] [Google Scholar]
- 13.Farina, S. F., G. P. Gao, Z. Q. Xiang, J. J. Rux, R. M. Burnett, M. R. Alvira, J. Marsh, H. C. Ertl, and J. M. Wilson. 2001. Replication-defective vector based on a chimpanzee adenovirus. J. Virol. 75:11603-11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flomenberg, P., V. Piaskowski, R. L. Truitt, and J. T. Casper. 1995. Characterization of human proliferative T cell responses to adenovirus. J. Infect. Dis. 171:1090-1096. [DOI] [PubMed] [Google Scholar]
- 15.Flomenberg, P. R., M. Chen, and M. S. Horwitz. 1988. Sequence and genetic organization of adenovirus type 35 early region 3. J. Virol. 62:4431-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flomenberg, P. R., M. Chen, G. Munk, and M. S. Horwitz. 1987. Molecular epidemiology of adenovirus type 35 infections in immunocompromised hosts. J. Infect. Dis. 155:1127-1134. [DOI] [PubMed] [Google Scholar]
- 17.Francki, R., C. Fauquet, D. Knudson, and F. Brown. 1991. Classification and nomenclature of viruses. Fifth report of the international committee on taxonomy of viruses. Arch. Virol. Suppl. 2:140-144.
- 18.Gahery-Segard, H., V. Molinier-Frenkel, C. Le Boulaire, P. Saulnier, P. Opolon, R. Lengagne, E. Gautier, A. Le Cesne, L. Zitvogel, A. Venet, C. Schatz, M. Courtney, T. Le Chevalier, T. Tursz, J. G. Guillet, and F. Farace. 1997. Phase I trial of recombinant adenovirus gene transfer in lung cancer. Longitudinal study of the immune responses to transgene and viral products. J. Clin. Investig. 100:2218-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goossens, P. H., M. J. Havenga, E. Pieterman, A. A. Lemckert, F. C. Breedveld, A. Bout, and T. W. Huizinga. 2001. Infection efficiency of type 5 adenoviral vectors in synovial tissue can be enhanced with a type 16 fiber. Arthritis Rheum. 44:570-577. [DOI] [PubMed] [Google Scholar]
- 20.Haanen, J. B., M. C. Wolkers, A. M. Kruisbeek, and T. N. Schumacher. 1999. Selective expansion of cross-reactive CD8+ memory T cells by viral variants. J. Exp. Med. 190:1319-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Havenga, M. J., A. A. Lemckert, J. M. Grimbergen, R. Vogels, L. G. Huisman, D. Valerio, A. Bout, and P. H. Quax. 2001. Improved adenovirus vectors for infection of cardiovascular tissues. J. Virol. 75:3335-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Havenga, M. J., A. A. Lemckert, O. J. Ophorst, M. van Meijer, W. T. Germeraad, J. Grimbergen, M. A. van Den Doel, R. Vogels, J. van Deutekom, A. A. Janson, J. D. de Bruijn, F. Uytdehaag, P. H. Quax, T. Logtenberg, M. Mehtali, and A. Bout. 2002. Exploiting the natural diversity in adenovirus tropism for therapy and prevention of disease. J. Virol. 76:4612-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawkins, L. K., and W. S. Wold. 1995. A 20,500-Dalton protein is coded by region E3 of subgroup B but not subgroup C human adenoviruses. Virology 208:226-233. [DOI] [PubMed] [Google Scholar]
- 24.Hierholzer, J., R. Wigand, L. Anderson, T. Adrian, and J. Gold. 1988. Adenoviruses from patients with AIDS: a plethora of serotypes and a description of five new serotypes of subgenus D (types 43-47). J. Infect. Dis. 158:804-813. [DOI] [PubMed] [Google Scholar]
- 25.Hofmann, C., P. Loser, G. Cichon, W. Arnold, G. W. Both, and M. Strauss. 1999. Ovine adenovirus vectors overcome preexisting humoral immunity against human adenoviruses in vivo. J. Virol. 73:6930-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong, J. S., K. G. Mullis, and J. A. Engler. 1988. Characterization of the early region 3 and fiber genes of Ad7. Virology 167:545-553. [PubMed] [Google Scholar]
- 27.Horwitz, M. S. 2001. Adenovirus immunoregulatory genes and their cellular targets. Virology 279:1-8. [DOI] [PubMed] [Google Scholar]
- 28.Kass-Eisler, A., L. Leinwand, J. Gall, B. Bloom, and E. Falck-Pedersen. 1996. Circumventing the immune response to adenovirus-mediated gene therapy. Gene Ther. 3:154-162. [PubMed] [Google Scholar]
- 29.Knowles, M. R., K. W. Hohneker, Z. Zhou, J. C. Olsen, T. L. Noah, P. C. Hu, M. W. Leigh, J. F. Engelhardt, L. J. Edwards, K. R. Jones, et al. 1995. A controlled study of adenoviral-vector-mediated gene transfer in the nasal epithelium of patients with cystic fibrosis. N. Engl. J. Med. 333:823-831. [DOI] [PubMed] [Google Scholar]
- 30.Kurimaya, S., K. Tominaga, M. Kikukawa, T. Nakatani, H. Tsujinoue, M. Yamazaki, S. Nagao, Y. Toyokawa, A. Mitoro, and H. Fukui. 1998. Inhibitory effects of human sera on adenovirus-mediated gene transfer into rat liver. Anticancer Res. 18:2345-2351. [PubMed] [Google Scholar]
- 31.Ma, L., H. A. Bluyssen, M. De Raeymaeker, V. Laurysens, N. van der Beek, H. Pavliska, A. J. van Zonneveld, P. Tomme, and H. H. van Es. 2001. Rapid determination of adenoviral vector titers by quantitative real-time PCR. J. Virol. Methods 93:181-188. [DOI] [PubMed] [Google Scholar]
- 32.Mastrangeli, A., B. G. Harvey, J. Yao, G. Wolff, I. Kovesdi, R. G. Crystal, and E. Falck-Pedersen. 1996. “Sero-switch” adenovirus-mediated in vivo gene transfer: circumvention of anti-adenovirus humoral immune defenses against repeat adenovirus vector administration by changing the adenovirus serotype. Hum. Gene Ther. 7:79-87. [DOI] [PubMed] [Google Scholar]
- 33.Miyazawa, N., R. G. Crystal, and P. L. Leopold. 2001. Adenovirus serotype 7 retention in a late endosomal compartment prior to cytosol escape is modulated by fiber protein. J. Virol. 75:1387-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moffatt, S., J. Hays, H. HogenEsch, and S. K. Mittal. 2000. Circumvention of vector-specific neutralizing antibody response by alternating use of human and non-human adenoviruses: implications in gene therapy. Virology 272:159-167. [DOI] [PubMed] [Google Scholar]
- 35.Molnar-Kimber, K. L., D. H. Sterman, M. Chang, E. H. Kang, M. ElBash, M. Lanuti, A. Elshami, K. Gelfand, J. M. Wilson, L. R. Kaiser, and S. M. Albelda. 1998. Impact of preexisting and induced humoral and cellular immune responses in an adenovirus-based gene therapy phase I clinical trial for localized mesothelioma. Hum. Gene Ther. 9:2121-2133. [DOI] [PubMed] [Google Scholar]
- 36.Morral, N., W. O'Neal, K. Rice, M. Leland, J. Kaplan, P. A. Piedra, H. Zhou, R. J. Parks, R. Velji, E. Aguilar-Cordova, S. Wadsworth, F. L. Graham, S. Kochanek, K. D. Carey, and A. L. Beaudet. 1999. Administration of helper-dependent adenoviral vectors and sequential delivery of different vector serotype for long-term liver-directed gene transfer in baboons. Proc. Natl. Acad. Sci. USA 96:12816-12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagao, S., S. Kuriyama, H. Okuda, K. Tominaga, T. Nakatani, H. Tsujinoue, H. Yoshiji, and H. Fukui. 2001. Adenovirus-mediated gene transfer into tumors: evaluation of direct readministration of an adenoviral vector into subcutaneous tumors of immunocompetent mice. Int. J. Oncol. 18:57-65. [PubMed] [Google Scholar]
- 38.Nichols, W. W., R. Lardenoije, B. J. Ledwith, K. Brouwer, S. Manam, R. Vogels, D. Kaslow, D. Zuijdgeest, A. J. Bett, L. Chen, M. van der Kaaden, S. M. Galloway, R. B. Hill, S. V. Machotka, C. A. Anderson, J. Lewis, D. Martinez, J. Lebron, C. Russo, D. Valerio, and A. Bout. 2002. Propagation of adenoviral vectors: use of PER.C6 cells. Elsevier Science, New York, N.Y.
- 39.Olive, M., L. Eisenlohr, N. Flomenberg, S. Hsu, and P. Flomenberg. 2002. The adenovirus capsid protein hexon contains a highly conserved human CD4+ T-cell epitope. Hum. Gene Ther. 13:1167-1178. [DOI] [PubMed] [Google Scholar]
- 40.Pilder, S., M. Moore, J. Logan, and T. Shenk. 1986. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol. Cell. Biol. 6:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rea, D., M. J. Havenga, M. van Den Assem, R. P. Sutmuller, A. Lemckert, R. C. Hoeben, A. Bout, C. J. Melief, and R. Offringa. 2001. Highly efficient transduction of human monocyte-derived dendritic cells with subgroup B fiber-modified adenovirus vectors enhances transgene-encoded antigen presentation to cytotoxic T cells. J. Immunol. 166:5236-5244. [DOI] [PubMed] [Google Scholar]
- 42.Roth, J. A., S. G. Swisher, J. A. Merritt, D. D. Lawrence, B. L. Kemp, C. H. Carrasco, A. K. El-Naggar, F. V. Fossella, B. S. Glisson, W. K. Hong, F. R. Khurl, J. M. Kurie, J. C. Nesbitt, K. Pisters, J. B. Putnam, D. S. Schrump, D. M. Shin, and G. L. Walsh. 1998. Gene therapy for non-small cell lung cancer: a preliminary report of a phase I trial of adenoviral p53 gene replacement. Semin. Oncol. 25:33-37. [PubMed] [Google Scholar]
- 43.Sanchez, M. P., D. D. Erdman, T. J. Torok, C. J. Freeman, and B. T. Matyas. 1997. Outbreak of adenovirus 35 pneumonia among adult residents and staff of a chronic care psychiatric facility. J. Infect. Dis. 176:760-763. [DOI] [PubMed] [Google Scholar]
- 44.Sarnow, P., P. Hearing, C. W. Anderson, D. N. Halbert, T. Shenk, and A. J. Levine. 1984. Adenovirus early region 1B 58,000-dalton tumor antigen is physically associated with an early region 4 25,000-dalton protein in productively infected cells. J. Virol. 49:692-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schulick, A. H., G. Vassalli, P. F. Dunn, G. Dong, J. J. Rade, C. Zamarron, and D. A. Dichek. 1997. Established immunity precludes adenovirus-mediated gene transfer in rat carotid arteries. Potential for immunosuppression and vector engineering to overcome barriers of immunity. J. Clin. Investig. 99:209-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shabram, P. W., D. D. Giroux, A. M. Goudreau, R. J. Gregory, M. T. Horn, B. G. Huyghe, X. Liu, M. H. Nunnally, B. J. Sugarman, and S. Sutjipto. 1997. Analytical anion-exchange HPLC of recombinant type-5 adenoviral particles. Hum. Gene Ther. 8:453-465. [DOI] [PubMed] [Google Scholar]
- 47.Shayakhmetov, D. M., Z. Y. Li, S. Ni, and A. Lieber. 2002. Targeting of adenovirus vectors to tumor cells does not enable efficient transduction of breast cancer metastases. Cancer Res. 62:1063-1068. [PubMed] [Google Scholar]
- 48.Shayakhmetov, D. M., Z. Y. Li, V. Ternovoi, A. Gaggar, H. Gharwan, and A. Lieber. 2003. The interaction between the fiber knob domain and the cellular attachment receptor determines the intracellular trafficking route of adenoviruses. J. Virol. 77:3712-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 50.Signäs, C., G. Akusjärvi, and U. Petersson. 1986. Region E3 of human adenoviruses: differences between the oncogenic adenovirus-3 and the non-oncogenic adenovirus-2. Gene 50:173-184. [DOI] [PubMed] [Google Scholar]
- 51.Smith, C. A., L. S. Woodruff, C. Rooney, and G. R. Kitchingman. 1998. Extensive cross-reactivity of adenovirus-specific cytotoxic T cells. Hum. Gene Ther. 9:1419-1427. [DOI] [PubMed] [Google Scholar]
- 52.Sullivan, N. J., A. Sanchez, P. E. Rollin, Z. Y. Yang, and G. J. Nabel. 2000. Development of a preventive vaccine for Ebola virus infection in primates. Nature 408:605-609. [DOI] [PubMed] [Google Scholar]
- 53.van Deutekom, J. C., B. Cao, R. Pruchnic, T. J. Wickham, I. Kovesdi, and J. Huard. 1999. Extended tropism of an adenoviral vector does not circumvent the maturation-dependent transducibility of mouse skeletal muscle. J. Gene Med. 1:393-399. [DOI] [PubMed] [Google Scholar]
- 54.Varnavski, A. N., Y. Zhang, M. Schnell, J. Tazelaar, J. P. Louboutin, Q. C. Yu, A. Bagg, G. P. Gao, and J. M. Wilson. 2002. Preexisting immunity to adenovirus in rhesus monkeys fails to prevent vector-induced toxicity. J. Virol. 76:5711-5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wadell, G. 1984. Molecular epidemiology of adenoviruses. Curr. Top. Microbiol. Immunol. 110:191-220. [DOI] [PubMed] [Google Scholar]
- 56.Wetterwald, A., G. van der Pluijm, I. Que, B. Sijmons, J. Buijs, M. Karperien, C. W. Lowik, E. Gautschi, G. N. Thalmann, and M. G. Cecchini. 2002. Optical imaging of cancer metastasis to bone marrow: a mouse model of minimal residual disease. Am. J. Pathol. 160:1143-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wold, W. S., K. Doronin, K. Toth, M. Kuppuswamy, D. L. Lichtenstein, and A. E. Tollefson. 1999. Immune responses to adenoviruses: viral evasion mechanisms and their implications for the clinic. Curr. Opin. Immunol. 11:380-386. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, Y., and R. J. Schneider. 1994. Adenovirus inhibition of cell translation facilitates release of virus particles and enhances degradation of the cytokeratin network. J. Virol. 68:2544-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]