Abstract
A final step in retrovirus assembly, particle release from the cell, is modulated by a small motif in the Gag protein known as a late domain. Recently, human immunodeficiency virus type 1 (HIV-1) and Moloney murine leukemia virus (M-MuLV) were shown to require components of the cellular vacuolar protein sorting (VPS) machinery for efficient viral release. HIV-1 interacts with the VPS pathway via an association of HIV-1 Gag with TSG101, a component of the cellular complexes involved in VPS. Equine infectious anemia virus (EIAV) is unique among enveloped viruses studied to date because it utilizes a novel motif, YPDL in Gag, as a late domain. Our analysis of EIAV assembly demonstrates that EIAV Gag release is blocked by inhibition of the VPS pathway. However, in contrast to HIV-1, EIAV Gag release is insensitive to TSG101 depletion and EIAV particles do not contain significant levels of TSG101. Finally, we demonstrate that fusing EIAV Gag directly with another cellular component of the VPS machinery, VPS28, can restore efficient release of an EIAV Gag late-domain mutant. These results provide evidence that retroviruses can interact with the cellular VPS machinery in several different ways to accomplish particle release.
The Gag polyprotein drives assembly and budding of retroviruses (reviewed in reference 45). Expression of viral Gag protein in the absence of any other viral protein or the viral genome results in the efficient formation and release of virus-like particles (VLPs) in a number of divergent cell types (13, 18, 50). For many retroviruses, including human immunodeficiency virus type 1 (HIV-1), Moloney murine leukemia virus (M-MuLV), and equine infectious anemia virus (EIAV), Gag assembles into spherical membrane-encapsulated particles at the plasma membrane (45). During the final stage of budding, a membrane fission event is required for efficient separation of the newly formed retrovirus from the cell. Concurrent with budding, the Gag polyprotein is processed by retroviral protease into matrix, capsid, nucleocapsid, and other virus-specific Gag-derived proteins.
Discrete regions within the Gag polyprotein mediate its ability to bind membrane, multimerize, and induce separation of nascent virus particles from the cell (45). This final separation event in retroviral egress is modulated by a motif within Gag commonly referred to as the late domain. Mutations within viral late domains lead to dramatic reductions in virion-associated Gag release (15, 19, 38, 50). Late-domain mutants characteristically accumulate as electron-dense particles at the plasma membrane that appear to be blocked from release at a very late stage, presumably at the final membrane fission event. Interestingly, late domains seem to function in a relatively context-independent manner as demonstrated by two important properties. First, late domains retain activity even when moved to atypical locations within Gag (26, 32). Second, late-domain motifs from heterologous viruses are able to functionally replace one another (26, 32).
Late-domain sequences have been identified in retroviruses, rhabdoviruses, and filoviruses (20, 21, 45). To date, three motifs that can function as late domains for viral release have been identified: PTAP (22), PPXY (49, 51), and YPDL (38). More recently, host proteins known to be involved in cellular membrane trafficking have been documented that interact with each of these motifs. The PTAP, PPXY, and YPDL motifs interact with TSG101 (17, 47), Nedd4-like ubiquitin ligases (20, 25), and adaptor protein 2 (AP-2), respectively (39). The best-characterized late-domain interaction is that of the PTAP motif in the p6 region of HIV-1 Gag with cellular TSG101 (17, 47). TSG101 is a component of ESCRT-1 (endosomal sorting complex required for transport), a 350-kDa cellular complex essential in the vacuolar protein sorting (VPS) pathway, which traffics proteins to the multivesicular body (MVB) and lysosome (6, 24). Small inhibitory RNA (siRNA)-mediated TSG101 depletion potently blocks HIV-1 release (17). Moreover, overexpression of the dominant-negative (dn) form of an ESCRT-1 recycling factor, VPS4, inhibits particle release of HIV-1 as well as the PPPY late-domain-encoding MuLV (17). Thus, VPS machinery is involved in the budding of both PTAP- and PPPY-encoding retroviruses.
EIAV is unique among retroviruses studied to date in that it utilizes a YPDL sequence as its late domain (38). The EIAV late domain resembles the well-characterized YXXφ motif that is recognized by the adaptor protein complexes, AP-1 and AP-2 (7, 11, 27). EIAV Gag has been reported to interact with the endocytic adaptor protein AP-2 in a late-domain-dependent manner (39); however, the functional significance of this interaction has not been examined.
There is no obvious link between AP-2 and cellular VPS factors, nor has a role for cellular factors apart from AP-2 been described for EIAV release. In this report we utilize a dnVPS4 mutant to demonstrate that EIAV Gag particle release requires a functional VPS pathway. Additionally, targeting of an EIAV late-domain mutant to the VPS pathway by fusion to an ESCRT-1 complex protein, VPS28, restores efficient budding. Finally, we demonstrate that EIAV release is insensitive to TSG101 depletion, suggesting that EIAV normally enters the VPS pathway downstream of the ESCRT-1 complex.
MATERIALS AND METHODS
Plasmids and siRNA.
Plasmids expressing the wild-type (wt) and L26A EIAV Gag were provided by Ronald Montelaro. To improve expression, EIAV gag was subcloned into the pCI expression vector containing a cytomegalovirus promoter and the hepatitis B virus (HBV) posttranscriptional regulatory element (PRE) (23). The HIV-1 gag expression vector, pCI HIV Gag, was a gift from Ted Pierson (8). Human VPS28 was obtained by PCR amplification from a human embryonic kidney cDNA library with primers that flank the coding region. EIAV Gag with a carboxyl-terminal fusion to human VPS28 was made by appending an XbaI site in frame at the 3′ end of the Gag coding region by standard PCR techniques. Similarly an XbaI site was added to the 5′ end of the VPS28 coding region. This fusion construct was cloned into the pCI expression vector. A similar strategy was used to construct EIAV Gag with carboxyl-terminal fusion to yellow fluorescent protein (YFP) by using YFP sequences derived from pEYFP-N1 (Clontech). The VPS4 open reading frame was amplified by PCR from a human embryonic kidney cDNA library (Gibco Life Technologies) and cloned into the mammalian expression plasmid TOPO pcDNA3.1 His/V5 (Invitrogen) upstream of the epitope tag. VPS4 K173A was made by Quickchange mutagenesis (Clontech). The pCMV hemagglutinin (HA) TSG101 expression vector was made by cloning of amplified TSG101 (from a kidney cDNA library) into pCMV HA (Clontech) downstream of an HA tag. All constructs made by PCR were confirmed by sequencing.
To perform siRNA-mediated TSG101 inhibition, 21-nucleotide RNA duplexes corresponding to TSG101 (nucleotides 413 to 433) were purchased from Dharmacon Inc. (sense, 5′ CCU CCA GUC UUC UCU CGU CTT; antisense, 5′ GAC GAG AGA AGA CUG GAG GTT). siRNA was resuspended in RNase-free water at 0.13 μg/μl and freeze-thawed a maximum of five times.
Cell lines and transfections.
293T cells were employed in all experiments shown except for the confocal fluorescence microscopy, where HeLa cells were used. 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% bovine calf serum and penicillin-streptomycin. HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and penicillin-streptomycin. Transfections that included siRNA were performed using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer's instructions. Lipofectamine 2000 transfections were carried out in six-well plates with 2.0 μg of each expression plasmid. Empty plasmid vector DNA was added to bring the total to 4.5 μg DNA per well. In experiments where siRNA was used, 0.2 μl (26 ng) of siRNA was added to each transfection mixture.
VLP release assays.
To examine Gag VLP release, 293T cells were transfected with the designated Gag expression plasmid and any other indicated plasmid or siRNA. Forty-eight hours posttransfection, culture medium was collected, clarified by filtration through a 0.45-μm-pore-size filter, loaded over a 20% sucrose cushion in phosphate-buffered saline buffer (0.14 M NaCl, 2.7 mM KCl, 12 mM sodium phosphate, pH 7.4), and centrifuged at 40,000 rpm in an SW41 rotor (Beckman) for 60 min. Pelleted VLPs were resuspended in Triton lysis buffer (1% Triton X-100, 150 mM NaCl, 50 mM Tris [pH 8.0], 5 mM EDTA). Cell lysates were prepared with Triton lysis buffer. Samples were prepared for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting with primary antibodies specific for EIAV Gag, HIV-1 Gag, or green fluorescent protein (GFP) (Covance, Inc.) and horseradish peroxidase-conjugated secondary antibodies (Sigma). Blots were developed using Super Signal West Pico (Pierce Biotechnology) and visualized by film or with a LAS-1000Plus luminescent image analysis system (FujiFilm). Ronald Montelaro and Mike Malim kindly provided anti-EIAV Gag rabbit polyclonal and anti-HIV-1 Gag monoclonal antibodies, respectively.
Fluorescence microscopy.
EIAV Gag-YFP was examined by confocal (University of Pennsylvania Morphology Core) and nonconfocal fluorescence microscopy as indicated to determine protein localization. For localization studies, HeLa cells were transfected as described above. At 24 h posttransfection cells were split onto coverslips, and at 48 h posttransfection cells were paraformaldehyde fixed, mounted, and examined for Gag-YFP localization.
RESULTS
Expression of EIAV Gag late-domain mutants and chimeric proteins.
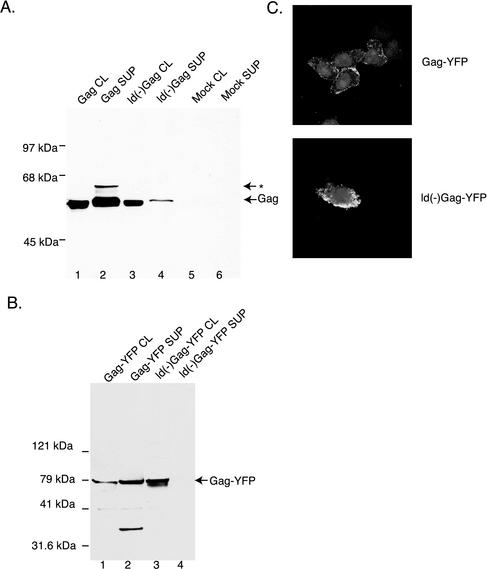
Expression and release of wt and a late-domain mutant EIAV Gag were examined by transient expression in 293T cells by using vectors encoding wt EIAV Gag (Gag) or a Gag late-domain mutant [ld(−)Gag] that altered the previously identified EIAV late-domain sequence YPDL to YPDA. Preliminary experiments revealed that EIAV Gag expression was markedly improved by including the HBV PRE (23) in the Gag transcript (data not shown). With the use of constructs containing the HBV PRE both wt and ld(−)Gag are expressed well in cell lysates (Fig. 1A, lanes 1 and 3). Analysis of the transfected cell supernatant demonstrated dramatically reduced levels of released ld(−)Gag particles compared to wt (Fig. 1A, lanes 2 and 4). These data are consistent with previous findings that EIAV Gag expression in the absence of other viral components is sufficient for the efficient production of VLPs and that efficient release occurs in a YXXL-dependent manner (38).
FIG. 1.
EIAV Gag VLP production and cellular localization. (A and B) Cell lysates and culture supernatants from transfected 293T cells were subjected to SDS-PAGE and Western blot analysis with anti-EIAV Gag (A) and anti-GFP (B) primary antibodies. Arrows denote Gag products. The asterisk denotes higher-molecular-mass species that frequently cross-react with antibody. Molecular masses are shown to the left of the blots, and lane numbers are shown below. (C) Confocal microscopy of Gag-YFP fusion-transfected HeLa cells. Both panels represent single optical sections. The membrane-localized fluorescence signal was similarly seen in unfixed cells.
To facilitate localization studies of EIAV Gag, a YFP carboxyl-terminal fusion with Gag was constructed. Upon transient expression in 293T cells Gag-YFP was well expressed and exhibited a particle release phenotype similar to that with Gag alone (Fig. 1B). As was observed with EIAV Gag, Gag-YFP particle release was blocked by the late-domain mutation (Fig. 1B). Fluorescence microscopy of cells expressing Gag-YFP showed punctate staining that was localized at the periphery of the cell at or near the plasma membrane (Fig. 1C). A similar fluorescence pattern was seen for the wt and ld(−) fusion proteins (Fig. 1C), suggesting that the L26A mutation does not cause a gross mislocalization of Gag. Because the localization and budding of Gag-YFP were similar to those of wt EIAV Gag, the chimeric proteins were used in the subsequent analysis of particle release requirements.
Effect of VPS inhibition on EIAV release.
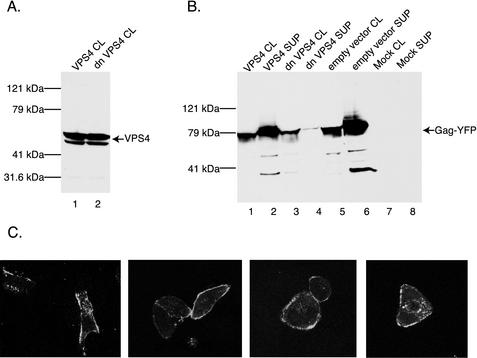
Recent evidence demonstrates that the cellular VPS pathway is required for release of virions that utilize PPPY or PTAP late-domain motifs (17, 28). Mutants affecting the cellular VPS pathway were used to examine whether EIAV Gag, which utilizes a YPDL late-domain motif (38), requires a functional VPS pathway for efficient particle release. To inhibit the VPS machinery, we used a dn mutant of VPS4, K173A, that is unable to bind ATP and has been shown to potently inhibit the VPS pathway presumably by blocking VPS4-mediated recycling of VPS complexes (3, 4). Upon transient transfection of 293T cells, wt VPS4 and dnVPS4 were expressed at similar levels (Fig. 2A, compare lanes 1 and 2). Analysis of cell lysates revealed that coexpression with Gag-YFP resulted in a modest decrease in the Gag-YFP expression levels with dnVPS4 but not with wt VPS4 (Fig. 2B, compare lanes 1 and 3). In contrast, release of Gag-YFP VLPs into the supernatant was dramatically inhibited by dnVPS4 but was unaffected by wt VPS4 (Fig. 2B, compare lanes 2 and 4). To exclude the possibility that the dnVPS4 virion release block was due to intracellular Gag protein mislocalization, cotransfected cells were examined by fluorescence microscopy. Gag-YFP localization was indistinguishable in cells expressing dnVPS4, wt VPS4, or empty vector and displayed a punctate, plasma membrane-localized pattern (Fig. 2C). These results demonstrate that inhibition of the VPS pathway via expression of dnVPS4 induces a potent block to EIAV Gag release. Furthermore, the fluorescence microscopy results suggest that the dnVPS4-induced particle release block is specific to a late event in EIAV Gag assembly and budding.
FIG. 2.
Effect of dnVPS4 K173A on EIAV Gag budding. (A) 293T cells were transfected with wt or dnVPS4 constructs and subsequently analyzed by SDS-PAGE and immunoblotting with an anti-V5 primary antibody. Equal volumes of cell lysates are loaded in both lanes. (B) 293T cells were transfected with 20 μg of indicated Gag construct and 10 μg of indicated VPS4 construct or empty vector. Cell lysates and culture supernatants were separated by SDS-PAGE and analyzed by immunoblotting with a mouse anti-GFP primary antibody. The arrow denotes Gag fusion proteins. Lane numbers and size markers are shown below and to the left of the blot, respectively. (C) Confocal microscopy of HeLa cells transfected with the indicated Gag and VPS4 constructs. Panels represent a single optical section of cells. Similar staining was visualized in unfixed cells also.
Effect of TSG101 inhibition on EIAV release.
HIV-1 Gag recruits an ESCRT-1 complex protein, TSG101, via a PTAP motif in the p6 region and possibly through ubiquitin conjugated to Gag which may interact with a ubiquitin interaction motif in TSG101 (17). HIV-1 is hypothesized to bind to TSG101 as a mechanism to enter the VPS pathway (17, 28). EIAV Gag lacks a PTAP motif but, like HIV-1 and many other retroviral Gag polyproteins, is modified by ubiquitin (31). It is unknown if EIAV Gag binds TSG101 or if TSG101 is functionally required during EIAV particle release.
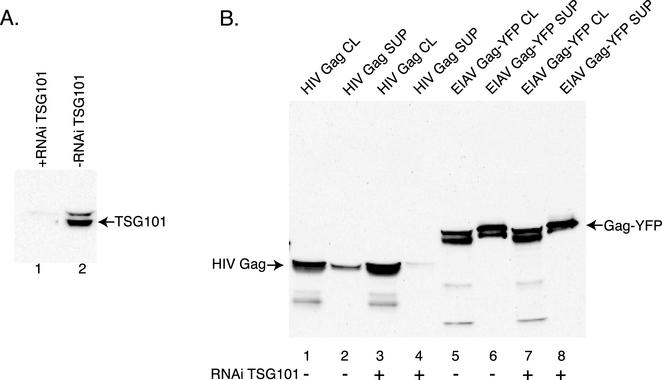
To examine this issue, we utilized siRNA to deplete TSG101 and determined the effect on EIAV Gag-YFP release. siRNA activity was examined by cotransfection of 293T cells with an HA epitope-tagged TSG101 and an siRNA specific for TSG101. Western blot analysis of cell lysates demonstrated that TSG101-HA expression was reduced to undetectable levels upon cotransfection of the siRNA (Fig. 3A, compare lanes 1 and 2). Analysis with a TSG101-specific antibody consistently demonstrated that the endogenous protein is also significantly depleted in the transfected cells with the low remaining amount of TSG101 most likely due to untransfected cells that did not receive siRNA (data not shown). To study the effect of TSG101 depletion on EIAV Gag release, 293T cells were cotransfected with the TSG101 siRNA and Gag-YFP, and then 48 h posttransfection cell lysates and supernatants were analyzed for EIAV Gag-YFP. As seen in Fig. 3B, TSG101 depletion had no significant effect on either Gag-YFP expression in cells or EIAV Gag release into the supernatant (compare lanes 5 and 6 to lanes 7 and 8). In a parallel control experiment, TSG101 depletion dramatically diminished HIV-1 Gag release without affecting intracellular expression (Fig. 3B, compare lanes 2 and 4). Thus, while EIAV Gag-YFP budding requires VPS recycling as demonstrated by dnVPS4, the ESCRT-1 complex protein TSG101 does not appear to be required by EIAV Gag for efficient particle release.
FIG. 3.
TSG101 inhibition does not affect EIAV Gag VLP production. (A) 293T cells were transfected with the TSG101 construct in the presence (lane 1) or absence (lane 2) of siRNA against TSG101. TSG101 expression in transfected cell lysates was assessed by SDS-PAGE and subsequent Western blot analysis with an anti-HA primary antibody. The arrow denotes TSG101 cross-reacting bands. (B) 293T cells were transfected with either HIV-1 Gag (lanes 1 to 4) or EIAV Gag (lanes 5 to 8) in the presence or absence of siRNA against TSG101 (denoted by a plus or minus sign below each lane marker). Cell lysates and culture supernatants were analyzed for HIV-1 and EIAV Gag content by SDS-PAGE and subsequent Western blotting with anti-HIV-1 and anti-GFP antibodies, respectively. Lanes are indicated below the blot.
Targeting of EIAV Gag to the VPS pathway overcomes a late-domain defect.
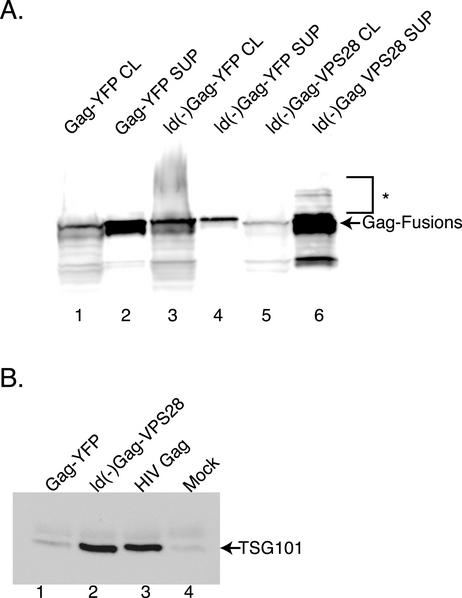
Our data support a role for the VPS pathway in normal EIAV assembly; however, the link between the EIAV Gag YPDL late-domain motif and the VPS machinery is unclear. To directly test whether recruitment of VPS proteins to the site of particle release is sufficient to mediate viral budding, we employed a chimeric Gag-VPS28 protein. Human VPS28 is a poorly characterized 28-kDa cytoplasmic protein that is a component of the ESCRT-1 complex along with TSG101 and VPS37 (6, 24). The VPS28 chimeras contained a late-domain mutant form of Gag. Analysis of cell lysates and supernatants from cells transiently expressing ld(−)Gag-VPS28 showed that the protein is expressed well and the ld(−)Gag-VPS28 particles are efficiently released (Fig. 4). As a control, release of ld(−)Gag-YFP was analyzed. Significant levels of ld(−)Gag-YFP were not detected in the supernatant (Fig. 4, compare lanes 3 and 4 to lanes 5 and 6). However, budding of ld(−)Gag-YFP could be rescued by coexpression of ld(−)Gag-VPS28 (data not shown). In addition, fusion of TSG101 to ld(−)EIAV Gag also resulted in release of particles into the supernatant (data not shown). The ability of a VPS28 or TSG101 fusion to rescue the EIAV Gag late-domain defect demonstrates that EIAV Gag can efficiently utilize the host VPS machinery for release and suggests that retroviral particles are capable of functionally recruiting cellular budding machinery by different mechanisms.
FIG. 4.
VPS28 fusion rescues budding phenotype in a late-domain mutant and incorporates endogenous TSG101 into particles. 293T cells were transfected with 20 μg of the indicated constructs and separately Western blotted with an anti-EIAV Gag (A) or anti-TSG101 (B) antibody. The arrow denotes EIAV fusion protein (A) or endogenous TSG101 (B) with lane numbers noted below. The bracket marked with an asterisk denotes higher-molecular-mass species that cross-react with the anti-EIAV Gag antibody.
Gag-VPS28 recruits TSG101 into particles.
VPS28 normally binds tightly to TSG101 (17, 37), suggesting that budded particles arising from expression of the chimeric ld(−)Gag-VPS28 might incorporate TSG101. To investigate this possibility, EIAV ld(−)Gag-VPS28, EIAV Gag-YFP, and HIV-1 Gag particles were examined by Western blotting for endogenous TSG101 incorporation. As shown in Fig. 4B, ld(−)Gag-VPS28 recruits significant amounts of TSG101 into particles. By comparison, HIV-1 Gag VLPs also contain significant amounts of TSG101 (Fig. 4B, lane 3). In contrast, analysis of VLPs derived from wt EIAV Gag-YFP demonstrates that the level of endogenous TSG101 in these particles was not significantly higher than background (compare lanes 1 and 4, Fig. 4B). This finding suggests that EIAV Gag does not specifically interact with TSG101 during budding or that the interaction is relative unstable. In addition, these findings support the TSG101 RNA inhibition data demonstrating that EIAV Gag is released by a TSG101-independent mechanism.
DISCUSSION
The cellular VPS pathway normally sorts endosomal membrane proteins to a late endosomal compartment, the MVB, for transport to the lysosome and degradation. After trafficking to the late endosome, membrane proteins are sorted from the limiting membrane of the endosome into membrane vesicles within the lumen of the organelle. Vesicles, and their included membrane proteins, are released into the lumen to create MVBs by a membrane fission event that is topologically analogous to viral budding. Sorting of proteins into the region of the endosomal membrane from which the vesicles derive and vesicular budding require specific cis-acting signals. In yeast and mammalian cells monoubiquitination is a key signal for VPS-mediated sorting. A currently accepted model hypothesizes that cellular VPS proteins, which normally carry out this membrane fission reaction or assemble factors that catalyze the reaction, are recruited to the site of viral particle release through interaction with signals encoded in Gag late domains where these cellular factors mediate the late stages of viral budding (17, 28, 37).
EIAV is unique among retroviruses studied to date in that it utilizes a YXXL motif in Gag to direct the late stage of viral release and not a PTAP or PPPY late-domain motif commonly found either alone or in tandem in other retroviral Gag proteins. Although different late-domain motifs are used by these retroviruses, based on the data presented here for EIAV Gag and previously for M-MuLV and HIV-1, it appears that all of the identified late domains require cellular VPS machinery for efficient particle release (17). Here we utilized a dn form of VPS4 to demonstrate that the YXXL late domain in EIAV Gag p9 requires an intact VPS pathway for efficient budding. Moreover, we have shown that the dnVPS4-mediated inhibition of EIAV Gag release is specific to a late viral budding event (Fig. 2C).
Distinct retroviral late domains recruit VPS factors in different ways. PTAP late domains bind to the VPS ESCRT-1 complex protein TSG101 (17, 47). PPPY-encoding Gag polyproteins enter the VPS pathway in a less defined manner. Gag polyproteins utilizing the PPPY late-domain motif have been shown to interact with the WW domain-containing Nedd4 family of ubiquitin ligases (25). Nedd4 and Nedd4-like proteins are implicated in the ubiquitination of plasma membrane proteins destined for lysosomal degradation via the MVB (14, 43). Monoubiquitination of target proteins appears to be a prerequisite for their entry into the VPS pathway (35). Indeed, numerous VPS proteins (including TSG101) contain ubiquitin binding domains (35, 36).
EIAV Gag, like all other retroviral Gag proteins studied to date, is modified by ubiquitin (31, 33). The relationship between Gag and ubiquitin remains to be fully elucidated. Gag polyproteins from a number of retroviruses are modified by ubiquitin (30, 34, 40, 48). Furthermore, investigators have reported that release of retroviruses utilizing either the PPPY (i.e., Rous sarcoma virus and MuLV) or PTAP (HIV-1) late domain are potently blocked by agents that inhibit ubiquitin modification (34, 42, 44). Interestingly, EIAV Gag release is relatively resistant to ubiquitin-blocking agents (31, 33). The reason for this disparity has not been determined but may be due to a region of EIAV Gag with homology to ubiquitin and which has been speculated to enable direct binding of EIAV Gag to ubiquitin binding factors (33).
The mechanism of EIAV Gag recruitment of VPS factors remains to be determined. However, data indicating that EIAV Gag does not require ubiquitination or TSG101 for particle egress suggest that Gag enters the VPS pathway at a point distinct from Gag polyproteins that utilize PTAP or PPPY late domains. EIAV Gag has been shown to bind to AP-2 via the YDPL motif that constitutes its late domain (39); however, it remains unclear if AP-2 is functionally required for EIAV budding. Unlike TSG101 or Nedd4, AP-2 has not yet been implicated as a player in the VPS pathway, and thus, it is unclear how AP-2 binding could promote entry into the VPS pathway. Further research into EIAV budding will likely reveal new information regarding associations between known VPS factors and other cellular proteins and trafficking pathways.
In yeast, protein sorting onto MVB vesicles requires the sequential action of a series of protein complexes called ESCRT-1, ESCRT-2, and ESCRT-3 (1, 2, 24). VPS23, the TSG101 ortholog, is an essential component, along with VPS28 and VPS37, of the ESCRT-1 complex (24). Unlike HIV-1, M-MuLV and EIAV Gag particle release is insensitive to TSG101 depletion, implying that the late domains in these retroviruses function independently of TSG101. The ability of EIAV and M-MuLV Gag to bud in a TSG101-independent manner may suggest that these Gag polyproteins are able to bind other ESCRT-1 components. However, this hypothesis would require either that TSG101 is not essential for ESCRT-1 function or, alternately, that another protein may substitute for TSG101 in the ESCRT-1 complex to facilitate M-MuLV and EIAV virion budding. Another hypothesis for the observed lack of TSG101 dependence is that EIAV accesses the VPS pathway downstream of ESCRT-1 possibly via ESCRT-2, ESCRT-3, or other factors. Our data demonstrating efficient release of EIAV ld(−)Gag-VPS28 suggest that entry into the VPS pathway is flexible and that viruses may therefore utilize numerous mechanisms to usurp the VPS machinery.
One possibility is Eps15, an adaptor complex binding protein that is implicated in trafficking of ubiquitinated proteins to the MVB (35). Another alternative is Hrs, an endosomal adaptor protein required for epidermal growth factor receptor sorting to the MVB (46) and a mammalian ortholog of the yeast class E protein VPS27p. Hrs associates with a range of adaptor proteins including Eps15 (reviewed in reference 10) and may contain a ubiquitin interaction motif (35). An enticing feature of invoking Eps15 and/or Hrs involvement in viral budding is their physical proximity to other known sorting proteins (ESCRT) and their potential ability to bind ubiquitinated cargo. The complete set of factors involved in MVB formation in mammalian cells is not clearly elucidated. Our results with a chimeric EIAV Gag suggest a unique method of identifying factors involved in MVB formation in higher eukaryotes. Although EIAV does not normally require the MVB ESCRT-1 complex component TSG101 for efficient virus budding, our data demonstrate that fusion to another protein from this complex, VPS28, can rescue budding of a late-domain-defective EIAV Gag. Previously it was shown that fusion of Gag to ubiquitin, a signal for sorting to the MVB, also restores efficient viral budding to a late-domain mutant (34). The work herein extends this finding to demonstrate that proteins of the VPS pathway can be appended to Gag and function as de facto late domains. Together these results imply that fusion to a late-domain-defective Gag may be a general method for confirming the identity of proteins that are thought to be involved in MVB formation and for mapping functional domains within these proteins. Additionally, our data demonstrating coincorporation of TSG101 into the chimeric VPS28-Gag particles indicate that this strategy might be used to identify novel MVB factors by employing viral budding to enrich and purify those proteins tightly associated with the chimeric protein. For example, the yeast ESCRT-1 complex contains three tightly associated proteins, TSG101, VPS28, and VPS37. However, the mammalian ortholog of VPS37 remains unidentified. Comparison of the proteins in chimeric VPS28-Gag VLPs with those in wt Gag VLPs may help to identify mammalian VPS37 and other proteins specifically associated with VPS28.
A number of retroviral envelope proteins contain a highly conserved YXXφ motif in their cytoplasmic tail (5, 16, 29, 41) that, like the EIAV Gag late-domain motif, is thought to interact with AP-2. Although several studies have demonstrated that for some retroviruses these motifs can function to direct endocytosis, in other viral glycoproteins the function of this motif was not clear (29, 41), and the role that these cytoplasmic tail motifs play in viral replication is vague. One intriguing possibility is that, similarly to the YPDL motif in EIAV Gag, these motifs in retroviral envelope proteins serve as signals that help to recruit the cellular machinery required for viral budding or to help concentrate the viral envelope proteins at sites of budding. In this regard, it is interesting that the effect of mutation in the EIAV Gag p9 YPDL motif is partially overcome when the complete viral genome is expressed (9), perhaps suggesting that the EIAV envelope protein is compensating for the defective assembly signal in Gag.
Nonretroviral enveloped viruses such as rhabdoviruses and filoviruses also encode PTAP and PPPY late-domain motifs (12, 20). It thus seems reasonable to presume that these viruses also recruit VPS factors during particle release. Indeed, TSG101 has been shown to be required for Ebola virus VP40 release (28). Thus, highly divergent enveloped viruses may utilize a common membrane fission mechanism during particle release. A conserved enveloped virus release mechanism is likely an excellent target for antiviral drugs. A further understanding of the mechanisms whereby viruses recruit these cellular factors should aid greatly in development of this class of therapeutics.
Acknowledgments
G.O.T. and A.J.P. contributed equally to this work.
We thank Graham Simmons for providing comments on the manuscript and Feng Li, Ronald Montelaro, Michael Malim, Patty Sanchez, Bridget Puffer, and Ted Pierson for providing reagents and technical assistance.
G.O.T. and A.J.P. were supported in part by NIH training grants T32-GM07229 and T32-NS07180, respectively.
REFERENCES
- 1.Babst, M., D. J. Katzmann, E. J. Estepa-Sabal, T. Meerloo, and S. D. Emr. 2002. ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell 3:271-282. [DOI] [PubMed] [Google Scholar]
- 2.Babst, M., D. J. Katzmann, W. B. Snyder, B. Wendland, and S. D. Emr. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3:283-289. [DOI] [PubMed] [Google Scholar]
- 3.Babst, M., T. K. Sato, L. M. Banta, and S. D. Emr. 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16:1820-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berlioz-Torrent, C., B. L. Shacklett, L. Erdtmann, L. Delamarre, I. Bouchaert, P. Sonigo, M. C. Dokhelar, and R. Benarous. 1999. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J. Virol. 73:1350-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop, N., and P. Woodman. 2001. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J. Biol. Chem. 276:11735-11742. [DOI] [PubMed] [Google Scholar]
- 7.Brett, T. J., L. M. Traub, and D. H. Fremont. 2002. Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure (Cambridge) 10:797-809. [DOI] [PubMed] [Google Scholar]
- 8.Buck, C. B., X. Shen, M. A. Egan, T. C. Pierson, C. M. Walker, and R. F. Siliciano. 2001. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 75:181-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C., F. Li, and R. C. Montelaro. 2001. Functional roles of equine infectious anemia virus Gag p9 in viral budding and infection. J. Virol. 75:9762-9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clague, M. J., and S. Urbe. 2001. The interface of receptor trafficking and signalling. J. Cell Sci. 114:3075-3081. [DOI] [PubMed] [Google Scholar]
- 11.Collins, B. M., A. J. McCoy, H. M. Kent, P. R. Evans, and D. J. Owen. 2002. Molecular architecture and functional model of the endocytic AP2 complex. Cell 109:523-535. [DOI] [PubMed] [Google Scholar]
- 12.Craven, R. C., R. N. Harty, J. Paragas, P. Palese, and J. W. Wills. 1999. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 73:3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delchambre, M., D. Gheysen, D. Thines, C. Thiriart, E. Jacobs, E. Verdin, M. Horth, A. Burny, and F. Bex. 1989. The GAG precursor of simian immunodeficiency virus assembles into virus-like particles. EMBO J. 8:2653-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunn, R., and L. Hicke. 2001. Multiple roles for Rsp5p-dependent ubiquitination at the internalization step of endocytosis. J. Biol. Chem. 276:25974-25981. [DOI] [PubMed] [Google Scholar]
- 15.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, F., S. G. Morrison, D. L. Robertson, C. L. Thornton, S. Craig, G. Karlsson, J. Sodroski, M. Morgado, B. Galvao-Castro, H. von Briesen, S. Beddows, J. Weber, P. M. Sharp, G. M. Shaw, B. H. Hahn, and the WHO and NIAID Networks for HIV Isolation and Characterization. 1996. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. J. Virol. 70:1651-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 18.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. De Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 19.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, Z. M., and T. S. Yen. 1995. Role of the hepatitis B virus posttranscriptional regulatory element in export of intronless transcripts. Mol. Cell. Biol. 15:3864-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katzmann, D. J., M. Babst, and S. D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145-155. [DOI] [PubMed] [Google Scholar]
- 25.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, F., C. Chen, B. A. Puffer, and R. C. Montelaro. 2002. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J. Virol. 76:1569-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marsh, M., and H. T. McMahon. 1999. The structural era of endocytosis. Science 285:215-220. [DOI] [PubMed] [Google Scholar]
- 28.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 29.Ochsenbauer, C., S. R. Dubay, and E. Hunter. 2000. The Rous sarcoma virus Env glycoprotein contains a highly conserved motif homologous to tyrosine-based endocytosis signals and displays an unusual internalization phenotype. Mol. Cell. Biol. 20:249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder II, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ott, D. E., L. V. Coren, R. C. Sowder II, J. Adams, K. Nagashima, and U. Schubert. 2002. Equine infectious anemia virus and the ubiquitin-proteasome system. J. Virol. 76:3038-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patnaik, A., V. Chau, F. Li, R. C. Montelaro, and J. W. Wills. 2002. Budding of equine infectious anemia virus is insensitive to proteasome inhibitors. J. Virol. 76:2641-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polo, S., S. Sigismund, M. Faretta, M. Guidi, M. R. Capua, G. Bossi, H. Chen, P. De Camilli, and P. P. Di Fiore. 2002. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416:451-455. [DOI] [PubMed] [Google Scholar]
- 36.Pornillos, O., S. L. Alam, D. R. Davis, and W. I. Sundquist. 2002. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 9:812-817. [DOI] [PubMed] [Google Scholar]
- 37.Pornillos, O., J. E. Garrus, and W. I. Sundquist. 2002. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 12:569-579. [DOI] [PubMed] [Google Scholar]
- 38.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puffer, B. A., S. C. Watkins, and R. C. Montelaro. 1998. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J. Virol. 72:10218-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Putterman, D., R. B. Pepinsky, and V. M. Vogt. 1990. Ubiquitin in avian leukosis virus particles. Virology 176:633-637. [DOI] [PubMed] [Google Scholar]
- 41.Sauter, M. M., A. Pelchen-Matthews, R. Bron, M. Marsh, C. C. LaBranche, P. J. Vance, J. Romano, B. S. Haggarty, T. K. Hart, W. M. Lee, and J. A. Hoxie. 1996. An internalization signal in the simian immunodeficiency virus transmembrane protein cytoplasmic domain modulates expression of envelope glycoproteins on the cell surface. J. Cell Biol. 132:795-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staub, O., S. Dho, P. Henry, J. Correa, T. Ishikawa, J. McGlade, and D. Rotin. 1996. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 15:2371-2380. [PMC free article] [PubMed] [Google Scholar]
- 44.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 46.Urbe, S., I. G. Mills, H. Stenmark, N. Kitamura, and M. J. Clague. 2000. Endosomal localization and receptor dynamics determine tyrosine phosphorylation of hepatocyte growth factor-regulated tyrosine kinase substrate. Mol. Cell. Biol. 20:7685-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogt, V. M. 2000. Ubiquitin in retrovirus assembly: actor or bystander? Proc. Natl. Acad. Sci. USA 97:12945-12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]