Abstract
Alphavirus core assembly proceeds along an assembly pathway involving a dimeric assembly intermediate. Several regions of the alphavirus capsid protein have been implicated in promoting and stabilizing this dimerization, including a putative heptad repeat sequence named helix I. This sequence, which spans residues 38 to 55 of the Sindbis virus capsid protein, was implicated in stabilizing dimeric contacts initiated through the C-terminal two-thirds of the capsid protein and nucleic acid. The studies presented here demonstrate that helix I can be functionally replaced by the corresponding sequence of a related alphavirus, western equine encephalitis virus, and also by an unrelated sequence from the yeast transcription activator, GCN4, that was previously shown to form a dimeric coiled coil. Replacing helix I with the entire leucine zipper domain of GCN4 (residues 250 to 281) produced a virus with the wild-type phenotype as determined by plaque assay and one-step growth analysis. However, replacement of helix I with a GCN4 sequence that favored trimer formation produced a virus that exhibited ∼40-fold reduction in virus replication compared to the wild-type Sindbis virus. Changing residues within the Sindbis virus helix I sequence to favor trimer formation also produced a virus with reduced replication. Peptides corresponding to helix I inhibited core-like particle assembly in vitro. On the basis of these studies, it is proposed that helix I favors capsid protein-capsid protein interactions through the formation of dimeric coiled-coil interactions and may stabilize assembly intermediates in the alphavirus nucleocapsid core assembly pathway.
The assembly of an icosahedral virus capsid is an accurate and strictly controlled process (2, 14). The subunits of the virus particle are arranged in an equivalent or quasi-equivalent manner to produce a shell that encapsidates the viral genome in order to transport it into other cells. The principles behind the assembly of proteins into these highly ordered structures like icosahedral virus particles have been investigated for several decades. These studies have focused on elucidating the various aspects of biological control that exist at different stages of virus assembly and the mechanism by which mishaps and errors in the assembly process are circumvented.
In the alphaviruses, the capsid protein (CP) is the major determinant of an accurately assembled nucleocapsid that is then enveloped by a glycoprotein shell to produce an icosahedrally symmetric virus particle (8, 36, 42). Molecular genetic, biochemical, and structural analyses of the CP have identified determinants important for the function of the CP in nucleocapsid assembly (39). However, a majority of these studies have focused on the C-terminal domain of the CP (residues 106 to 264) for which the atomic structure has been solved (3). The remaining N-terminal residues have been predicted to have a random coil with the exception of residues 38 to 55. This sequence (referred to as helix I) is comprised of 18 amino acids and has a heptad repeat organization of leucine residues predicted to form a coiled coil (32). It was predicted that this region of the CP also played an important role in nucleocapsid assembly (38).
Helix I is flanked by positively charged, low-complexity sequences. The residues within helix I are well conserved and predominantly hydrophobic, with three leucine residues, L38, L45, and L52 (Sindbis virus [SINV] numbering) forming two complete heptad repeats and an N-terminally truncated heptad. Two of these leucines (L45 and L52) are completely conserved among all alphaviruses, while L38 is conserved among the New World alphaviruses. A helical wheel representation of the residues within this region places leucines exclusively at the d position of the heptads and places β-branched residues at the a position of the heptads. This distribution of amino acids is similar to the hydrophobic core residues found in proteins known to form dimeric parallel coiled coils.
Molecular genetic analyses of helix I have previously shown that complete or partial deletion of helix I caused a significant decrease in virus replication (32). Furthermore, these mutant viruses displayed an increased sensitivity to elevated temperatures compared to the wild-type virus. The mutants also failed to accumulate stable nucleocapsid cores (NCs) in the cytoplasm of infected cells, although protein translation and processing remained unaffected. Viruses with even single site substitutions at the conserved leucine residues had similar defective phenotypes. Analysis of these mutants in in vitro assembly studies indicated that they were defective in core-like particle (CLP) assembly. When SINV CP(19-264)(L52D), which contains residues 19 to 264 of the SINV CP and contains a mutation at residue 52 in helix I that renders it assembly defective, was cross-linked in the presence of nucleic acid, it was able to form a CP dimer, which could be purified and assembled into a CLP (38). However, when cross-linking was reduced, the CP dimers in the CLPs dissociated into monomeric CPs and caused the CLPs to fall apart. A similar phenotype was observed for all the mutants with substitutions within helix I and for N-terminally truncated CP missing the first 80 residues (38). It had been previously shown that the regions flanking helix I were not important for NC assembly but that they played a role in nonspecific neutralization of the negative charge on the nucleic acid (6-8, 32). Helix I, however, was shown to have a prominent role in NC assembly and was proposed to function as a stability factor during CP oligomerization. Furthermore, it was suggested that this occurred through coiled-coil interactions mediated through its leucine zipper-like region.
In this study, it is shown that helix I can be functionally replaced by the corresponding sequence from a related alphavirus, western equine encephalitis virus (WEEV), and by sequences from several unrelated, well-characterized, leucine zipper motifs. It is also demonstrated that helix I has a propensity for sequences that tend to form dimers rather than trimers. This observation supports the previous model that alphavirus assembly follows a dimer-dependent assembly pathway (32, 38).
MATERIALS AND METHODS
Viruses and cells.
All viruses were grown at 37°C in BHK-15 cells propagated in Eagle minimal essential medium (MEM) (GIBCO BRL) supplemented with 10% fetal bovine serum, unless otherwise noted.
Construction of chimeric viruses and mutants. (i) SINV/GCN4(250-281).
Oligonucleotide-directed mutagenesis was used to replace the helix I region and flanking residues (nucleotides [nt] 7748 to 7817) in the SINV CP with the dimerization domain of the yeast transcriptional activator, GCN4. The pJH370 plasmid containing the dimerization domain of GCN4 (13) was used as a template to amplify the GCN4 leucine zipper sequence (corresponding to amino acids 250 to 281 of the dimerization domain of GCN4) using oligonucleotide primers 5′-GCGTGGGTGCCGGCACATATGAAA-3′ and 3′-CACTTGCATGCGCAAGGCCGCG-5′. The resulting 128-bp PCR product (includes nt 501 to 601 of pJH370) was digested with restriction enzymes NgoMI and MluI (restriction sites designed into the primers and shown as underlined sequences) to yield a 102-bp product. This product was used in a three-fragment ligation with an MluI-ClaI-digested DNA fragment (8,756 bp) and a ClaI-NgoMI-digested DNA fragment (5,030 bp) from pToto71 to produce the full-length, chimeric cDNA clone, SINV/GCN4(250-281). pToto71 is the full-length cDNA clone encoding wild-type SINV (32).
(ii) SINV/GCN4(255-277) and SINV/WEEV(34-56).
The SINV/GCN4(255-277) and SINV/WEEV(34-56) clones were constructed in a manner similar to that described above. The SINV/GCN4(255-277) clone has only part of the GCN4 dimerization domain corresponding to residues 255 to 277 in place of nt 7748 to 7817 of the SINV CP. The SINV/WEEV(34-56) clone has the helix I region of WEEV corresponding to nt 7574 to 7640 of the WEEV cDNA in place of nt 7748 to 7817 of the SINV sequence.
(iii) SINV tri(38-55).
The SINV tri(38-55) clone was constructed using overlap PCR to substitute L38V, I42V, L45V, and L52V in the SINV CP. The amplified DNA corresponded to nt 7467 to 8541 of the SINV cDNA and was inserted into pToto71 using XbaI and BstEII.
(iv) SINV/GCN4 tri(250-281).
Residues 250 to 281 of the GCN4 dimerization domain, containing substitutions L253I, V257I, L260I, N264I, L267I, V271I, L274I, and V278I, were cloned into pToto71 using the same procedure as described above for SINV tri(38-55). The template for PCR was the SINV/GCN4(250-281) cDNA clone.
(v) SINV/GCN4(255-277)(L68D).
A mutation of L268D (CTC to GAT) at nt 7785 to 7787 was introduced into the SINV/GCN4(255-277) clone using a three-fragment ligation involving a SacI-XbaI-digested PCR product, which corresponds to nt 7785 to 8541 of SINV/GCN4(255-277), and ClaI-SacI (5,049-bp) and XbaI-ClaI (8,045-bp) fragments from pToto71, to yield the chimeric full-length cDNA clone, SINV/GCN4(255-277)(L68D).
In vitro transcription and transfection of chimeras.
Following sequencing and verification of the inserted mutations, the full-length SINV-derived cDNA clones were linearized with SacI and transcribed in vitro, and the resulting RNA was transfected into BHK cells using DEAE-dextran as previously described (34). However, due to an internal SacI site in the CP coding region in chimeric clones containing GCN4 sequences, all plasmids containing this sequence were partially linearized with SacI to produce the intact genome. For this purpose, the enzyme was diluted 1:80 in distilled water before use, to give a final concentration of 0.005 U/μl. The linearized products were then used in in vitro transcription reactions, and RNA was transfected as previously described.
Thermal inactivation studies.
Thermal inactivation studies were performed as described previously with some modifications (35). Briefly, phosphate-buffered saline (PBS) supplemented with Ca2+, Mg2+, and 1% fetal bovine serum was used to dilute virus stocks to give a final concentration of 10,000 PFU/ml. This diluted virus (500 μl, which corresponds to 5,000 PFU) were incubated at 56°C for up to 20 min in a circulating water bath. Thirty microliters of sample was removed at time zero and every 5 min thereafter and added to 270 μl of ice-cold PBS supplemented with Ca2+, Mg2+, and 1% fetal bovine serum. Immediately following the last time point, virus titers were determined on BHK cell monolayers.
Accumulation of intracellular NCs.
NC accumulation studies were performed as previously described with a small modification (21, 31). Due to an internal SacI site (this enzyme was used for linearizing SINV-derived cDNA clones for in vitro transcription) in the SINV/GCN4(255-277), SINV/GCN4(250-281) (L268D), and SINV/GCN4(250-281) CP coding region, authentic full-length in vitro-transcribed RNA could not be produced for these clones. Therefore, infection rather than transfection was used for these viruses and the wild-type SINV (Toto71) in this assay. A total of 2.6 × 106 BHK cells were infected with the indicated virus at a multiplicity of infection (MOI) of 1. At 5 h postinfection, the cells were treated with actinomycin D to a final concentration of 1 μg/ml. At 6 h postinfection, 40 μCi/ml of [5,6-3H]uridine (Amersham Pharmacia Biotech, Piscataway, N.J.) was added to the cells to label replicating RNA. At 12 h postinfection, cytoplasmic extracts were harvested and layered onto a 22% freeze-thaw sucrose gradient, which was centrifuged at 32,000 rpm in an SW41 rotor (Beckman, Palo Alto, Calif.) for 2.5 h. The gradient was prepared by freezing a 22% sucrose solution in a centrifuge tube and allowing it to thaw to room temperature. This process establishes a concentration gradient that is useful for fractionation of virus-size particles. The gradients were fractionated into 600-μl aliquots, and 50 μl of each fraction was counted in 8 ml of Cytoscint liquid scintillant (ICN Biomedicals, Costa Mesa, Calif.).
One-step growth analysis.
BHK cells were infected with the indicated virus at a MOI of 1. This was the highest MOI that could be achieved with all of the viruses tested, as some of the mutants failed to produce high-titer stocks. The medium over the cells was replaced every 30 min for the first 2 h and then every hour for 12 h postinfection. Supernatant was collected at the indicated times and assayed for released virus by plaque titration on BHK cell monolayers at 37°C.
In vitro CLP assembly.
Recombinant wild-type and chimeric CPs used in in vitro assembly reactions were expressed in Escherichia coli and purified as previously described for the wild-type CP (37). In vitro assembly reactions using E. coli-expressed CP in the presence of a 48-mer oligonucleotide were also performed as previously described (37).
For inhibition studies, the wild-type CP (400 μg/ml) and chimeric CPs (400 μg/ml) were mixed together in various molar ratios in assembly buffer and incubated at room temperature for 15 min. The mixed protein sample (final concentration, 400 μg/ml) was then added to the 48-mer oligonucleotide (240 μg/ml) to give a final protein/oligonucleotide molar ratio of either 1:1 or 1:2. The helix I peptide (Research Genetics, Inc., Huntsville, Ala.) used in the in vitro assembly inhibition studies had the sequence NH3+-LASQIQQLTTAVSALVIGQ-CO2− corresponding to helix I of the SINV CP. The peptide was purified using standard reverse-phase high-performance liquid chromatography techniques (Laboratory for Macromolecular Research, Purdue University, West Lafayette, Ind.).
For assembly studies, the peptide was solubilized in 100% dimethyl sulfoxide (DMSO) and diluted in assembly buffer as needed. The final concentration of DMSO in the assembly reaction mixtures was <20%. The wild-type SINV CP(19-264) had been previously tested for its sensitivity to DMSO by the addition of increasing amounts of DMSO (2 to 30%) to the assembly reaction mixtures and assaying for the ability of the CP to assemble CLPs. The maximum tolerance was observed to be 20% DMSO. A peptide corresponding to the C-terminal 24 amino acids of the cytoplasmic domain of the SINV E2 glycoprotein (+H3N-YALAPNAVIPTSLALLCCVRSANA-CO2−) was used as a control peptide in inhibition studies. Purification and inhibition assays for the E2 peptide were identical to those described for the helix I peptide.
RESULTS
Helix I in SINV can be functionally replaced by the helix I region of WEEV CP.
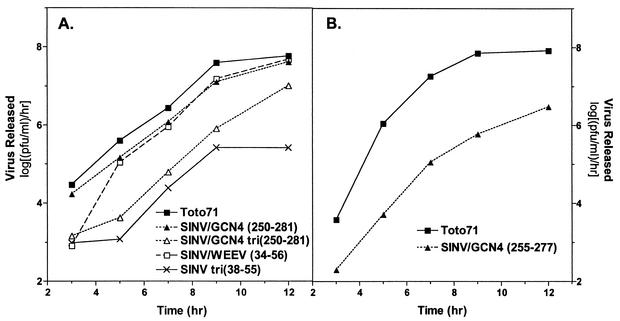
Secondary structure prediction analyses of the alphavirus CPs have predicted an overall helical propensity for the region called helix I of ∼60% (Fig. 1) (32). These predictions also indicated that the helix I region of the CP of WEEV had a stronger helical propensity than SINV. Therefore, it was of interest to see whether this conserved helix I region of WEEV was interchangeable with the helix I region of SINV. For this purpose, residues 34 to 56 of the WEEV CP were used to replace residues 35 to 57 (which included helix I and 2 flanking residues) in the SINV CP. In vitro-transcribed RNA from the chimera was transfected into BHK cells, and the resulting virus was isolated and characterized by plaque assay and one-step growth analysis. The plaque phenotype and replication efficiency determined by these assays were compared to the wild-type SINV, Toto71. As shown in Fig. 1 and Table 1, the SINV/WEEV(34-56) chimeric virus produced a large plaque phenotype (∼3.5 to 4 mm in diameter) similar to the wild-type virus. Furthermore, as shown in Fig. 2A, the replication efficiency of the virus, as determined by one-step growth analysis, was consistently only ca. threefold lower than the wild-type virus, Toto71. These results indicated that the entire 23 amino acids in this conserved region of the CP could be exchanged between two viruses in the same genus without significant effects on virus replication.
FIG. 1.
Schematic of the Sindbis virus capsid protein and sequences of helix I mutants. (A) The SINV CP is shown schematically. Residues 1 to 76 are predominantly positively charged, except for residues 38 to 55 which are uncharged and form helix I. Residues 76 to 132 (hatched box) are implicated in specific binding of the genomic RNA. This region is also important for CP-CP interactions during core assembly. Residues 114 to 264 form the protease domain responsible for autocatalytic cleavage of the CP from the structural polyprotein. This domain is also involved in CP-CP and CP-glycoprotein interactions. (B) Sequences for chimeric viruses and mutants within helix I and their corresponding plaque phenotypes determined 48 h postinfection on BHK cells. The amino acids in bold type are the amino acids of the foreign sequence inserted instead of amino acids 35 to 57 of the SINV CP (helix I). The numbers in parentheses in the virus designations indicate the amino acids of the foreign sequence that was inserted or substituted. Identical amino acids at a given position are indicated by dashes. SINV/GCN4(255-277)(L268D) is identical in sequence to SINV/GCN4(255-277) except for the change at residue 268. Plaque phenotypes are as follows: VSP, very small plaque (<2.0 mm in diameter); SP, small plaque (2.0 to 2.5 mm); MP, medium plaque (2.5 to 3.5 mm); LP, large plaque (3.5 to 4 mm); NR, not recovered.
TABLE 1.
Characteristics of chimeric viruses
| Virus | Residue at positiona
|
No. of amino acidsb | No. of heptad repeatsc | Plaque phenotyped | |
|---|---|---|---|---|---|
| a | d | ||||
| SINV (wild type) | V | L | 2.5 | LP | |
| SINV/WEEV(34-56) | V/I | L | 23 | 2.5 | LP |
| SINV/GCN4(255-277) | V/N | L | 23 | 3.3 | MP |
| SINV/GCN4(255-277)(L268D) | V/N | L(L268D) | 23 | 3.3 | NR |
| SINV/GCN4(250-281) | V/N | L | 33 | 4.5 | LP |
| SINV/GCN4(250-281)(L268D) | V/N | L(L268D) | 33 | 4.5 | SP |
| SINV/GCN4 tri(250-281) | I | I | 33 | 4.5 | SP |
| SINV tri(38-55) | V | V | 4 | 2.5 | VSP |
Residue at position a or d of the helical wheel.
Number of amino acids substituted for the wild-type SINV helix I and flanking region.
One heptad repeat defined from the a position of the helical wheel to the next a position. It is not equivalent to the number of amino acids substituted, since flanking residues were included in certain cases.
The plaque phenotype was determined at 48 h postinfection. Abbreviations: LP, large plaque (3.5 to 4.0 mm in diameter); MP, medium plaque (2.5 to 3.5 mm); SP, small plaque (2.0 to 2.5 mm); VSP, very small plaque (<2.0 mm); NR, not recovered.
FIG. 2.
One-step growth analysis of chimeric viruses in BHK cells. BHK cells were infected with the indicated virus at a MOI of 1. The medium over the cells was replaced every 30 min for the first 2 h and then every hour for 12 h postinfection. Supernatant was collected at the indicated times and assayed for released virus by titration on BHK cell monolayers at 37°C. Values are averages from two independent experiments.
Helix I of SINV can be replaced by an unrelated coiled-coil sequence.
The dimerization domain of the yeast transcription factor GCN4 is a leucine zipper sequence that has been well characterized structurally and biochemically (5, 10-12, 18, 22, 28, 29). It has been shown to form a parallel dimeric coiled coil and upon substitution of only the hydrophobic core residues at the a and d positions, this dimerization domain can be converted into a domain that forms trimeric or tetrameric coiled-coil interactions (10, 11). Therefore, the GCN4 leucine zipper was chosen as a model sequence for coiled-coil proteins and used to replace helix I in the SINV CP to probe the possibility that helix I also participated in coiled-coil interactions.
As shown in Fig. 1 and Table 1, residues 35 to 57 (which include helix I) of the SINV CP was replaced by 3.3 heptad repeats (23 residues) of the GCN4 dimeric, coiled-coil sequence (residues 255 to 277). The residues inserted, residues 255 to 277, correspond to residues 6 to 28 of the dimerization domain. Following transfection of in vitro-transcribed RNA and isolation of virus, the plaque phenotype and replication efficiency of the chimeric virus was determined. This virus, named SINV/GCN4(255-277), had a medium plaque phenotype (∼2.5 to 3.5 mm in diameter) compared to the large plaque phenotype of the wild-type virus (Fig. 1) and ∼100-fold decrease in the rate of virus release compared to the wild-type virus, Toto71 (Fig. 2B). This phenotype is quantitatively similar to the phenotype with a single site substitution at the conserved L45 or L52 in the SINV helix I sequence (32). Furthermore, an L268D mutation constructed in the SINV/GCN4(255-277) chimera failed to produce infectious virus (Fig. 1), suggesting that the leucine residues within the inserted heptad repeat sequence were an important feature of the helix.
Due to our attempts to keep the length of the chimeric CP the same as the wild-type CP (264 amino acids), it was possible that the chosen heptad repeat sequence of GCN4 was insufficient to allow the inserted residues (residues 255 to 277) to fold and function optimally. To address this problem, another chimeric virus was constructed with the full-length dimerization domain of GCN4 (residues 250 to 281) in place of SINV residues 35 to 57 (Fig. 1 and Table 1). The chimeric virus SINV/GCN4(250-281) was assayed for its plaque phenotype and replication efficiency. Interestingly, the virus produced a large plaque phenotype (∼3.5 to 4.0 mm in diameter) and its replication was reduced only ca. threefold (∼0.4 to 1.0 log unit lower) compared to the wild-type virus, Toto71 (Fig. 2A). The only common features between the unrelated GCN4 sequence and helix I are the periodicity of the leucine residues and the arrangement of hydrophobic core amino acids in the a and d positions of the sequence. Comparison of the above data to the results of the single site substitutions at L45 in a previous study by Perera et al. (32) suggested that the GCN4 sequence was successful in functionally replacing helix I. The data also indicated that variability in the length of the coiled-coil sequence was tolerated. Furthermore, a single site mutation of L268D in the latter chimera [SINV/GCN4(250-281)] also produced a virus with a reduction of ∼100-fold in replication efficiency compared to the wild-type virus (Fig. 1). This phenotype was consistent with previously mentioned single site leucine mutations within the helix I region and further supported the importance of leucine residues in this region of the CP.
Dimeric sequences are preferred over trimeric sequences in helix I.
Analysis of alphavirus core assembly using a well-established in vitro core assembly system has suggested that the CP forms a dimer intermediate before it assembles into a T=4 CLP (37, 38). The dimerization of the CP is dependent on nucleic acid, and helix I in the alphavirus CPs has been implicated as a major participant in this dimerization process. It has also been shown that assembly-defective CPs that retain the ability to bind nucleic acid are capable of dimerization, yet they cannot proceed along the assembly pathway unless the dimer intermediates are stabilized by chemical cross-linking reagents (40). It has also recently been shown that this cross-link mimics the function of helix I in the CP (32, 40).
To test the hypotheses that helix I functions as a dimerization motif and that the dimer formed is the preferred oligomer for capsid assembly, the sequence of helix I in the SINV CP was replaced with the sequence of the GCN4 leucine zipper that promotes trimeric coiled-coil formation (11). Several studies have shown that sequences that form dimeric coiled-coil interactions prefer β-branched residues in the a position of the hydrophobic interface of the coiled-coil helix and prefer leucine residues in the d position (23, 28). Sequences that form trimeric coiled-coil interactions, however, prefer β-branched residues in both the a and d positions of the hydrophobic interface (10, 11). The trimeric sequence of GCN4 which was inserted in place of helix I of the SINV CP had exclusively isoleucine residues in the a and d positions of the heptads (Fig. 1 and Table 1). The rescued chimeric virus, SINV/GCN4 tri(250-281), displayed a small plaque phenotype (∼2.0 to 2.5 mm in diameter) compared to the plaque phenotype of the wild-type virus and was impaired in the rate of virus release by ∼40-fold compared to the wild-type virus. A similar virus containing only the SINV sequence was also constructed by changing the residues in the a and d positions in the SINV helix I (Fig. 1). This mutant, SINV tri(38-55), had exclusively valine residues in the a and d positions of the heptads in helix I. The rescued mutant virus displayed a very small plaque phenotype (<2.0 mm in diameter) compared to the phenotype of the wild-type virus, Toto71. This phenotype was more severe than the L45V single site substitution (which produced a small-plaque virus) previously constructed in SINV helix I (32). SINV tri(38-55) also had a smaller plaque phenotype than that observed for the SINV/GCN4 tri(250-281). Furthermore, SINV tri(38-55) had a reduction of ∼160-fold in the rate of virus release compared to the rate for the wild-type virus (Fig. 2A). It is possible that the longer helix in the SINV/GCN4 tri(250-281) chimera allowed it to be more efficient in virus replication than the SINV sequence. These data indicated that a sequence favoring trimeric coiled-coil interactions in place of helix I had a more detrimental effect on virus replication than a sequence favoring dimeric coiled-coil interactions. In all cases, the mutants displayed wild-type levels of RNA synthesis and thus are not defective in uncoating.
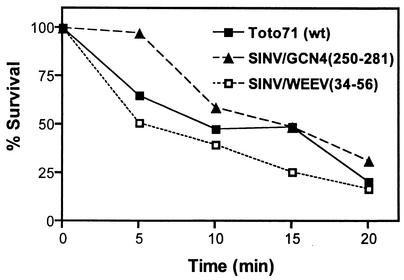
Length of helix I affects virion stability.
As mentioned above, replacing the helix I region of the SINV CP with the full-length dimerization domain of GCN4 resulted in a virus [SINV/GCN4(250-281)] with a growth phenotype similar to that of the wild-type virus (Toto71). This chimeric virus was used to further investigate whether the longer helix provided greater stability to the virus particles. It was previously shown that the stability of virus particles can be assayed indirectly by their sensitivity to elevated temperatures (35). Thus, the wild-type and chimeric viruses were assayed for their sensitivity to elevated temperatures. Viruses were incubated at 56°C for various times up to 20 min, and samples were assayed for infectivity by plaque titration. Analysis of the results shown in Fig. 3 indicated that the chimeric virus with the full-length dimerization domain of GCN4 was slightly more stable than the wild-type virus, Toto71. In comparison, the thermal stability of the chimeric virus containing the shorter GCN4 helix [SINV/GCN4(255-277)] was similar to that of the wild-type virus (data not shown). The stability of the chimera containing the WEEV helix was slightly reduced compared to those of the wild-type virus and SINV/GCN4(255-277), although all three viruses had similar values at the end point of the experiment.
FIG. 3.
Thermal inactivation of chimeric viruses. Thermal stability of chimeric virus particles was determined by incubation of 5,000 PFU (diluted in PBS supplemented with Ca2+ and Mg 2+) at 56°C for the times indicated. Aliquots were taken at the indicated time points, and virus titers were determined by plaque titration on BHK cell monolayers. The wild-type (wt) virus, Toto71, was included for comparison. Values are averages from four independent experiments.
Analysis of NC assembly in chimeric viruses.
During alphavirus assembly, newly synthesized CP encapsidates the viral genomic RNA and assembles into icosahedral, stable NCs in the cytoplasm of infected cells (1, 4). In the wild-type virus, these NCs have been observed to accumulate near internal membranes prior to migrating towards the plasma membrane and budding out of the cell (9, 30). The extent of accumulation of these NCs in the cytoplasm can be monitored by sucrose gradient sedimentation (21, 31). This assay was performed on the SINV/GCN4(255-277), SINV/GCN4(250-281), SINV/WEEV(34-56), and SINV/GCN4 tri(250-281) chimeric viruses to determine the ability of these viruses to assemble NCs in the cytoplasm of infected cells.
As shown in Table 2, the SINV/GCN4(250-281) and SINV/WEEV(34-56) chimeric viruses accumulated NCs in the cytoplasm; however, the SINV/GCN4(255-277) chimera did not accumulate NCs to detectable levels. SINV/GCN4 tri(250-281) showed a peak of radioactivity that sedimented slightly faster than cores, indicating the presence of possible aggregated material (data not shown). To investigate this possibility, SINV/GCN4 tri(250-281) was examined by thin-section electron microscopy, and the resulting micrographs were compared to those for SINV/GCN4(250-281) and wild-type SINV. All three viruses showed similar levels of NCs assembled in the cytoplasm, but SINV/GCN4 tri(250-281) showed aggregated material in the cytoplasm not observed in the SINV/GCN4(250-281) and wild-type SINV micrographs (data not shown). Together with the virus replication data (Fig. 1 and 2), these data suggest that SINV/GCN4 tri(250-281) may assemble NCs in the cytoplasm but that these NCs may not be as stable as those assembled by the SINV/GCN4(250-281) and wild-type viruses. Comparison of purified virus particles from SINV/GCN4(250-281), SINV/GCN4 tri(250-281), and the wild-type virus using electron microscopy and negative staining indicated no observable morphological difference between the particles from the three viruses (data not shown).
TABLE 2.
NC assembly by chimeric viruses
| Virus | In vivo NC assemblya | In vitro CLP assemblyb | Inhibition of WT CLP assembly (in vitro) |
|---|---|---|---|
| SINV (wild type) | Yes | Yes | |
| SINV/WEEV(34-56) | Yes | NDc | ND |
| SINV/GCN4(255-277) | No | No | Yes |
| SINV/GCN4(250-281) | Yes | Yes | No |
| SINV/GCN4 tri(250-281) | Yes, with aggregation | Mainly aggregation | No |
NC assembly was determined by sucrose gradient sedimentation of cytoplasmic extracts from infected cells and by thin-section electron microscopy of infected cells.
CLP assembly was determined by agarose gel analysis and electron microscopy.
ND, not determined.
In vitro core assembly of chimeric viruses.
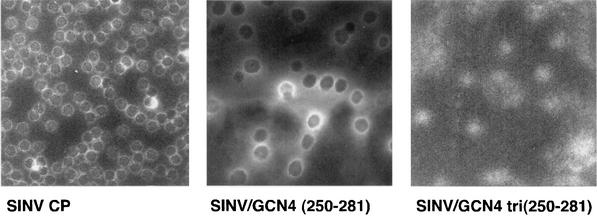
An in vitro CLP assembly system has been previously established for alphaviruses (41). This assay was used to analyze the assembly of NCs by the chimeric CPs described in this study. The in vitro assembly system utilizes recombinant, purified CP expressed in E. coli and single-stranded DNA more than 12 nt long (37). The chimeric CPs from SINV/GCN4(255-277), SINV/GCN4(250-281), and SINV/GCN4 tri(250-281) were expressed in E. coli, purified to homogeneity, and tested for their ability to assemble CLPs. Assembly reactions were performed by mixing chimeric CP (400 μg/ml) and single-stranded DNA oligonucleotide (240 μg/ml) in a molar ratio of 1:1 or 1:2 and assaying CLP formation using protocols previously established for the wild-type CP (37). CLP assembly was not observed for the SINV/GCN4(255-277) chimera, but it was observed for SINV/GCN4(250-281) as determined by the mobility of CLPs in 0.8% agarose gels (Table 2). Negative staining and electron microscopy of assembly reaction mixtures indicated that the SINV/GCN4(250-281) CLPs were not as spherical as those observed for the wild-type virus (Fig. 4) and may be influenced by the differences in length between the helices of SINV/GCN4(250-281) and wild-type SINV. The SINV/GCN4 tri(250-281) showed protein-nucleic acid complexes that comigrated with CLPs on an agarose gel but showed only aggregated material when assembly reaction mixtures were analyzed by negative staining and electron microscopy (Fig. 4).
FIG. 4.
Electron microscopy of in vitro-assembled CLPs. E. coli-expressed capsid protein was mixed with a 48-mer DNA oligonucleotide at a molar ratio of 1:1 or 1:2 in assembly buffer and incubated at room temperature for 15 min. The assembly reaction mixtures without further purification were negatively stained and examined by electron microscopy by the method of Mukhopadhyay et al. (27).
It was previously shown that mutant proteins defective in CLP assembly were capable of inhibiting wild-type CLP assembly (37). The mechanism behind this inhibition is unclear, but it has been proposed that assembly-defective CPs sequester wild-type CP into dead-end protein-nucleic acid complexes. The proteins produced by chimeric viruses SINV/GCN4(255-277), SINV/GCN4(250-281), and SINV/GCN4 tri(250-281) were tested in this CLP inhibition assay. As expected, SINV/GCN4(255-277), which was shown to be defective in CLP assembly, was able to inhibit wild-type CLP assembly when present in wild-type assembly reaction mixtures at a molar ratio of 1:1 (chimeric CP/wild-type CP) or greater (Table 2). However, neither SINV/GCN4(250-281) nor SINV/GCN4 tri(250-281) inhibited wild-type CLP assembly.
Inhibition of in vitro assembly with a helix I peptide.
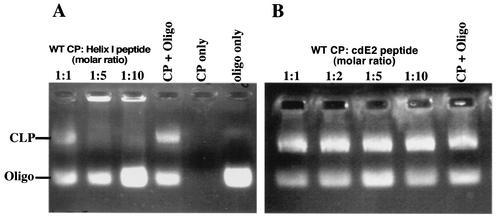
To determine whether helix I of the alphavirus CP could function independently of the CP, a synthetic peptide corresponding to SINV helix I was synthesized and purified by standard reverse-phase high-performance liquid chromatographic methods. The 19-mer peptide corresponding to residues 38 to 56 of the SINV CP had the sequence +H3N-LASQIQQLTTAVSALVIGQ-CO2−. Due to its hydrophobic character, the peptide had low solubility in aqueous solution, so the peptide was solubilized in 100% DMSO for use in assembly reactions. Recombinant wild-type CP(19-264) expressed in E. coli and helix I peptide were mixed at various molar ratios and incubated at room temperature for 5 min. The final protein/nucleic acid molar ratio was 1:1. The assembly reaction mixtures were incubated at room temperature for another 5 min and assayed on a 0.8% agarose gel as previously described (37). The results are shown in Fig. 5A. The helix I peptide inhibited wild-type CLP assembly when present in molar ratios greater than 1:1 (CP/peptide). It should be noted that assembly of the wild-type CP in various organic solvents was tested previously (data not shown), and the results indicated that for DMSO, inhibition of assembly was observed only when the DMSO concentration was above 20%. In the reactions mentioned above, the DMSO concentration was maintained below 20% to ensure that the presence of DMSO was not contributing to the inhibition observed.
FIG. 5.
Inhibition of wild-type CLP assembly in vitro with peptides corresponding to helix I and the cytoplasmic domain of E2. (A) In vitro assembly with the helix I peptide. A 0.8% agarose gel stained for nucleic acid with ethidium bromide shows in vitro-assembled CLPs and unincorporated oligonucleotides (Oligo). E. coli-expressed wild-type (WT) CP and helix I peptide were incubated together at the indicated molar ratios and allowed to equilibrate at room temperature. The samples were then mixed with a 48-mer oligonucleotide and incubated at room temperature for 15 to 30 min. CLP assembly was assayed by agarose gel analysis as previously described (37). The data show that increasing amounts of helix I peptide (left to right) inhibit wild-type CLP assembly. (B) In vitro assembly with a peptide corresponding to the C-terminal 24 residues of the cytoplasmic domain of E2 (cdE2) of SINV. The data show that increasing amounts of the cdE2 peptide (left to right) do not inhibit wild-type CLP assembly.
As a control, a 24-amino-acid peptide with characteristics similar to those of the helix I peptide (and which corresponded to the C terminus of the cytoplasmic domain of E2 in SINV) was also tested in wild-type assembly reactions. As shown in Fig. 5B, even at high concentrations, the E2 peptide did not inhibit wild-type CLP assembly. This indicated that the inhibition observed by the helix I peptide was specific for the sequence used and suggested a interaction between the peptide and the CP. Although the mechanism of inhibition is unknown, the peptide could be forming coiled-coil interactions with monomeric wild-type CP, preventing the formation of dimers necessary for CLP assembly to proceed (38).
DISCUSSION
The sequence between residues 38 to 55 (helix I) of the alphavirus CP has prominent features that differentiate it from the low-complexity flanking sequences in the N-terminal region of the CP. It has two and a half highly conserved leucine heptad repeats consisting of predominantly hydrophobic residues which are organized in a 4-3 repeat in the a and d positions of the heptad (32). This 4-3 repeat of hydrophobic residues is a hallmark of coiled-coil sequences (23, 26). Furthermore, the heptad repeats in the alphavirus CPs are centrally located between two highly charged random coil regions of the CP that were previously shown to be dispensable for NC assembly. Helix I, however, is important for NC assembly. On the basis of its striking similarity to other protein sequences containing a coiled coil, it has been proposed that helix I in the alphavirus CP may be involved in coiled-coil interactions during NC assembly.
The length of the predicted helix I sequence (18 residues) is low compared to the lengths of other dimeric coiled-coil sequences (an average of 30 residues) (23). However, this abbreviated size does not make it unique. Several crystal structures of proteins containing short, parallel dimeric coiled-coil sequences have been determined (23). For instance, the GAL4 DNA-binding domain has a coiled-coil structure consisting of only 15 residues. This coiled coil, however, forms only in the presence of DNA and may require other sequences for stable dimerization (24). The Gal6 DNA-binding domain (bleomycin hydrolase) has a coiled-coil structure of 11 residues that clamps two subunits into a tight dimer (15). The catabolite activator protein has a coiled-coil structure of 21 residues, functions in dimerization, and forms part of the cyclic AMP-binding pocket (25). It has been shown by studies on synthetic peptides derived from tropomyosin designed to have 8, 16, 29, and 36 residues that shorter sequences are capable of folding into coiled coils. However, these sequences may require an interaction with additional sequences in the protein for stable dimerization (19). Therefore, although helix I is shorter than optimal but similar to other short coiled-coil structures, it is proposed that helix I may function in the oligomerization of the CP by folding into a coiled-coil structure during alphavirus NC assembly. However, due to its length, helix I may require additional sequences in the CP for proper coiled-coil formation or for stability of oligomers formed through the coiled-coil interactions.
The most compelling evidence that helix I functions as a coiled coil has been attained through the biological analyses described here. It has been demonstrated that helix I can be replaced by (i) a helix I sequence from a related alphavirus (WEEV) or (ii) a completely unrelated coiled-coil sequence of GCN4 (either in its full-length form or as a truncated sequence). Chimeric viruses containing residues 34 to 56 of the WEEV CP or the entire dimerization domain of the GCN4 transcription factor (residues 250 to 281) instead of SINV helix I exhibited the wild-type phenotype as determined by plaque assay and one-step growth analyses (Fig. 1 and 2A). Furthermore, in the case of SINV/GCN4(250-281), it was demonstrated that the longer helix rendered the virus more thermostable than the wild type (Fig. 3). The ability to substitute 33 amino acids in the case of SINV/GCN4(250-281) and 23 amino acids in the case of SINV/WEEV(34-46) without affecting virus replication seems remarkable, when compared to the phenotypes of mutants with single site substitutions at selected leucine residues within helix I (this work) (32). It was previously shown that a single site substitution of the conserved leucine residues within helix I of SINV affected virus replication by 10- to 100-fold compared to that of the wild-type virus (32). These observations were further supported in the present study through the analyses of the two single site mutants, SINV/GCN4(250-281) (L268D), and SINV/GCN4(255-277)(L68D). These mutants also showed a decrease in replication efficiency of about ∼100-fold (Fig. 1) compared to their parent viruses, SINV/GCN4(250-281) and SINV/GCN4(255-277), respectively. Together, these data have demonstrated that although the substitution of an entirely unrelated leucine heptad repeat sequence in the SINV CP is tolerated by the virus, a single site substitution at individual leucine residues is detrimental. This emphasizes the requirement for a functional leucine heptad repeat sequence in the region of helix I in the SINV CP.
As shown in Fig. 2B, the SINV/GCN4(255-277) chimera had a replication defect compared to the wild-type virus. The replication defect observed for SINV/GCN4(255-277) has been attributed to the lack of detectable NC assembly (also confirmed by studies in vitro). The leucine heptad repeat sequence corresponding to a part of the GCN4 dimerization domain in this chimera (residues 255 to 277) was chosen randomly from the entire dimerization domain to maintain the relative length and periodicity of the helix I sequence in the SINV CP. The sequence consisting of residues 255 to 277 in the native GCN4 dimerization domain is N-terminally truncated by six residues and C-terminally truncated by four residues. On the basis of studies on N-terminally and C-terminally truncated GCN4 peptides (22), it is possible that the length of the GCN4 sequence chosen to replace helix I (residues 255 to 277) was not optimal to facilitate its folding and proper function in the context of the CP.
Mutations within helix I of the SINV CP do not interfere with RNA replication, protein translation, or RNA encapsidation, but they do affect the ability of CP to assemble stable nucleocapsids (32). This assembly defect has been demonstrated in nucleocapsid accumulation studies in vivo and in CLP assembly assays in vitro (Table 2). Furthermore, it has been established that the assembly defect in these helix I mutants was due to the inability of the mutated CPs to form assembly-competent intermediates (32, 38). These assembly-competent intermediates were shown to be CP dimers. The present study has obtained additional evidence supporting these observations by comparing chimeric viruses with helix I sequences that favor trimeric coiled-coil interactions to those that favor dimeric coiled-coil interactions. The chimeric virus SINV/GCN4 tri(250-281), which is expected to favor a trimeric coiled-coil interaction, had a replication efficiency ∼160-fold less than that of SINV/GCN4(250-281), which is expected to favor a dimeric coiled-coil interaction. Similarly, SINV tri(38-55), which was also expected to favor trimeric coiled-coil interactions, had a replication efficiency more than 100-fold less than that of wild-type SINV.
Analysis of in vitro-assembled CLPs by negative staining and electron microscopy indicated that SINV/GCN4 tri(250-281) formed predominantly aggregated protein-nucleic acid complexes but that SINV/GCN4(250-281) and wild-type SINV formed stable CLPs (Fig. 4). However, the SINV/GCN4 tri(250-281) CP was capable of forming protein-nucleic acid complexes that were similar to those of the wild-type CP as determined by the migration of these complexes on an agarose gel. Therefore, the lack of CLP assembly in this chimera may indicate that these protein-nucleic acid complexes have an assembly defect. It must be noted, however, that the CLPs formed in vitro by the SINV/GCN4(250-281) CP were not as spherical as those formed by wild-type SINV or those observed for SINV/GCN4(250-281) CP in infected cells. This observation may reflect differences between the in vitro and in vivo systems. In contrast to CLPs or NCs, the morphology of purified virus particles from SINV/GCN4 tri(250-281) and SINV/GCN4(250-281) was similar to that of wild-type SINV. Therefore, defects observed in nucleocapsid assembly for SINV/GCN4 tri(250-281) were not sufficient to completely eliminate virus particle assembly or affect particle morphology. It has previously been proposed that the viral glycoproteins play a significant role in the final assembly steps of the virus particle and are key contributors to the icosahedral symmetry of these particles (6, 8, 20, 27, 33, 42). Therefore, it is possible that glycoproteins may mediate the assembly of the SINV/GCN4 tri(250-281) chimera and allow a reduced number of completely assembled virus particles to bud out of the cell. Alternatively, these assembly-competent virus particles could be a subpopulation that are the result of a reversion event or second site mutations in the CP. Such reversion events were previously shown to rescue defects in the tick-borne encephalitis virus CP (16, 17). However, on the basis of plaque phenotypes and sequence analysis of progeny virion RNA, no such revertant event was observed for the chimeras and single site mutants discussed in this study.
In conclusion, the data presented here have demonstrated that helix I in the SINV CP can be functionally replaced by a helix I sequence of a related virus, WEEV, or by a well-characterized unrelated coiled-coil leucine zipper sequence. This demonstrated that the only requirement for the helix I region was a functional leucine heptad repeat sequence. Furthermore, it has also been demonstrated that helix I is required to have a sequence that favors dimeric coiled-coil interactions over trimeric coiled-coil interactions. In the absence of structural data, these studies have supported the hypothesis that helix I functions as a coiled-coil oligomerization motif during alphavirus NC assembly and supports the previously established nucleocapsid assembly model that proposes an assembly pathway mediated by a dimeric assembly intermediate.
Acknowledgments
We thank Dagmar Sedlak for excellent technical assistance in cell culture. We acknowledge James Strauss for kindly providing us with the WEEV cDNA clone. We also acknowledge Peter Kim and James Hu for providing us with the plasmids encoding the GCN4 dimerization domains.
This research was supported by Public Health Service grant GM56279 from the National Institutes of Health.
REFERENCES
- 1.Brown, D. T., and J. F. Smith. 1975. Morphology of BHK-21 cells infected with Sindbis virus temperature-sensitive mutants in complementation groups D and E. J. Virol. 15:1262-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casjens, S. 1997. Principles of virion structure, function, and assembly, p. 3-37. In W. Chiu, R. M. Burnett, and R. Garcia (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 3.Choi, H.-K., L. Tong, W. Minor, P. Dumas, U. Boege, M. G. Rossmann, and G. Wengler. 1991. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature 354:37-43. [DOI] [PubMed] [Google Scholar]
- 4.Coombs, K., and D. T. Brown. 1987. Organization of the Sindbis virus nucleocapsid as revealed by bifunctional cross-linking agents. J. Mol. Biol. 195:359-371. [DOI] [PubMed] [Google Scholar]
- 5.DeLano, W. L., and A. T. Brunger. 1994. Helix packing in proteins: prediction and energetic analysis of dimeric, trimeric, and tetrameric GCN4 coiled coil structures. Proteins 20:105-123. [DOI] [PubMed] [Google Scholar]
- 6.Forsell, K., G. Griffiths, and H. Garoff. 1996. Preformed cytoplasmic nucleocapsids are not necessary for alphavirus budding. EMBO J. 15:6495-6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forsell, K., M. Suomalainen, and H. Garoff. 1995. Structure-function relation of the NH2-terminal domain of the Semliki Forest virus capsid protein. J. Virol. 69:1556-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forsell, K., L. Xing, T. Kozlovska, R. H. Cheng, and H. Garoff. 2000. Membrane proteins organize a symmetrical virus. EMBO J. 19:5081-5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garoff, H., and K. Simons. 1974. Location of the spike glycoproteins in the Semliki Forest virus membrane. Proc. Natl. Acad. Sci. USA 71:3988-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harbury, P. B., P. S. Kim, and T. Alber. 1994. Crystal structure of an isoleucine-zipper trimer. Nature (London) 371:80-83. [DOI] [PubMed] [Google Scholar]
- 11.Harbury, P. B., T. Zhang, P. S. Kim, and T. Alber. 1993. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262:1401-1407. [DOI] [PubMed] [Google Scholar]
- 12.Holtzer, M. E., E. G. Lovett, D. A. d'Avignon, and A. Holtzer. 1997. Thermal unfolding in a GCN4-like leucine zipper: 13C alpha NMR chemical shifts and local unfolding curves. Biophys. J. 73:1031-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu, J. C., E. K. O'Shea, P. S. Kim, and R. T. Sauer. 1990. Sequence requirements for coiled-coils: analysis with lambda repressor-GCN4 leucine zipper fusions. Science 250:1400-1403. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, J., and A. J. Fisher (ed.). 1994. Principles of virus structure. Academic Press, London, United Kingdom.
- 15.Joshua-Tor, L., H. E. Xu, S. A. Johnston, and D. C. Rees. 1995. Crystal structure of a conserved protease that binds DNA: the bleomycin hydrolase, Gal6. Science 269:945-950. [DOI] [PubMed] [Google Scholar]
- 16.Kofler, R. M., F. X. Heinz, and C. W. Mandl. 2002. Capsid protein C of tick-borne encephalitis virus tolerates large internal deletions and is a favorable target for attenuation of virulence. J. Virol. 76:3534-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kofler, R. M., A. Leitner, G. O'Riordain, F. X. Heinz, and C. W. Mandl. 2003. Spontaneous mutations restore the viability of tick-borne encephalitis virus mutants with large deletions in protein C. J. Virol. 77:443-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landshultz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759-1764. [DOI] [PubMed] [Google Scholar]
- 19.Lau, S. Y., A. K. Taneja, and R. S. Hodges. 1984. Synthesis of a model protein of defined secondary and quaternary structure. Effect of chain length on the stabilization and formation of two-stranded alpha-helical coiled-coils. J. Biol. Chem. 259:13253-13261. [PubMed] [Google Scholar]
- 20.Lescar, J., A. Roussel, M. W. Wein, J. Navaza, S. D. Fuller, G. Wengler, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 21.Lopez, S., J. S. Yao, R. J. Kuhn, E. G. Strauss, and J. H. Strauss. 1994. Nucleocapsid-glycoprotein interactions required for assembly of alphaviruses. J. Virol. 68:1316-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lumb, K. J., C. M. Carr, and P. S. Kim. 1994. Subdomain folding of the coiled coil leucine zipper from the bZIP transcriptional activator GCN4. Biochemistry 33:7361-7367. [DOI] [PubMed] [Google Scholar]
- 23.Lupas, A. 1996. Coiled coils: new structures and new functions. Trends Biochem. Sci. 21:375-382. [PubMed] [Google Scholar]
- 24.Marmorstein, R., M. Carey, M. Ptashne, and S. C. Harrison. 1992. DNA recognition by GAL4: structure of a protein-DNA complex. Nature 356:408-414. [DOI] [PubMed] [Google Scholar]
- 25.McKay, D. B., and T. A. Steitz. 1981. Structure of catabolite gene activator protein at 2.9 Å resolution suggests binding to left-handed B-DNA. Nature 290:744-749. [DOI] [PubMed] [Google Scholar]
- 26.McLachlan, A. D., and M. Stewart. 1975. Tropomyosin coiled-coil interactions: evidence for an unstaggered structure. J. Mol. Biol. 98:293-304. [DOI] [PubMed] [Google Scholar]
- 27.Mukhopadhyay, S., P. R. Chipman, E. M. Hong, R. J. Kuhn, and M. G. Rossmann. 2002. In vitro-assembled alphavirus core-like particles maintain a structure similar to that of nucleocapsid cores in mature virus. J. Virol. 76:11128-11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Shea, E. K., J. D. Klemm, P. S. Kim, and T. Alber. 1991. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254:539-544. [DOI] [PubMed] [Google Scholar]
- 29.O'Shea, E. K., R. Rutkowski, and P. S. Kim. 1989. Evidence that the leucine zipper is a coiled coil. Science 243:538-542. [DOI] [PubMed] [Google Scholar]
- 30.Owen, K. E., and R. J. Kuhn. 1997. Alphavirus budding is dependent on the interaction between the nucleocapsid and hydrophobic amino acids on the cytoplasmic domain of the E2 envelope glycoprotein. Virology 230:187-196. [DOI] [PubMed] [Google Scholar]
- 31.Owen, K. E., and R. J. Kuhn. 1996. Identification of a region in the Sindbis virus nucleocapsid protein that is involved in specificity of RNA encapsidation. J. Virol. 70:2757-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perera, R., K. E. Owen, T. L. Tellinghuisen, A. E. Gorbalenya, and R. J. Kuhn. 2001. Alphavirus nucleocapsid protein contains a putative coiled coil alpha-helix important for core assembly. J. Virol. 75:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pletnev, S. V., W. Zhang, S. Mukhopadhyay, B. R. Fisher, R. Hernandez, D. T. Brown, T. S. Baker, M. G. Rossmann, and R. J. Kuhn. 2001. Locations of carbohydrate sites on Sindbis virus glycoproteins show that E1 forms an icosahedral scaffold. Cell 105:127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strauss, E. G., E. M. Lenches, and J. H. Strauss. 1976. Mutants of Sindbis virus. I. Isolation and partial characterization of 89 new temperature-sensitive mutants. Virology 74:154-168. [DOI] [PubMed] [Google Scholar]
- 36.Strauss, J. H., E. G. Strauss, and R. J. Kuhn. 1995. Budding of alphaviruses. Trends Microbiol. 3:346-350. [DOI] [PubMed] [Google Scholar]
- 37.Tellinghuisen, T. L., A. E. Hamburger, B. R. Fisher, R. Ostendorp, and R. J. Kuhn. 1999. In vitro assembly of alphavirus cores by using nucleocapsid protein expressed in Escherichia coli. J. Virol. 73:5309-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tellinghuisen, T. L., and R. J. Kuhn. 2000. Nucleic acid-dependent cross-linking of the nucleocapsid protein of Sindbis virus. J. Virol. 74:4302-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tellinghuisen, T. L., R. Perera, and R. J. Kuhn. 2001. Genetic and biochemical studies on the assembly of an enveloped virus. Genet. Eng. 23:83-112. [DOI] [PubMed] [Google Scholar]
- 40.Tellinghuisen, T. L., R. Perera, and R. J. Kuhn. 2001. In vitro assembly of Sindbis virus core-like particles from cross-linked dimers of truncated and mutant capsid proteins. J. Virol. 75:2810-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wengler, G., U. Boege, G. Wengler, H. Bischoff, and K. Wahn. 1982. The core protein of the alphavirus Sindbis virus assembles into core-like nucleoproteins with the viral genome RNA and with other single-stranded nucleic acids in vitro. Virology 118:401-410. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, W., S. Mukhopadhyay, S. V. Pletnev, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2002. Placement of the structural proteins in Sindbis virus. J. Virol. 76:11645-11658. [DOI] [PMC free article] [PubMed] [Google Scholar]