Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) shows a very restricted tropism for cells of the monocyte/macrophage lineage. It enters cells via receptor-mediated endocytosis. A monoclonal antibody (MAb) that is able to block PRRSV infection of porcine alveolar macrophages (PAM) and that recognizes a 210-kDa protein (p210) was described previously (MAb41D3) (X. Duan, H. Nauwynck, H. Favoreel, and M. Pensaert, J. Virol. 72:4520-4523, 1998). In the present study, the p210 protein was purified from PAM by immunoaffinity using MAb41D3 and was subjected to internal peptide sequencing after tryptic digestion. Amino acid sequence identities ranging from 56 to 91% with mouse sialoadhesin, a macrophage-restricted receptor, were obtained with four p210 peptides. Using these peptide data, the full p210 cDNA sequence (5,193 bp) was subsequently determined. It shared 69 and 78% amino acid identity, respectively, with mouse and human sialoadhesins. Swine (PK-15) cells resistant to viral entry were transfected with the cloned p210 cDNA and inoculated with European or American PRRSV strains. Internalized virus particles were detected only in PK-15 cells expressing the recombinant sialoadhesin, demonstrating that this glycoprotein mediated uptake of both types of strains. However, nucleocapsid disintegration, like that observed in infected Marc-145 cells as a result of virus uncoating after fusion of the virus with the endocytic vesicle membrane, was not observed, suggesting a block in the fusion process. The ability of porcine sialoadhesin to mediate endocytosis was demonstrated by specific internalization of MAb41D3 into PAM. Altogether, these results show that sialoadhesin is involved in the entry process of PRRSV in PAM.
Porcine reproductive and respiratory syndrome virus (PRRSV) induces respiratory problems in pigs and severe reproductive failure in sows, leading to late-term abortions or the birth of stillborn and weak pigs. It is a member of the Arteriviridae family (order Nidovirales), which so far includes three other members: Equine arteritis virus, Mouse lactate dehydrogenase-elevating virus (LDV), and Simian hemorrhagic fever virus (6, 35). These enveloped, single-stranded RNA viruses share morphological and genomic similarities, can establish a persistent infection in their natural host, and have a predilection for cells of the monocyte/macrophage lineage. Of the porcine cells tested, only porcine alveolar macrophages (PAM) and some cultivated peripheral blood monocytes support productive replication of PRRSV in vitro (19, 60). Additional studies showed that the virus also infects macrophages in the spleen, tonsils, lymph nodes, liver, Peyer's patches, and thymus, whereas peritoneal macrophages, freshly isolated blood monocytes, and progenitor cells in the bone marrow are refractory to infection (18, 19, 51). A few nonmacrophage cells are also susceptible to PRRSV, including porcine testicular germ cells (spermatids and spermatocytes) (52), the African green monkey kidney cell line MA-104, and cells derived from MA-104 (Marc-145 and CL-2621) (28).
Receptor molecules mediating arterivirus entry into target cells have not been characterized until now. Several indications suggest that the restricted tropism of LDV and PRRSV for macrophages can be attributed to the presence of a macrophage-specific surface receptor. (i) Using pseudotype virions consisting of LDV RNA and the mouse hepatitis virus envelope, a productive LDV infection was obtained in cells that are refractory to LDV but susceptible to mouse hepatitis virus (23). (ii) Transfection of nonpermissive cells with genomic RNA of PRRSV or LDV sufficed to allow virus replication (25, 31, 38). After attachment to cellular receptors, PRRSV enters cells by a process of receptor-mediated endocytosis through clathrin-coated pits and vesicles (30, 42). A subsequent drop in pH in the endosome is required for proper virus replication. Heparan sulfate glycosaminoglycans were shown to mediate PRRSV attachment to Marc-145 cells and PAM (15, 26, 57). Although infection of PAM was reduced by the addition of heparin, a complete inhibition of infection could not be obtained, indicating the presence of other receptor molecules. A monoclonal antibody (MAb) (MAb41D3) that is able to abolish PRRSV infection of PAM while only reducing the binding of the virus was described (20, 21). This antibody colocalized with biotinylated PRRSV on the membrane of PAM and with PRRSV antigen-positive cells in experimentally infected pigs. It immunoprecipitated a 210-kDa protein (p210) from PAM. Nevertheless, the identity of this protein was not elucidated. In the present study, we identified the p210 protein and investigated the ability of a recombinant form of the p210 to mediate PRRSV infection of nonsusceptible cells. Since PRRSV enters into PAM by endocytosis (42), we also investigated the ability of the p210 to mediate endocytosis in PAM using MAb41D3.
MATERIALS AND METHODS
Cells and viruses.
PAM were obtained and cultivated as described by Delputte et al. (15) in minimum essential medium-Eagle with Earle's salts (MEM). The PK-15 cell line free of porcine circovirus was a kind gift of G. M. Allan (Department of Veterinary Science, The Queen's University of Belfast, Belfast, United Kingdom) and was maintained in MEM supplemented with 5% fetal bovine serum, 2 mM l-glutamine, and a mixture of antibiotics in a humidified 5% CO2 atmosphere at 37°C. The Lelystad strain of PRRSV (LV) was kindly provided by G. Wensvoort (Institute for Animal Science and Health, Lelystad, The Netherlands). Strains 94V360 (19) and 96V198 are Belgian isolates, and strain VR2332 is the American prototype (7). The European PRRSV strains were first passaged on PAM and subsequently cultivated on Marc-145 cells for four or six (96V198) passages, while for the American strain a fourth passage on Marc-145 cells was used.
Purification of p210.
PAM were seeded in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum and a mixture of antibiotics at a density of 50 × 106 cells/175 cm2. Twenty-four hours after seeding, fresh medium supplemented with recombinant porcine alpha interferon (50 U/ml) (33) was added, and PAM were maintained in culture for an additional 48 to 72 h. This treatment increases surface expression of the p210 (H. J. Nauwynck, unpublished data). Cells were pooled, washed with phosphate-buffered saline (PBS) and collected by centrifugation (700 × g, 4°C, 7 min). The cell pellet was resuspended in Tris buffer (10 mM Tris-HCl, pH 7.4, supplemented with a cocktail of protease inhibitors [Complete; Boehringer Mannheim]) and lysed using a Dounce homogenizer. Cell nuclei were pelleted by centrifugation (800 × g, 4°C, 10 min) and supernatant was further centrifuged (100,000 × g, 4°C, 1 h) to pellet the membrane fraction that was further resuspended in 70% saccharose in Tris buffer and subjected to a 60%-30%-0% saccharose step gradient (100,000 × g, 4°C, 1 h). The fraction at the 60%-30% interface was collected, diluted three times in Tris buffer, and pelleted by centrifugation (100,000 × g, 4°C, 1 h). The purified membrane fraction corresponding to 1.5 × 108 PAM was resuspended in 1 ml of Tris buffer and kept at −70°C. After the addition of Triton X-100 (0.1% final) and Tris-HCl (pH 7.4) (25 mM final), incubation for 1 h at 37°C, and centrifugation (13,000 × g, 4°C, 30 min), the supernatant containing the solubilized membrane fraction was incubated 16 h at 4°C with protein-G Sepharose (Pharmacia) that had been preincubated first with rabbit anti-mouse polyclonal serum (Dako A/S) and subsequently with MAb41D3.
Immunoprecipitates were washed four times with PBS-0.1% Tween 20 and once with water. A modified Laemmli buffer (15.5 mM Tris-HCl [pH 6.8], 1% sodium dodecyl sulfate [SDS], 1.25% 2-mercaptoethanol) was added to the immunoprecipitates before boiling. After a short spin, the supernatant was concentrated eightfold using a vacuum centrifuge (Jouan), 10% glycerol was added, and proteins were separated using SDS-7% polyacrylamide gel electrophoresis (PAGE) under reducing conditions. The 210-kDa band was excised from the gel. Three preparations of the p210 purified from 4 × 108 PAM in total were pooled and subjected to protein sequence analysis.
Protein sequence analysis.
An overnight in-gel tryptic digestion was performed on the 210-kDa protein band in 0.1 M Tris-HCl (pH 8.2) buffer containing 2 M urea. The resulting peptides were extracted once in 5 μl of 10% formic acid in water and four times in 5 μl of 0.1% formic acid and loaded on a C18 reversed-phase column (1 mm [inner diameter] by 100 mm [catalog no. 218TP5105]; Vydac). Peptides were eluted with a linear acetonitrile gradient (from 5% acetonitrile in 0.1% trifluoroacetic acid [TFA] to 90% acetonitrile in 0.1% TFA over 50 min) at a flow rate of 50 μl/min. Eluting peptides were collected automatically in 18 fractions of 50 μl each. Aliquots of 0.5 μl of each fraction were removed, mixed with 0.5 μl of matrix solution (3.2 mg of alpha-cyano-4-hydroxycinnamic acid and 0.8 mg of 2,5-hydroxy benzoic acid dissolved in 50% acetonitrile on 0.1% TFA), and analyzed by mass spectrometry (MS) (matrix-assisted laser desorption ionization-time of flight [TOF] MS) using a Bruker Reflex III mass spectrometer. Fractions containing peptides were pooled in three groups, lyophilized, and redissolved. Each of these peptide pools was loaded onto a capillary reversed-phase column (0.3 mm [inner diameter] by 250 mm; PepMap C18; LC-Packings) and eluted with an acetonitrile gradient in 0.05% (vol/vol) formic acid in water at a flow rate of 3 μl/min. The eluting peptides were detected at a wavelength of 214 nm and manually collected in fractions of 2 to 5 μl.
Aliquots of UV-absorbing fractions were loaded into metal nanospray probe tips (Micromass) and subjected to static nanospray electospray-TOF MS) (Q-TOF mass spectrometer running with Masslynx software package [version 3.2; Micromass]). From MS surveys, doubly or triply charged ions were selected and fragmentation was obtained by collision-induced decomposition, optimized by tuning the collision energy. The fragmentation spectra containing multiple ions were converted into spectra containing only singly charged ions by using MaxEnt3 (Micromass). The latter fragmentation spectra (nine) were subjected to SEQUEST-based analysis (version B22 [J. Eng and J. Yates]) using NRDB and EST databases. Three SEQUEST searches resulted in significant hits: one keratin and two immunoglobulins, which are common contaminations. Of the six remaining spectra, amino acid sequences were deduced manually and by aid of Pepseq (Micromass), making no distinction between isobaric amino acids leucine and isoleucine or lysine and glutamine. The deduced amino acids sequences were subjected to BLAST homology searches using the NRDB database.
Cloning and sequencing of the p210 cDNA.
DNA was prepared from pig whole blood (27) and served as the initial target for PCR experiments using nondegenerate primers based on the nucleotide sequence corresponding to the p210 peptides in mouse or in human sialoadhesins (EMBL accession numbers, Z36293 and AL109804, respectively). Two primers derived from the mouse sialoadhesin sequence and p210 peptides (forward, 5′-TCCTCAACTGCAGCCTCTGT-3′; reverse, 5′-AGTGAGGCAGCCGTTCCCTC-3′) amplified a 340-nucleotide fragment in porcine genomic DNA corresponding to the end of exon 14, an intron, and exon 15 in the mouse sialoadhesin gene. Specific porcine sialoadhesin primers were derived from this first sequence and used to screen a swine bacterial artificial chromosome (BAC) library (49) by PCR. One positive clone (BAC634C10) was used for further analysis. A major part of the sequence of the p210 gene (exons 4 to 9 and exons 11 to 15) was obtained from PCR products amplified using purified (Qiagen) genomic BAC DNA, primers chosen in conserved regions between the mouse and human gene homologs, and a Mastercycler Gradient PCR apparatus (Eppendorf) to allow a gradient of annealing temperatures. In addition, a 4-kb EcoRI/KpnI fragment of the BAC clone spanning exons 8 to 11 was identified by PCR, cloned and sequenced. The genomic sequence spanning exons 14 to 18 was also directly sequenced from the BAC clone. In parallel, reverse transcription (RT)-PCR was performed on total RNA extracted (Qiagen) from alveolar macrophages, and products covering exons 4 to 18 were sequenced. 5′ and 3′ rapid amplification of cDNA ends (RACE) was performed using the same RNA according to the manufacturer's instructions (Gibco BRL [RACE protocols for GC-rich cDNA]) to allow the determination of the sequence of the cDNA ends. For the 5′ RACE, the first-strand cDNA was synthesized using a reverse primer derived from exon 4 (EX4B4: 5′-TCTGGTCTTTGAGCTTCGTC-3′) and TdT tailed with dCTP. Second-strand synthesis was performed using supplied 5′ RACE abridged anchor primer and a nested primer derived from exon 4 (EX4B3: 5′-ACCTGAGGGTTGCTGCTATT-3′). A seminested PCR was performed using supplied abridged universal amplification primer and a nested primer also derived from exon 4 (EX4B1RACE: 5′-CACCTGGCAGCTGAGGGTGACCAGATC-3′). For the 3′ RACE, we used the supplied adapter primer for making the first-strand cDNA and a forward primer derived from exon 18 (EX18F1: 5′-GACGCCCACCATGACTGTTTTTG-3′) together with the supplied abridged universal amplification primer for the seminested PCR. Amplification products of 0.9 kb spanning a 5′ untranslated region of approximately 130 bp and of 2 kb spanning a 3′ untranslated region of approximately 1.45 kb were obtained, respectively, by 5′ and 3′ RACE. The complete coding sequence of porcine sialoadhesin, including 29 nucleotides of the 3′ untranslated region next to the stop codon, was amplified by RT-PCR starting from random primed cDNA and total alveolar macrophage RNA. For this RT-PCR, the ThermalAce polymerase (Invitrogen) was used in combination with forward primer 5′-CACCATGGACTTCCTGCTCCTGCTCCTC-3′ and reverse primer 5′-CTTGGGGTTTGAAGCTAGGTCATAA-3′. The amplified PCR product was cloned into the pcDNA3.1D/V5-HisTOPO (Invitrogen) to yield plasmid pcDNAp210, in which the recombinant sialoadhesin cDNA was expressed as a nonfusion protein under the control of the constitutive human cytomegalovirus promoter. The sequence of the cloned cDNA was verified by sequencing.
Cell transfections and infections.
PK-15 cells were transfected with pcDNAp210 using the CellPhect transfection kit (Amersham Pharmacia Biotech) and selected for Geneticin resistance (1 mg/ml). Three weeks after transfection, a population of Geneticin-resistant cells (rPK) was generated, and stocks of this population were used in this study. Cells were infected at a multiplicity of infection of 1 to 10 50% tissue culture infective doses/cell. Twenty hours postinoculation with the different PRRSV strains, cells were washed with PBS, fixed with methanol, incubated with a 1:20 dilution of antinucleocapsid MAb P3/27 (62) in PBS containing 10% heat-inactivated goat serum (PBS-G) for 1 h at 37°C, and washed with PBS before incubation with a 1:100 dilution of fluorescein isothiocyanate (FITC)-conjugated goat polyclonal anti-mouse immunoglobulins (Molecular Probes) in PBS-G for 1 h at 37°C. After washing with PBS, cells were incubated with a 1:30 dilution of biotinylated antisialoadhesin MAb41D3 followed, after washing, by a 1:100 dilution in PBS of Texas Red-labeled streptavidin (Molecular Probes) and mounted in a glycerin-PBS solution (0.9/0.1, vol/vol) with 2.5% 1,4-diazabicyclo(2.2.2)octane (DABCO; Janssen Chimica). Samples were observed with a DM IRBE inverted fluorescent microscope (Leica) using FITC filter L5 and Texas Red filter TX2. For the kinetics experiment, the rPK or Marc-145 cells were inoculated for 1 h with the PRRSV LV strain, washed with PBS, and further incubated at 37°C for different times before being fixed with methanol and stained for detecting the nucleocapsid as described above. Samples were examined with a TCS SP2 laser scanning spectral confocal system linked to a DM IRBE inverted microscope (Leica). An argon laser was used to excite FITC (488 nm) fluorochrome. Analysis of the images was performed with Leica confocal software.
Internalization assay.
PAM were incubated for 1 h at 37°C in the presence or absence of 100 μM amantadine (Sigma-Aldrich) with antisialoadhesin MAb41D3 or the irrelevant, isotype-matched MAb 13D12 (directed against glycoprotein gD of pseudorabies virus [41]) or macrophage-specific anti-CD14 MAb MIL-2 (53). The amantadine concentration was chosen based on previous studies (58). Cells were fixed in a 3% solution of paraformaldehyde in PBS for 10 min, washed with PBS, and permeabilized in a 0.1% saponin solution in PBS. As a control, cells were also fixed before the 1 h incubation with the MAbs and subsequently washed and permeabilized. Cells were incubated with FITC-conjugated goat polyclonal anti-mouse immunoglobulins (Molecular Probes) for 1 h at 37°C in the presence of 0.1% saponin. Cells were washed with PBS containing 0.1% saponin, once with PBS, mounted and analyzed by confocal microscopy. The number of cells showing internalized MAb was evaluated by examining z-sections through at least 100 cells for each condition.
Flow cytometric analysis of 41D3 endocytosis.
PAM were preincubated on ice for 30 min, then medium supplemented with purified MAb41D3 (20 μg/ml) was added and cells were further incubated for 1 h on ice. After washing, prewarmed medium was added and cells were further incubated at 37°C with 5% CO2. At different time points (0, 30, 60, 90, and 180 min) post-temperature shift, cells were washed with warm medium and the endocytosis process was stopped by adding ice-cold PBS and incubating the cells on ice. Cells were detached after addition of 50 mM EDTA and were centrifuged (460 × g, 6 min, 4°C), washed with ice-cold PBS, centrifuged, resuspended in a 1:100 dilution of FITC-labeled goat anti-mouse antibodies (Molecular Probes) in PBS, incubated for 1.5 h on ice, washed with ice-cold PBS, resuspended in 0.4 ml ice-cold PBS, and analyzed by flow cytometry (FACScalibur; Becton Dickinson). Forward-scattered light versus side-scattered light signals were used to identify macrophages, and FITC fluorescence intensity was determined for 5,000 cells per sample.
Double immunofluorescence staining of sialoadhesin and clathrin in macrophages.
PAM were incubated for 10 min at 37°C in medium with 20 μg/ml purified MAb41D3 prior to being washed and subsequently fixed in 3% paraformaldehyde in PBS. After washing, cells were permeabilized in methanol before being washed extensively with Tris-buffered saline supplemented with sucrose and goat serum (TBS-SG) (58) and incubated with a mixture of 1:100 FITC-labeled immunoglobulin G1 (IgG1)-specific rat anti-mouse antibodies (Serotec Ltd.) and 1:10 mouse anticlathrin heavy chain IgM antibodies (ICN Biomedicals Inc.) in PBS with 0.3% gelatin for 1 h at 37°C. Afterwards, cells were washed with TBS-SG and incubated with a 1:100 dilution of rat anti-mouse IgM-biotin (Serotec Ltd.) in PBS with 0.3% gelatin for 1 h at 37°C, washed with TBS-SG, and further incubated with a 1:50 dilution of Texas Red-X-labeled streptavidin (Molecular Probes) for 1 h at 37°C. Finally, cells were washed with TBS-SG, mounted, and analyzed by confocal microscopy.
Characterization of the porcine sialoadhesin protein.
Surface proteins of 7 × 106 PAM cells were biotinylated (Amersham). The cell pellet was lysed in 25 mM Tris (pH 7.4)-0.1% Triton X-100 and protease inhibitor cocktail (Complete; Boehringer Mannheim) for 1 h at 37°C. After centrifugation (13,000 × g, 4°C, 30 min), supernatant was added to protein G-Sepharose preincubated with MAb41D3 and incubated for 2 h at 20°C. Immunoprecipitates were washed four times with PBS-0.1% Tween 20 and once with water. Treatment (16 h at 37°C) with N-glycosidase F (Boehringer Mannheim) in PBS without predenaturation using 4 U of enzyme was subsequently performed. Western blotting was carried out as recommended (Amersham) except that the detection step was performed using 3,3′diaminobenzidine (Sigma).
Computer analysis.
Nucleotide and amino acid comparisons were performed using Blast (version 2; National Center for Biotechnology Information, National Institutes of Health, Bethesda, Md.) (2). Protein motif searches were carried out using the Genetics Computer Group (GCG) motifs software (16) that uses the Prosite Dictionary of Protein Sites and Patterns (A. Bairoch, University of Geneva, Geneva, Switzerland) and the HmmerPfam software run under GCG that uses the Pfam database (4, 22). The alignment of the amino acid sequences of the three sialoadhesins was obtained using the GCG PILEUP and PRETTY programs.
Nucleotide sequence accession number. The cDNA sequence of porcine sialoadhesin has been submitted to the GenBank database under accession number AF509585.
RESULTS
Identification of p210
The p210 protein was purified from PAM by immunoaffinity using MAb41D3 and SDS-PAGE. The amino acid sequence of six peptides obtained after tryptic digestion of the p210 protein was obtained using mass spectrometry (Table 1) and used to search protein databanks for similarities. Sequence identities ranging from 56 to 91% with mouse sialoadhesin (11) were found with peptides 1 to 4 (Table 1), indicating that the p210 is the porcine homolog of mouse sialoadhesin. No significant homology with any known proteins was observed for peptides 5 and 6.
TABLE 1.
Peptide masses and partial sequences deduced from the porcine p210 protein compared with the cDNA sequence from mouse sialoadhesina
| Peptide | Observed/calculated monoisotopic mass of the peptides | Sequence derived from MS MS | Homology with mouse sialoadhesin | Corresponding region in porcine cDNA-derived amino acid sequence |
|---|---|---|---|---|
| 1 | 1,587.87/1,587.84 | sesiPPAQLQLLHR | Yes | 626-639 |
| 2 | 1,499.83/1,499.83 | xxASSTAASVPxxxR | Yes | 1211-1225 |
| 3 | 1,790.96/1,790.89 | WLQEGSAASLSFsrpr | Yes | 1300-1315 |
| 4 | 1,281.61/1,281.60 | daVLSSFWDsR | Yes | 1348-1358 |
| 5 | 1,125.60/1,125.65 | ALLLGQVeqR | No | 87-96 |
| 6 | 1,462.78/1,462.74 | QATLTTLMDsqlgR | No | 985-998 |
Porcine sialoadhesin was purified by immunoaffinity from alveolar macrophages and subjected to internal peptide sequencing. Amino acids in capital letters are assigned with a high degree of confidence. Lowercase letters refer to tentative amino acid sequences.
Sequencing of the porcine sialoadhesin cDNA and characterization of the protein.
PCR amplification of parts of the porcine sialoadhesin gene was performed using porcine genomic DNA and primers that were based on the deduced nucleic acid sequence of the peptides and mouse sialoadhesin cDNA. Specific primers were subsequently derived from the obtained amplification products. These specific primers were then used for a PCR-based screening of a porcine bacterial artificial chromosome library representing a fivefold coverage of the swine haploid genome (49). Purified DNA from one positive BAC clone was used for direct cloning and sequencing and as target for PCR amplification of additional portions of the gene. In parallel, RT-PCR and 5′ and 3′ RACE were performed on PAM RNA, and amplification products were sequenced to determine the cDNA sequence of the complete sialoadhesin open reading frame. Unidentified peptides 5 and 6 (Table 1) were found in the translated 5,193-bp cDNA (Fig. 1), confirming the identity of the p210.
FIG.1.
Alignment of the amino acid sequence of porcine sialoadhesin with its homologous sequences in human and mouse (SWISS-PROT accession numbers, Q9BZZ2 and Q62230, respectively). The predicted hydrophobic leader peptides and transmembrane regions are underlined. The conserved cysteine residues characteristic of Siglecs are shaded. Residues that interact with sialic acid in mouse sialoadhesin are boxed in white in the consensus sequence. The six sequenced peptides are also boxed. Residues that are identical between the three proteins are indicated by stars and are given below in the consensus sequence. Gaps are indicated by a dot.
Comparison of the porcine sialoadhesin coding sequence with its mouse and human counterparts revealed 69 and 78% amino acid identities, respectively. It also showed characteristics of a type I transmembrane glycoprotein with a predicted N-terminal signal peptide of 16 amino acids, a large extracellular domain of 1,626 amino acids with 15 potential N-linked glycosylation sites, a transmembrane region (24 residues), and a cytoplasmic tail of 64 amino acids. The extracellular region of porcine sialoadhesin consisted of 17 immunoglobulin-like domains, as determined using the HmmerPfam software, like its mouse and human counterparts (11, 24). The structural basis for sialic acid binding by mouse sialoadhesin was resolved by X-ray crystallography (34) in conjunction with site-directed mutagenesis (59) and nuclear magnetic resonance analysis (12). A highly conserved arginine residue at position 116 forms a salt bridge with the carboxylate group of sialic acid, and two tryptophan residues at positions 21 and 125 make hydrophobic interactions with its N-acetyl and glycerol moieties. These three invariant residues important for sialic acid binding as well as the pattern of cysteine residues characteristic of the Siglec family were conserved in porcine sialoadhesin (Fig. 1). The cytoplasmic tail was poorly conserved and was 20 or 32 residues longer than its human or mouse counterparts, respectively. It contained five potential phosphorylation sites, one for cyclic AMP-dependent protein kinase at position 1686, two for casein kinase-2 at positions 1685 and 1694, and two for protein kinase C at positions 1685 and 1725.
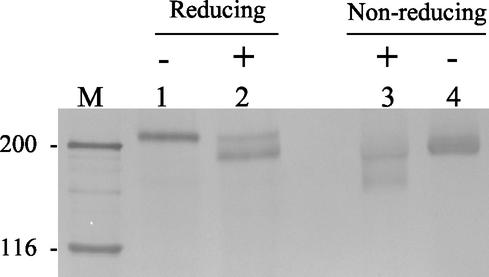
Further, we characterized the sialoadhesin protein present on the surface of PAM. These cells were surface labeled with biotin, and porcine sialoadhesin was immunoprecipitated using MAb41D3. After SDS-PAGE in reducing conditions, a band of approximately 210 kDa was observed (Fig. 2, lane 1) while under nonreducing conditions, a major diffuse band of approximately 180 kDa was detected (Fig. 2, lane 4). The difference in apparent molecular mass observed between the reduced and the nonreduced protein is consistent with the presence of multiple intramolecular disulfide bonds, as observed with rodent and human sialoadhesins (10, 24, 55). Treatment of the immunoprecipitated, nondenatured, porcine sialoadhesin with N-glycosidase F partially increased the mobility of the protein (Fig. 2, lanes 2 and 3), indicating that the p210 is modified with N-linked glycans, as expected from the amino acid sequence data. Complete digestion with N-glycosidase F was obtained when the immunoprecipitated protein was denatured before treatment (data not shown), indicating that local protein structure may protect some N-glycosidic linkages from the enzyme.
FIG. 2.
Porcine sialoadhesin immunoprecipitated from alveolar macrophages is modified with N-linked glycans. Alveolar macrophages were surface-labeled with biotin, and lysates were subjected to immunoprecipitation with MAb41D3. Immunoprecipitates were either treated with N-glycosidase F (lanes 2 and 3) or untreated (lanes 1 and 4) and run under reducing (lanes 1 and 2) or nonreducing (lanes 3 and 4) conditions on SDS-7% polyacrylamide gels. After transfer to polyvinylidene difluoride membrane, proteins on Western blots were detected with streptavidin-horseradish peroxidase, and this was followed by staining with 3,3′-diaminobenzidine as the substrate. M, apparent molecular mass markers (kilodaltons).
Porcine sialoadhesin mediates internalization of PRRSV into transfected cells.
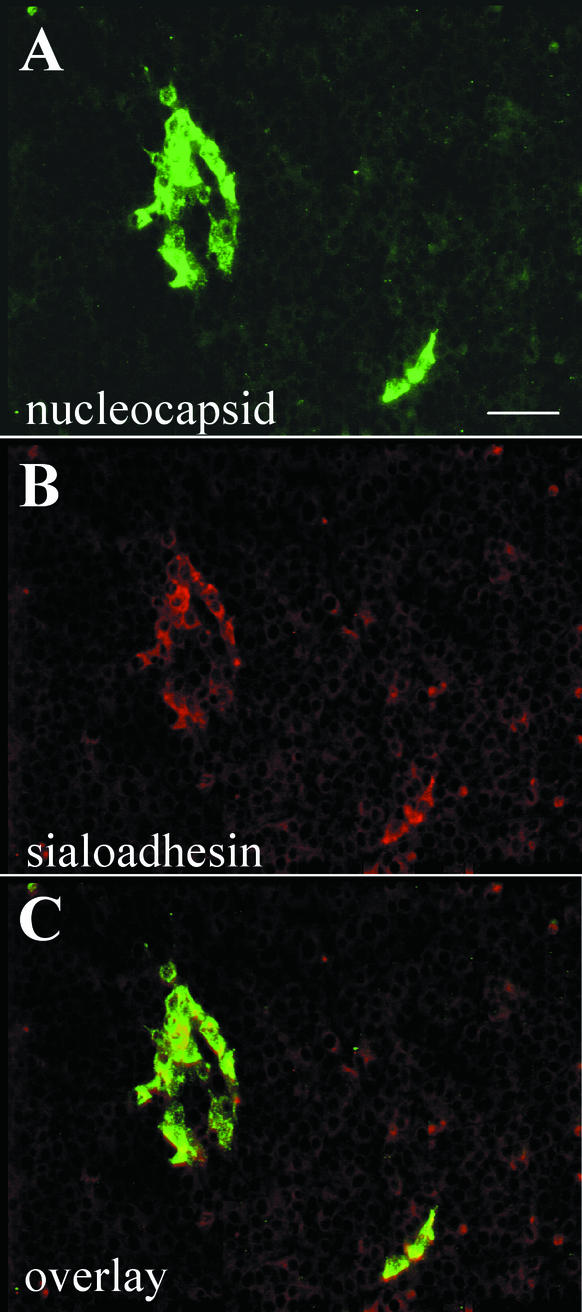
The full cDNA sequence of the porcine sialoadhesin coding sequence was amplified by RT-PCR using PAM RNA and cloned into an eukaryotic expression vector to yield plasmid pcDNAp210. Porcine (PK-15) cells, resistant to viral entry (31, 54), were transfected with the pcDNAp210 plasmid and selected for Geneticin resistance. Generation of a cell line originating from a single positive cell was not achieved, but instead, we used a noncloned, Geneticin-resistant cell population (designated rPK) in which about 6% of the cells expressed the recombinant p210 on the plasma membrane and in the cytoplasm, as determined by an indirect immunofluorescence assay using MAb41D3. The rPK and parental PK-15 cells were exposed to PRRSV (LV strain), and 20 h postinoculation, their permissivity to infection was assessed using double immunofluorescence staining. The PRRSV nucleocapsid protein was detected exclusively in rPK cells that also expressed the recombinant porcine sialoadhesin (Fig. 3). No staining was observed in the parental PK-15 cells. The rPK cells were inoculated with different strains of PRRSV, including the two Belgian isolates 94V360 and 96V198 and the American prototype strain VR2332, and colocalization of viral nucleocapsid antigen and porcine sialoadhesin in rPK cells was also observed with these strains (data not shown).
FIG. 3.
Colocalization of PRRSV nucleocapsid protein and recombinant porcine sialoadhesin in rPK cells. rPK cells were infected with PRRSV (LV strain), and 20 h postinoculation, cells were fixed and a double immunofluorescence staining was performed to detect the nucleocapsid protein (A) and the recombinant porcine sialoadhesin (B). (C) Overlay of panels A and B. Bar: 75 μm.
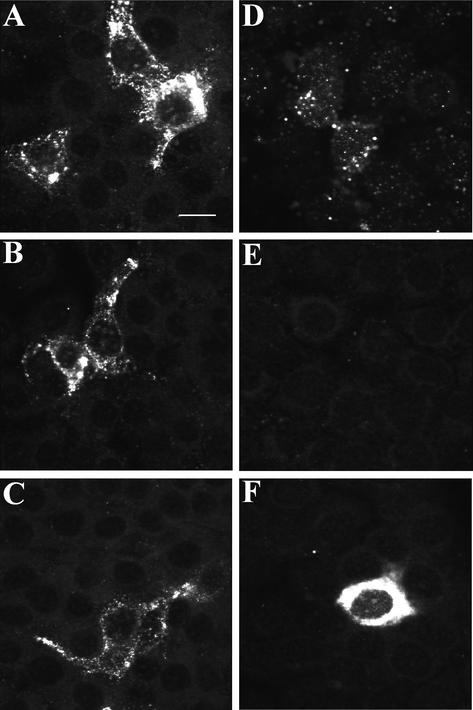
A time kinetic of infection of the rPK cells, followed by immunostaining and confocal microscopy, was performed. Unexpectedly, the cytoplasmic nucleocapsid staining observed 20 h postinoculation in the experiments described above was already detected 1 h postinoculation (Fig. 4A), suggesting it was not the result of transcription and translation of the viral nucleocapsid gene, which is only detectable starting at 6 h postinoculation in Marc-145 cells (63). The same nucleocapsid staining performed 1 h postinoculation in Marc-145 cells showed that it labeled viral particles, before and after internalization (Fig. 4D). The observed cytoplasmic staining in the rPK cells indicated that viral particles were internalized. At 3 h postinoculation, no nucleocapsid staining could be detected in Marc-145 cells (Fig. 4E), indicating that nucleocapsid disassembly, as a result of virus uncoating, had taken place. At 12 h postinoculation, an intense and diffuse cytoplasmic staining was observed in Marc-145 cells (Fig. 4F), indicating that the nucleocapsid protein was synthesized, as a result of transcription and translation of the viral genome. In contrast with Marc-145 cells, the staining patterns in the rPK cells 1, 3, and 12 h postinoculation showed no major differences (Fig. 4A to C). These results suggest that nucleocapsid dissasembly, as a result of virus uncoating, was not achieved after virus internalization in the rPK cells. Furthermore, after virus titration on Marc-145 cells, no significant increase in intracellular or extracellular virus titers was detected in the rPK 12 or 24 h postinoculation.
FIG. 4.
Detection of the viral nucleocapsid at different time points after inoculation of rPK (A, B, and C) and Marc-145 (D, E, and F) cells with PRRSV. (A and D) 1 h, (B and E) 3 h, and (C and F), 12 h postinoculation. After the selected time points, cells were fixed, stained for the nucleocapsid, and analyzed by confocal microscopy. Bar: 16 μm.
Porcine sialoadhesin mediates endocytosis of specific antibodies into PAM.
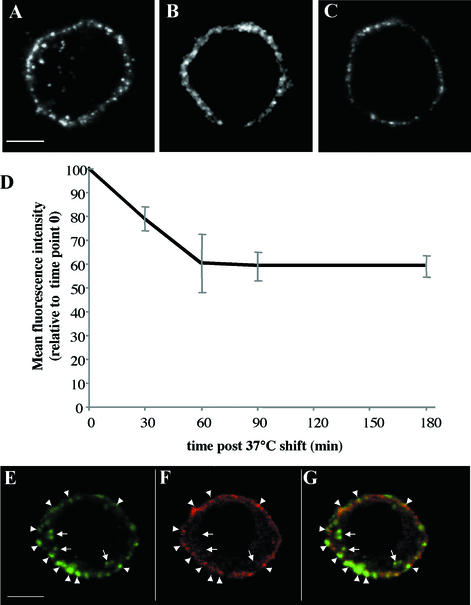
PRRSV enters into PAM through clathrin-dependent, receptor-mediated endocytosis (42). With the knowledge that porcine sialoadhesin mediated virus uptake, we investigated its involvement in the endocytosis process using monoclonal antibodies. PAM were incubated for 1 h at 37°C with MAb41D3 (recognizing porcine sialoadhesin) or with control MAbs 13D12 (isotype-matched, irrelevant MAb) or MIL-2 (anti-CD14, a macrophage-specific marker). After cell fixation, washes, and permeabilization, MAbs were detected using a FITC-conjugated polyclonal serum. PAM incubated with MAb41D3 showed membrane staining as well as bright fluorescent vesicles inside the cell, as determined by confocal microscopy (Fig. 5A). PAM incubated with MAb 13D12 displayed no staining (data not shown), while cells incubated with MAb MIL-2 showed membrane staining only (Fig. 5B). When PAM were fixed before the addition of MAb41D3, and subsequently washed and permeabilized, only membrane staining was detected (Fig. 5C). Addition of amantadine, a drug that inhibits clathrin-coated pit invagination at the plasma membrane (45, 46), reduced the percentage of PAM with internalized MAb41D3-labeled vesicles from 80.4 to 15.1%.
FIG. 5.
Porcine sialoadhesin mediates endocytosis of MAb41D3. Alveolar macrophages were incubated for 1 h at 37°C with MAb41D3 (A) or with control MAb MIL-2 anti-CD14 (B). After fixation, washings, and permeabilization, cells were incubated with FITC-conjugated goat polyclonal anti-mouse immunoglobulins and analyzed by confocal microscopy. (C) As a control, cells were fixed, incubated for 1 h at 37°C with MAb41D3, washed, permeabilized, and incubated with the same conjugated polyclonal antiserum as above. (D) Macrophages were incubated at 4°C with MAb41D3 before being shifted to 37°C to allow endocytosis. At different time points (0, 30, 60, 90, and 180 min), cells were shifted back to 4°C, stained with FITC-labeled goat anti-mouse antibodies, and analyzed by flow cytometry. Each value reflects the means ± standard deviation (error bars) of three experiments. (E to G) Macrophages were incubated with MAb41D3 and incubated at 37°C for 10 min before fixation and permeabilization. Cells were analyzed by confocal microscopy after performing a double immunofluorescence staining to detect porcine sialoadhesin (E) and clathrin (F). (G) Overlay of panels E and F. Symbols: Arrowheads, colocalization; arrows, no colocalization. Bar: 10 μm.
In a separate experiment, PAM were incubated at 4°C in the presence of MAb41D3, then transferred at 37°C to allow endocytosis of the antibodies, and again shifted to 4°C to block the process after different time periods. Unfixed cells were subsequently stained with FITC-labeled goat anti-mouse antibodies, and FITC fluorescence intensity was analyzed by flow cytometry. A reduction of 40% in the fluorescence intensity was observed 1 h after the 37°C shift (Fig. 5D), whereas no reduction could be observed when cells were paraformaldehyde fixed and permeabilized with 0.1% Triton prior to incubation with FITC-labeled goat anti-mouse antibodies (data not shown). Further, MAb41D3 patches at the plasma membrane were often associated with accumulation of clathrin (arrowheads, Fig. 5E). In internalized, MAb41D3-positive vesicles, clathrin was uncoated (arrows, Fig. 5F and G).
Together, these results demonstrate the involvement of porcine sialoadhesin in clathrin-mediated endocytosis.
DISCUSSION
Sialoadhesin (Siglec-1), the prototypic member of the Siglec family of sialic acid binding immunoglobulin-like lectins (P. R. Crocker et al., Letter, Glycobiology 8:v, 1998), is a macrophage-restricted receptor (8). In mice and humans, sialoadhesin is only expressed on discrete subsets of tissue macrophages that are found mostly in spleen, lymph node, bone marrow, liver, colon, and lung (9, 24). It is also expressed by inflammatory macrophages, such as those found in rheumatoid arthritis, and by tumor-infiltrating macrophages in breast cancer (24, 40). Although sialoadhesin is involved in cell-to-cell interactions as it binds to granulocytes, monocytes, natural killer cells, lymphocytes, and also breast cancer cells, its exact biological functions are poorly understood (10, 40). The experimental results presented here, together with previous data, demonstrate that porcine sialoadhesin is involved in the entry process of the PRRS arterivirus into porcine macrophages. This conclusion is based on the following observations. First, MAb41D3 that was shown here to be directed against porcine sialoadhesin, is able to abolish PRRSV infection of PAM (20, 21). Second, accumulation of viral particles in the cytoplasm of PK-15 cells was only observed in cells that also expressed recombinant porcine sialoadhesin. Both American and European strains of PRRSV, despite their extensive genetical divergence (1), were internalized in cells expressing recombinant sialoadhesin. Third, all PRRSV-infected cells observed in lungs, thymus, tonsils, spleen, and lymph nodes from pigs experimentally infected with PRRSV also expressed sialoadhesin, as they were recognized by MAb41D3 (20). PRRSV-nonpermissive peripheral blood mononuclear cells and peritoneal macrophages did not express sialoadhesin on their membranes, as shown by flow cytometry and fluorescence microscopy using MAb41D3 (20). Furthermore, we observed that porcine sialoadhesin may be involved in endocytosis since MAb41D3, upon binding with sialoadhesin, was specifically internalized into PAM. This process was inhibited by amantadine, a drug that blocks clathrin-coated pit invagination at the plasma membrane. In addition, clathrin accumulated with MAb41D3 patches at the plasma membrane during the internalization process. Altogether, these data show that porcine sialoadhesin is able to function as an endocytic receptor mediating clathrin dependent endocytosis. This receptor is thus likely to be used by PRRSV to enter into PAM, because PRRSV was shown to enter into cells by a process of receptor mediated endocytosis through clathrin-coated pits and vesicles (42). Interestingly, the ultrastructural localization of sialoadhesin in intracellular vesicles of rat spleens that were distinct from Golgi, lysosomes, or phagolysosomes led Schadee-Eestermans et al. (50) to propose its involvement in a receptor-mediated uptake process. This new function attributed to sialoadhesin is not in conflict with previous data describing sialoadhesin as a nonphagocytic receptor (8). Although both endocytosis and phagocytosis make use of several common protein components, these two cellular functions are clearly distinct and have different requirements (5, 14, 46). Further, porcine sialoadhesin shares high amino acid identity with its mouse (69%) and human (78%) counterparts but also exhibits unique features that could be crucial for mediating viral uptake. One such feature is a potential internalization signal, namely Phe1671-Tyr-Lys-Leu (32, 61) in its cytoplasmic tail, close to the transmembrane domain. Such peptides are recognized by the endocytic adapter complex AP-2 that is associated with clathrin, as a first step in the formation of clathrin-coated vesicles (reviewed in reference 29). The cytoplasmic tail of sialoadhesin is encoded by two short exons (39), exon 20 (24 residues) that shows 37 or 58% amino acid identity between pig and, respectively, mouse or human, and exon 21 that is not conserved, in size or sequence, in the three species. This lack of conservation might be resulting from alternative splicing events already described for the extracellular domain of mouse sialoadhesin (11). Although porcine sialoadhesin mediated virus internalization, other factors, absent in the PK-15 cell line, seem necessary to allow subsequent steps in the viral infection process.
Mouse and human sialoadhesins specifically recognize the terminal oligosaccharide sequence NeuAcα2-3Gal in N- and O-glycans and in glycolipids (10, 24). The sialomucin CD43 on T lymphocytes and mucin MUC1 on breast cancer cells have been identified as sialoadhesin-binding glycoproteins (40, 56). An interesting issue is now to determine how PRRSV binds to porcine sialoadhesin and if putative viral sialic acids are involved in this interaction or not. Although involvement of sialic acids is likely, one member of the Siglec family, Siglec 6, is known to bind a nonglycosylated protein ligand, i.e., leptin in addition to binding sialic acids (43). Four structural PRRSV proteins, namely, GP2, GP3, GP4, and GP5, are glycosylated (36, 37) and could possibly carry sialic acids.
Despite its sialic acid binding function, sialoadhesin in rodents and human has never been shown to mediate cellular entry of microorganisms with exposed sialic acids (13). Whether sialoadhesin is also involved in the entry process of other arteriviruses remains to be determined. Macrophages appear to be the primary target cell for all arteriviruses (47), with equine arteritis virus also targeting endothelial cells (17). Some indications suggest that sialoadhesin could also be involved in LDV entry. First, LDV-infected cells are observed in those mouse tissues that also possess the subsets of sialoadhesin-expressing macrophages. In the spleen and lymph nodes of mice, two tissues that are productively infected with LDV, infected cells are localized in a well-defined region (the marginal zone of both tissues and the paracortex of the lymph nodes) that also contains the sialoadhesin-expressing macrophages (3, 9, 44, 55). Further, both structural and molecular similarities suggest that the recently emerged PRRSV has evolved from LDV and further diverged independently on the European and American continents (48). In this case, it is likely that the entry mechanism used by PRRSV is derived from its ancestor virus.
The discovery of the involvement of porcine sialoadhesin in pathogenic (virus entry) and physiological (endocytosis) processes opens the way to new research, including the study of the possible endocytosis function of the sialoadhesin homologs in other mammals, the physiological role of sialoadhesin, and the potential role of this protein as an endocytic receptor for other microorganisms.
Acknowledgments
We thank C. Vanmaercke, V. Van Hoorde, D. Defever, and K. van der Heijden-Liefkens for excellent technical assistance. We also thank K. Haverson for supplying us with MAb MIL-2, G. M. Allan for the PK-15 cell line, P. Chardon for the BAC clone, and C. La Bonnardiere for recombinant porcine interferon. We are very grateful to L. Peelman and M. Van Poucke for their support for the screening of the porcine BAC library and to the Belgian EMBnet Node facility (http://ben.vub.ac.be) for access to updated sequence analysis software.
N. Vanderheijden was supported by the Belgian Ministry of Agriculture and Intervet (AkzoNobel), and P. L. Delputte was supported by a grant from the Flemish Institute for the Promotion of Innovation by Science and Technology (I.W.T.-Flanders).
REFERENCES
- 1.Allende, R., T. L. Lewis, Z. Lu, D. L. Rock, G. F. Kutish, A. Ali, A. R. Doster, and F. A. Osorio. 1999. North American and European porcine reproductive and respiratory syndrome viruses differ in non-structural protein coding regions. J. Gen. Virol. 80:307-315. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, G. W., R. R. Rowland, G. A. Palmer, C. Even, and P. G. Plagemann. 1995. Lactate dehydrogenase-elevating virus replication persists in liver, spleen, lymph node, and testis tissues and results in accumulation of viral RNA in germinal centers, concomitant with polyclonal activation of B cells. J. Virol. 69:5177-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booth, J. W., M.-K. Kim, A. Jankowski, A. D. Schreiber, and S. Grinstein. 2002. Contrasting requirements for ubiquitylation during Fc receptor-mediated endocytosis and phagocytosis. EMBO J. 21:251-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 7.Collins, J. E., D. A. Benfield, W. T. Christianson, L. Harris, J. C. Hennings, D. P. Shaw, S. M. Goyal, S. McCullough, R. B. Morrison, H. S. Joo, D. Gorcyca, and D. Chladek. 1992. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J. Vet. Diagn. Investig. 4:117-126. [DOI] [PubMed] [Google Scholar]
- 8.Crocker, P. R., and S. Gordon. 1986. Properties and distribution of a lectin-like hemagglutinin differentially expressed by murine stromal tissue macrophages. J. Exp. Med. 164:1862-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crocker, P. R., and S. Gordon. 1989. Mouse macrophage hemagglutinin (sheep erythrocyte receptor) with specificity for sialylated glycoconjugates characterized by a monoclonal antibody. J. Exp. Med. 169:1333-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crocker, P. R., S. Kelm, C. Dubois, B. Martin, A. S. McWilliam, D. M. Shotton, J. C. Paulson, and S. Gordon. 1991. Purification and properties of sialoadhesin, a sialic acid-binding receptor of murine tissue macrophages. EMBO J. 10:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crocker, P. R., S. Mucklow, V. Bouckson, A. S. McWilliam, A. C. Willis, S. Gordon, G. Milon, S. Kelm, and P. Bradfield. 1994. Sialoadhesin, a macrophage sialic acid binding receptor for haemopoietic cells with 17 immunoglobulin-like domains. EMBO J. 13:4490-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crocker, P. R., M. Vinson, S. Kelm, and K. Drickamer. 1999. Molecular analysis of sialoadhesin-sialoside interactions by NMR and site-directed mutagenesis. Biochem J. 341:355-361. [PMC free article] [PubMed] [Google Scholar]
- 13.Crocker, P. R., and A. Varki. 2001. Siglecs, sialic acids and innate immunity. Trends Immunol. 22:337-342. [DOI] [PubMed] [Google Scholar]
- 14.Davis, W., P. T. Harrison, M. J. Hutchinson, and J. M. Allen. 1995. Two distinct regions of FcγRI initiate separate signalling pathways involved in endocytosis and phagocytosis. EMBO J. 14:432-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delputte, P. L., N. Vanderheijden, H. J. Nauwynck, and M. B. Pensaert. 2002. Involvement of the matrix protein in the attachment of porcine reproductive and respiratory syndrome virus to a heparinlike receptor on porcine alveolar macrophages. J. Virol. 76:4312-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Vries, A. A., P. J. Rottier, A. L. Glaser, and M. C. Horzinek. 1996. Equine viral arteritis, p. 171-200. In M. J. Studdert (ed.), Virus infections of equines. Elsevier Science, Amsterdam, The Netherlands.
- 18.Duan, X., H. J. Nauwynck, and M. B. Pensaert. 1997. Virus quantification and identification of cellular targets in the lungs and lymphoid tissues of pigs at different time intervals after inoculation with porcine reproductive and respiratory syndrome virus (PRRSV). Vet. Microbiol. 56:9-19. [DOI] [PubMed] [Google Scholar]
- 19.Duan, X., H. Nauwynck, and M. Pensaert. 1997. Effects of origin and state of differentiation and activation of monocytes/macrophages on their susceptibility to PRRSV. Arch. Virol. 142:2483-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan, X., H. Nauwynck, H. Favoreel, and M. Pensaert. 1998. Identification of a putative receptor for porcine reproductive and respiratory syndrome virus on porcine alveolar macrophages. J. Virol. 72:4520-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan, X., H. Nauwynck, H. Favoreel, and M. Pensaert. 1998. Porcine reproductive and respiratory syndrome virus infection of alveolar macrophages can be blocked by monoclonal antibodies against cell surface antigens. Adv. Exp. Med. Biol. 440:81-88. [DOI] [PubMed] [Google Scholar]
- 22.Eddy, S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755-763. [DOI] [PubMed] [Google Scholar]
- 23.Even, C., and P. G. Plagemann. 1995. Pseudotype virions formed between mouse hepatitis virus and lactate dehydrogenase-elevating virus (LDV) mediate LDV replication in cells resistant to infection by LDV virions. J. Virol. 69:4237-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartnell, A., J. Steel, H. Turley, M. Jones, D. Jackson, and P. R. Crocker. 2001. Characterization of human sialoadhesin, a sialic acid binding receptor expressed by resident and inflammatory macrophage populations. Blood 97:288-296. [DOI] [PubMed] [Google Scholar]
- 25.Inada, T., H. Kikuchi, and S. Yamazaki. 1993. Comparison of the ability of lactate dehydrogenase-elevating virus and its virion RNA to infect murine leukemia virus-infected or -uninfected cell lines. J. Virol. 67:5698-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jusa, E., Y. Inaba, M. Kouno, and O. Hirose. 1997. Effect of heparin on infection of cells by porcine reproductive and respiratory syndrome virus. Am. J. Vet. Res. 58:488-491. [PubMed] [Google Scholar]
- 27.Kawasaki, E. S. 1990. Sample preparation from blood, cells, and other fluids, p. 146-152. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols. A guide to methods and applications. Academic Press, San Diego, Calif.
- 28.Kim, H. S., J. Kwang, I. J. Yoon, H. S. Joo, and M. L. Frey. 1993. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in homogeneous subpopulation of MA-104 cell line. Arch. Virol. 133:477-483. [DOI] [PubMed] [Google Scholar]
- 29.Kirchhausen, T., J. S. Bonifacino, and H. Riezman. 1997. Linking cargo to vesicle formation: receptor tail interactions with coat proteins. Curr. Opin. Cell Biol. 4:488-495. [DOI] [PubMed] [Google Scholar]
- 30.Kreutz, L. C., and M. R. Ackermann. 1996. Porcine reproductive and respiratory syndrome virus enters cells through a low pH-dependent endocytic pathway. Virus Res. 42:137-147. [DOI] [PubMed] [Google Scholar]
- 31.Kreutz, L. C. 1998. Cellular membrane factors are the major determinants of porcine reproductive and respiratory syndrome virus tropism. Virus Res. 53:121-128. [DOI] [PubMed] [Google Scholar]
- 32.Ktistakis, N. T., D. Thomas, and M. G. Roth. 1990. Characteristics of the tyrosine recognition signal for internalization of transmembrane surface glycoproteins. J. Cell Biol. 111:1393-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lefevre, F., R. L'Haridon, F. Borras-Cuesta, and C. La Bonnardiere. 1990. Production, purification and biological properties of an Escherichia coli-derived recombinant porcine alpha interferon. J. Gen. Virol. 71:1057-1063. [DOI] [PubMed] [Google Scholar]
- 34.May, A. P., R. C. Robinson, M. Vinson, P. R. Crocker, and E. Y. Jones. 1998. Crystal structure of the N-terminal domain of sialoadhesin in complex with 3′ sialyllactose at 1.85 A resolution. Mol. Cell 1:719-728. [DOI] [PubMed] [Google Scholar]
- 35.Meulenberg, J. J., M. M. Hulst, E. J. De Meijer, P. L. Moonen, A. Den Besten, E. P. De Kluyver, G. Wensvoort, and R. J. Moormann. 1993. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology 192:62-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meulenberg, J. J., A. Petersen-den Besten, E. P. De Kluyver, R. J. Moormann, W. M. Schaaper, and G. Wensvoort. 1995. Characterization of proteins encoded by ORFs2. to 7 of Lelystad virus. Virology 206:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meulenberg, J. J., and A. P. Den Besten. 1996. Identification and characterization of a sixth structural protein of Lelystad virus: the glycoprotein GP2 encoded by ORF2 is incorporated in virus particles. Virology 225:44-51. [DOI] [PubMed] [Google Scholar]
- 38.Meulenberg, J., J. Bos-de Ruijter, R. van de Graaf, G. Wensvoort, and R. Moormann. 1998. Infectious transcripts from cloned genome-length cDNA of porcine reproductive and respiratory syndrome virus. J. Virol. 72:380-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mucklow, S., S. Gordon, and P. R. Crocker. 1997. Characterization of the mouse sialoadhesin gene, SN. Mamm. Genome 8:934-937. [DOI] [PubMed] [Google Scholar]
- 40.Nath, D., A. Hartnell, L. Happerfield, D. W. Miles, J. Burchell, J. Taylor-Papadimitriou, and P. R. Crocker. 1999. Macrophage-tumour cell interactions: identification of MUC1 on breast cancer cells as a potential counter-receptor for the macrophage-restricted receptor, sialoadhesin. Immunology 98:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nauwynck, H. J., and M. B. Pensaert. 1995. Effect of specific antibodies on the cell-associated spread of pseudorabiesvirus in monolayers of different cell types. Arch. Virol. 140:1137-1146. [DOI] [PubMed] [Google Scholar]
- 42.Nauwynck, H. J., X. Duan, H. W. Favoreel, P. Van Oostveldt, and M. B. Pensaert. 1999. Entry of porcine reproductive and respiratory syndrome virus into porcine alveolar macrophages via receptor-mediated endocytosis. J. Gen. Virol. 80:297-305. [DOI] [PubMed] [Google Scholar]
- 43.Patel, N., E. C. Brinkman-Van der Linden, S. W. Altmann, K. Gish, S. Balasubramanian, J. C. Timans, D. Peterson, M. P. Bell, J. F. Bazan, A. Varki, and R. A. Kastelein. 1999. OB-BP1/Siglec-6. A leptin-and sialic acid-binding protein of the immunoglobulin superfamily. J. Biol. Chem. 274:22729-22738. [DOI] [PubMed] [Google Scholar]
- 44.Perry, V. H., P. R. Crocker, and S. Gordon. 1992. The blood-brain barrier regulates the expression of a macrophage sialic acid-binding receptor on microglia. J. Cell Sci. 101:201-207. [DOI] [PubMed] [Google Scholar]
- 45.Phonphok, Y., and K. S. Rosenthal. 1991. Stabilization of clathrin coated vesicles by amantadine, tromantadine and other hydrophobic amines. FEBS Lett. 281:188-190. [DOI] [PubMed] [Google Scholar]
- 46.Perry, D. G., G. L. Daugherty, and W. J. Martin. 1999. Clathrin-coated pit-associated proteins are required for alveolar macrophage phagocytosis. J. Immunol. 162:380-386. [PubMed] [Google Scholar]
- 47.Plagemann, P. G. 1996. Lactate dehydrogenase-elevating virus and related viruses, p. 1105-1120. In B. N. Fields, D. N. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 48.Plagemann, P. G., Z. Chen, and K. Li. 1999. Polylactosaminoglycan chains on the ectodomain of the primary envelope glycoprotein of an arterivirus determine its neuropathogenicity, sensitivity to antibody neutralization and immunogenicity of the neutralization epitope. Curr. Top. Virol. 1:27-43. [DOI] [PubMed] [Google Scholar]
- 49.Rogel-Gaillard, C., N. Bourgeaux, A. Billault, M. Vaiman, and P. Chardon. 1999. Construction of a swine BAC library: application to the characterization and mapping of porcine type C endoviral elements. Cytogenet. Cell. Genet. 85:205-211. [DOI] [PubMed] [Google Scholar]
- 50.Schadee-Eestermans, I. L., E. C. Hoefsmit, M. van de Ende, P. R. Crocker, T. K. van den Berg, and C. D. Dijkstra. 2000. Ultrastructural localisation of sialoadhesin (Siglec-1) on macrophages in rodent lymphoid tissues. Immunobiology 202:309-325. [DOI] [PubMed] [Google Scholar]
- 51.Sur, J. H., V. L. Cooper, J. A. Galeota, R. A. Hesse, A. R. Doster, and F. A. Osorio. 1996. In vivo detection of porcine reproductive and respiratory syndrome virus RNA by in situ hybridization at different times postinfection. J. Clin. Microbiol. 34:2280-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sur, J. H., A. R. Doster, J. S. Christian, J. A. Galeota, R. W. Wills, J. J. Zimmerman, and F. A. Osorio. 1997. Porcine reproductive and respiratory syndrome virus replicates in testicular germ cells, alters spermatogenesis, and induces germ cell death by apoptosis. J. Virol. 71:9170-9179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thacker, E., A. Summerfield, K. McCullough, A. Ezquerra, J. Dominguez, F. Alonso, J. Lunney, J. Sinkora, and K. Haverson. 2001. Summary of workshop findings for porcine myelomonocytic markers. Vet. Immunol. Immunopathol. 80:93-109. [DOI] [PubMed] [Google Scholar]
- 54.Therrien, D., Y. St-Pierre, and S. Dea. 2000. Preliminary characterization of protein binding factor for porcine reproductive and respiratory syndrome virus on the surface of permissive and non-permissive cells. Arch. Virol. 145:1099-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van den Berg, T. K., J. J. Brevé, J. G. Damoiseaux, E. A. Döpp, S. Kelm, P. R. Crocker, C. D. Dijkstra, and G. Kraal. 1992. Sialoadhesin on macrophages: its identification as a lymphocyte adhesion molecule. J. Exp. Med. 176:647-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van den Berg, T. K., D. Nath, H. J. Ziltener, D. Vestweber, M. Fukuda, I. van Die, and P. R. Crocker. 2001. Cutting edge: CD43 functions as a T cell counterreceptor for the macrophage adhesion receptor sialoadhesin (Siglec-1). J. Immunol. 166:3637-3640. [DOI] [PubMed] [Google Scholar]
- 57.Vanderheijden, N., P. Delputte, H. Nauwynck, and M. Pensaert. 2001. Effects of heparin on the entry of porcine reproductive and respiratory syndrome virus into alveolar macrophages. Adv. Exp. Med. Biol. 494:683-689. [DOI] [PubMed] [Google Scholar]
- 58.Van de Walle, G. R., H. W. Favoreel, H. J. Nauwynck, P. Van Oostveldt, and M. B. Pensaert. 2001. Involvement of cellular cytoskeleton components in antibody-induced internalization of viral glycoproteins in pseudorabies virus-infected monocytes. Virology 288:129-138. [DOI] [PubMed] [Google Scholar]
- 59.Vinson, M., P. A. van der Merwe, S. Kelm, A. May, E. Y. Jones, and P. R. Crocker. 1996. Characterization of the sialic acid-binding site in sialoadhesin by site-directed mutagenesis. J. Biol. Chem. 271:9267-9272. [DOI] [PubMed] [Google Scholar]
- 60.Voicu, I. L., A. Silim, M. Morin, and M. A. Elazhary. 1994. Interaction of porcine reproductive and respiratory syndrome virus with swine monocytes. Vet. Rec. 134:422-423. [DOI] [PubMed] [Google Scholar]
- 61.Weixel, K. M., and N. A. Bradbury. 2000. The carboxyl terminus of the cystic fibrosis transmembrane conductance regulator binds to AP-2 clathrin adaptors. J. Biol. Chem. 275:3655-3660. [DOI] [PubMed] [Google Scholar]
- 62.Wieczorek-Krohmer, M., F. Weiland, K. Conzelmann, D. Kohl, N. Visser, P. Van Woensel, H. J. Thiel, and E. Weiland. 1996. Porcine reproductive and respiratory syndrome virus (PRRSV): monoclonal antibodies detect common epitopes on two viral proteins of European and U.S. isolates. Vet. Microbiol. 51:257-266. [DOI] [PubMed] [Google Scholar]
- 63.Wootton, S. K., R. R. Rowland, and D. Yoo. 2002. Phosphorylation of the porcine reproductive and respiratory syndrome virus nucleocapsid protein. J. Virol. 76:10569-10576. [DOI] [PMC free article] [PubMed] [Google Scholar]