Abstract
The very-low-density lipoprotein receptor (VLDL-R) is a receptor for the minor-group human rhinoviruses (HRVs). Only two of the eight binding repeats of the VLDL-R bind to HRV2, and their footprints describe an annulus on the dome at each fivefold axis. By studying the complex formed between a selection of soluble fragments of the VLDL-R and HRV2, we demonstrate that it is the second and third repeats that bind. We also show that artificial concatemers of the same repeat can bind to HRV2 with the same footprint as that for the native receptor. In a 16-Å-resolution cryoelectron microscopy map of HRV2 in complex with the VLDL-R, the individual repeats are defined. The third repeat is strongly bound to charged and polar residues of the HI and BC loops of viral protein 1 (VP1), while the second repeat is more weakly bound to the neighboring VP1. The footprint of the strongly bound third repeat extends down the north side of the canyon. Since the receptor molecule can bind to two adjacent copies of VP1, we suggest that the bound receptor “staples” the VP1s together and must be detached before release of the RNA can occur. When the receptor is bound to neighboring sites on HRV2, steric hindrance prevents binding of the second repeat.
Human rhinoviruses (HRVs) belong to the Picornaviridae family and are the most frequent cause of the common cold. They have an icosahedral capsid that is only 300 Å in diameter. The capsid is composed of 60 copies each of four viral-coat proteins—VP1, VP2, VP3, and VP4—arranged on a T=1 or P=3 (pseudo-T=3) icosahedral lattice (31). VP1 has a hydrophobic pocket, or hollow, which is accessible from the exterior and located at the base of the canyon, a depression that encircles the fivefold axes of symmetry. The pocket is frequently occupied by a natural pocket factor, a fatty acid-like molecule that is believed to stabilize the virus during its spread from cell to cell (12). Several antiviral capsid-binding compounds, which inhibit rhinoviral infection, have been described previously (2). Due to higher affinity, they replace the natural pocket factor and render the capsid more resistant to uncoating (33).
The HRVs are classified into two groups depending on their cellular receptors. The intercellular adhesion molecule 1 (ICAM-1) is the receptor for the major group (36), and members of the low-density lipoprotein receptor (LDL-R) family act as receptors for the minor group (16). The binding sites of ICAM-1 on the major-group HRV16 and HRV14 have been located at the base of the canyon by Kolatkar et al. and Olson et al. (20, 28) by using cryoelectron microscopy and X-ray crystallography. Employing the same techniques, we recently located the binding site of the very-low-density lipoprotein receptor (VLDL-R) on the minor-group HRV2 (15). A model of the complex was produced by combining the cryoelectron microscopy map, the atomic structure of HRV2, and similarity-based structures of the three VLDL-R repeats. The footprint of the VLDL-R on HRV2 lies on an annulus on the dome at the fivefold axes. It does not overlap the footprint of ICAM-1 in the canyon, nor does it overlap the entrance to the hydrophobic pocket of VP1 located at the base of the canyon (15).
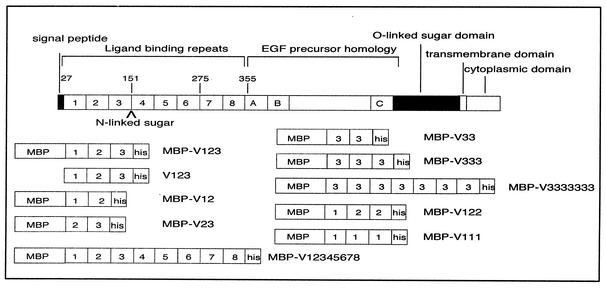
The VLDL-R is a member of the LDL-R family of cell surface receptors, which mediate the transport of structurally and functionally diverse protein ligands into cells by receptor-mediated endocytosis. The VLDL-R consists of eight imperfect ligand-binding repeats of approximately 40 amino acids at its N terminus, followed by an epidermal growth factor precursor domain, an O-linked sugar domain, a transmembrane segment, and a cytoplasmic domain containing coated-pit internalization signals (Fig. 1). Each ligand-binding repeat forms a cage containing a Ca2+ ion and six cysteine residues that form three disulfide bridges (10). These rigid ligand-binding domains are linked by four to five amino acids that confer some flexibility. This was indicated by nuclear magnetic resonance analysis of concatemers of repeats 1 to 2 and of repeats 5 to 6 of the LDL-R, which suggested that the repeats could move almost independently from each other (22, 26). Combinations of several repeats are probably necessary for ligand recognition, as single repeats alone do not bind any of the natural ligands (1). Based on visualization of vesicle-reconstituted LDL-R by cryoelectron microscopy, Jeon and Shipley (18) proposed that the two N-terminal repeats of LDL-R fold back onto the fourth and fifth repeats, with the third repeat being on the apex of the turn. However, the packing arrangement of VLDL-R is not known. The LDL-Rs appear to bind their ligands by electrostatic interactions of their negatively charged ligand-binding domains (4, 9), although some of the carboxylates previously thought to be exposed actually point inward, thereby chelating the Ca2+ ion (10).
FIG. 1.
Schematic model of human VLDL-R and the recombinant minireceptors studied. Numbers refer to individual repeats and amino acids (top). (A through C) Epidermal growth factor-related cysteine-rich repeats. MBP was fused to the amino terminus and a hexa-His tag to the carboxy terminus. In the case of V123, MBP was removed by cleavage with factor Xa.
The differences in the receptor-binding site of the major- and minor-group HRVs correlate well with the differences in their biochemical properties. The presence of hydrophobic antiviral compounds in the VP1 pocket modifies the canyon floor of certain major-group HRVs and so inhibits receptor binding (33). In contrast, receptor binding of the minor-group HRV1A is not affected by the presence of these compounds (19), a fact that is in accord with the absence of interaction between the LDL-R and the entrance to the hydrophobic pocket of VP1. Also, binding of ICAM-1 to major-group HRVs serves to catalyze uncoating (11, 17), while binding of the LDL-R to the minor-group HRVs does not directly initiate uncoating—which occurs after internalization in the low-pH environment of the late endosomes (29). For major-group HRVs, Kolatkar and colleagues (20) proposed a two-step entry mechanism in which the ICAM-1 first binds to one side of the canyon. Then, by expelling the natural pocket factor from the VP1 pocket, ICAM-1 binds to the other side of the canyon as well, thus causing a hinge movement of VP1 that opens a channel on the fivefold axis. Since the LDL-R does not interact with the base of the canyon, no such mechanism involving a competition between binding of the pocket factor and the LDL-R can be invoked. It was proposed (13) that the pocket factor is expelled in the low-pH (i.e., ≤5.6) conditions of the late endosome, thus allowing a movement of VP1 that results in the opening of a channel on the fivefold axes to allow the RNA to exit. The role of the LDL-R is seen as attaching the virion to the host cell for internalization.
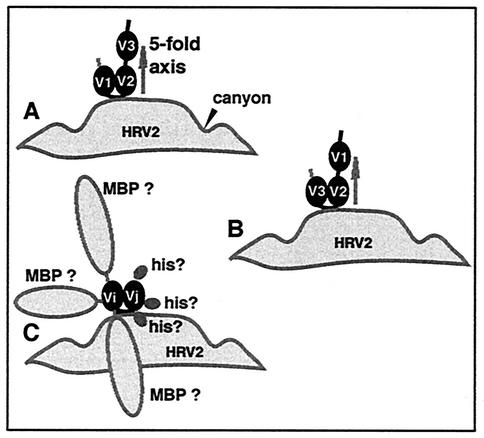
Based on the similarity-based model of the complex between HRV2 and a recombinant soluble VLDL-R fragment that encompasses repeats 1 to 3 (V123), it became clear that only two of the three VLDL-R repeats are attached per asymmetric unit of HRV2 (15). Simultaneous binding of repeats 1 and 3 could be excluded, since this would result in immobilization of repeat 2, thereby making it visible in the map; this was not the case. However, it was not possible to distinguish between the attachment of repeats 1 and 2 or 2 and 3 (Fig. 2). With the aim of identifying the repeats that are in contact with HRV2, we determined the structures of virus-receptor complexes for eight different soluble fragments of the VLDL-R—including concatemers composed of multiple copies of repeat 3 (summarized in Fig. 1; Moser et al., unpublished data). Also, in order to better define the HRV2-receptor interaction and to determine the individual footprint of each repeat, we used a field emission gun (FEG) cryoelectron microscope to improve the resolution of the HRV2-MBP-V123 reconstruction.
FIG. 2.
Schematic diagrams of the two possible models for the binding of V123 on HRV2 proposed by our previous work (15) with V1 and V2 bound (A) and with V2 and V3 bound (B). Schematic diagram of the possible steric interference of the hexa-His tag and MBP for the binding of the minireceptors (C). For example, binding of the receptor or of a repeat can be prevented if the MBP physically interferes.
MATERIALS AND METHODS
Preparation and purification of HRV2.
HRV2 was grown in Rhino-HeLa cells in suspension culture and purified as described previously (32) with minor modifications (described in reference 15). Purified HRV2 was suspended in 50 mM Tris-HCl (pH 7.4) at a concentration of about 4 mg/ml as determined by capillary electrophoresis (27).
Preparation and purification of the VLDL-R fragments.
Recombinant VLDL minireceptors that encompassed different ligand-binding repeats were cloned into a pMAL-c2x vector (New England BioLabs) with a maltose binding protein (MBP) fused to the N terminus and a hexa-His tag fused to the C terminus. The receptor fragments were then purified over Ni-nitrilotriacetic acid columns (Qiagen), oxidized, and folded in the presence of glutathione S-transferase-receptor-associated protein immobilized on Sepharose as described previously (30). Correctly folded protein remained bound to the resin and was eluted with 1 N NH3 in 0.5× Tris-buffered saline-2 mM CaCl2. Ammonia was removed in a Speedvac concentrator, and, in parallel, the volume was reduced by a factor of 2. V111 did not bind to glutathione S-transferase-receptor-associated protein, and this step was therefore omitted. Samples were further purified by size exclusion chromatography on S200 Superdex by using a fast-pressure liquid chromatography system from Amersham Pharmacia. For preparation of the receptor fragment without MBP, the material was cleaved with factor Xa (New England BioLabs) as described previously (15).
Calcium binding assay.
Approximately 1 μg of MBP-V111 or MBP-V123 was run on an SDS-12% polyacrylamide gel in the absence or presence of reducing agent and transferred electrophoretically onto a polyvinylidene difluoride membrane by using 25 mM Tris-HCl, 195 mM Glycin (pH 7.5), and 20% methanol in a semidry-blotting apparatus. The membrane was then washed two times for 20 min each with 10 ml of (15 mM Tris-HCl, pH 7.5, 50 mM NaCl, 5 mM MgCl2) supplemented with 10 mM EDTA followed by washing with blotting buffer (without EDTA). The membrane was then incubated with 10 μCi of 45CaCl2 (Amersham Pharmacia) in 5 ml of blotting buffer for 20 min. After washing two times with blotting buffer, the membrane was air dried and exposed to Kodak X-ray film overnight. Blotting with 35S-labeled HRV2 was carried out as described previously (29).
Preparation of complexes of HRV2 and the VLDL-R fragments.
HRV2 and Vi-j or MBP-Vi-j were incubated at a molar ratio of approximately 1 to 120 for 1 h at 4°C to form stable complexes. The quantities employed for each complex are summarized in Table 1. The cryoelectron microscope specimens of the complex were prepared immediately without additional concentration.
TABLE 1.
Quantities of materials used for complex formationa
| Component | Mol mass (kDa) | Concn (mg/ml) | Vol (μl) | Excess times |
|---|---|---|---|---|
| HRV2 | 100 | 4 | 4 | |
| V12 | 54 | 0.05 | 100 | >∼0.5 |
| V23 | 54 | 0.5 | 40 | ∼2.5 |
| V33 | 54 | 0.37 | 40 | <∼2 |
| V123 | 59 | 0.5 | 40 | >∼2 |
| V123 (FEG) | 59 | 0.5 | 40 | >∼2 |
| V123 (no MBP) | 17 | 0.1 | 60 | ∼2 |
| V333 | 59 | 0.4 | 40 | <∼2 |
| V3333333 | 76 | 0.35 | 40 | >∼2 |
| V12345678 | 81 | 0.3 | 70 | <∼2 |
| V122 | 59 | 0.7 | 40 | ∼3 |
| V111 | 59 | NAb | NA | NA |
One asymmetric unit of HRV2 is ∼100 kDa.
NA, not applicable (because V111 does not bind).
Preparation of frozen hydrated specimens.
Frozen hydrated specimens were prepared on holey carbon grids as previously described (14). Samples of the virus-receptor suspension (3 μl) were applied to grids, blotted immediately with filter paper for 1 to 2 s, and rapidly plunged into liquid ethane cooled by nitrogen gas at −175°C. Specimens were photographed at a temperature close to −175°C by using an Oxford cryoholder CT3200 either in a Philips CM200 twin (LaB6 gun) or in a JEOL 2010F electron microscope with a high-resolution photography pole piece and FEG operating at 200 kV. Defocused image pairs were obtained under low-dose conditions (<10 electrons/Å2) at nominal magnifications of ×38,000 (for the LaB6) and ×40,000 (for the FEG), with underfocus values ranging from 1 to 3 μm.
Image analysis.
Preliminary selection of micrographs and preparation of virus particle images for analysis were performed as described previously (14). Images were digitized on a Zeiss scanner. The pixel size of 7 μm on the micrograph corresponds to pixel sizes of 1.77 and 1.76 Å for the LaB6 and FEG, respectively, as calibrated with the 23.0-Å layer line of tobacco mosaic virus. Further image analysis was performed on SGI and Alpha workstations. The HRV2 map obtained in a previous study (15) was used as a starting model for the analyses. All subsequent refinement of particle origin and orientation was performed by using the model-based polar Fourier transform (PFT) programs (3). The program CTFMIX (6) was used to correct for contrast transfer function effects and to combine defocused pairs for orientation and origin refinement; it was also used for the final Fourier-Bessel reconstruction. The statistics for the different reconstructions are given in Table 2. The resolutions were estimated by Fourier shell correlation of reconstructions from half-data sets by using the criterion of 0.5 correlation. Isosurface representations of the reconstructed density were visualized with Amira on a Silicon Graphics workstation.
TABLE 2.
Statistics for the HRV2-minireceptor reconstructionsa
| Complex of HRV2 with: | Underfocus (μm) | No. of particles selected | No. of particles included | Resolution (Å) |
|---|---|---|---|---|
| V23 | 1.03→2.77 | 1,535 | 369 | 20 |
| V33 | 1.09→1.89 | 1,074 | 363 | 18 |
| V123 | 1.24→3.68 | 1,983 | 1,077 | 19 |
| V123 (FEG) | 1.18→2.90 | 4,228 | 912 | 16 |
| V123 (no MBP) | 1.44→3.26 | 1,483 | 782 | 19 |
| V333 | 1.16→2.05 | 1,060 | 268 | 20 |
| V3333333 | 1.41→2.48 | 850 | 443 | 21 |
| V12345678 | 0.72→2.34 | 1,646 | 501 | 21 |
| V122 | 1.11→2.15 | 1,248 | 641 | 19 |
The ranges of underfocus of the defocused pairs of photomicrographs used in the reconstructions are indicated together with the total number of particles selected, the number of particles (particle image pairs) included in the reconstruction, and the resolution (0.5 criterion). Note that the resolutions given in reference 15 used the more generous 0.1 criterion rather than the 0.5 criterion used here.
Fitting the X-ray structures of HRV2 and the VLDL-R repeats to the cryoelectron microscope-reconstructed density.
The cryoelectron microscopy map and the atomic structure of HRV2 were visualized together by using the program “O” on an SGI workstation. Thus, the residues included in the footprints were determined. The similarity-based structure of V2 and V3, determined as described previously (15), was placed visually in the higher-resolution HRV2-MBP-V123 map in order to compare the volume assigned to these repeats with the atomic model.
Figures were produced with Amira (Fig. 4 and 5) and Bobscript (8) with Raster3D (24) (Fig. 5).
FIG. 4.
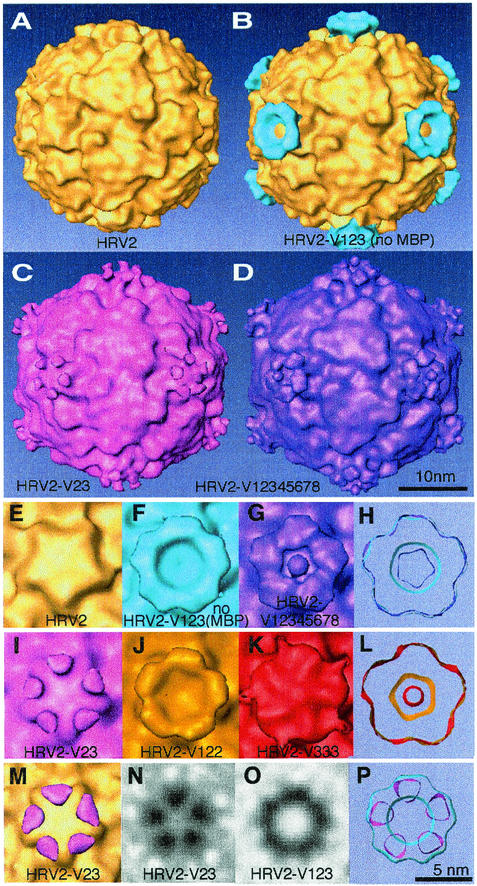
Reconstruction (from LaB6 data) of the complex between HRV2 and selected minireceptors. Reconstructions of native HRV2 alone (A), in complex with V123 without MBP (B) (difference image), in complex with V23 (C), and in complex with V12345678 (D). Panels A and B are derived from results found in reference 14. Close-up views down the fivefold axis of the native virus (E) and some of the complexes studied are shown. (F) HRV-V123 without MBP; (G) HRV2-V12345678; (I) HRV2-V23; (J) HRV2-V122; (K) HRV2-V333; (H) comparison of the footprints of V123 without MBP (light blue) and V12345678 (purple); (L) comparison of the footprints of V122 (gold) and V333 (red); (P) comparison of the footprints of V123 without MBP (light blue) and V23 (pink). Panels H, L, and P are thick sections made just above the interface between the virus and the receptor. (M) Difference map of the HRV2-V23 complex and HRV2 (pink) and the HRV2 simulation from X-ray data to 15 Å (gold). The threshold of 0.8σ for the difference map is just above the highest noise peak, and the volume represented corresponds to 80% of the volume of V3. (N and O) Sections through V23 and V123 at roughly 10 Å above the viral surface at the fivefold axis. In panel N, the density represents only one repeat, while in panel O there is density shown that represents two repeats per asymmetric unit. These reconstructions (from LaB6 data) each have a resolution of about 20 Å (Table 2) and were calculated including data to 15 Å.
FIG. 5.
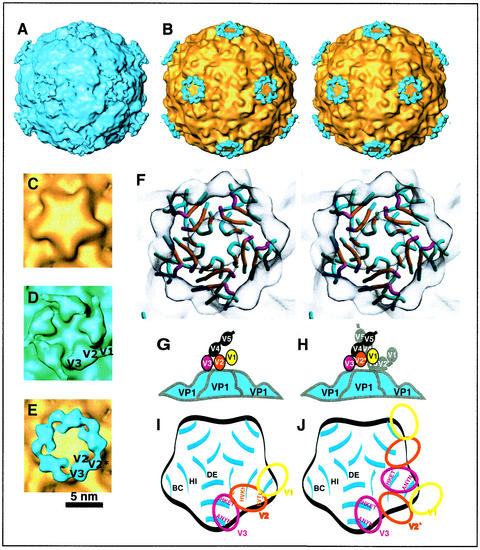
Reconstruction (from FEG data) of the complex between HRV2 and V123 with the individual repeats resolved. For comparison, we used the X-ray map of HRV2 rendered at a resolution of 15 Å with a temperature factor of 500 Å2 (shown in gold) (15). (A) The HRV2-V123 complex; (B) a stereo view down a twofold axis of a difference map with HRV2 shaded in gold and the receptor in blue. A close-up view down the fivefold axis of the native HRV2 (C), a less selective map of the HRV2-V123 complex with the V1 visible at low density but not bound to the viral surface (D), and the more selective map of the HRV2-V123 complex (difference map) (E) are shown in the same orientation. The second repeat when bound to the virus is labeled V2 and when not in contact with the virus is labeled V2* in panel E. Panel E includes only half the data included in panel D, and both have the same resolution of 16 Å. (F) A stereo view down a fivefold axis with the cryoelectron microscopy map shown in gray. The footprint of the receptor repeats on VP1 (colored blue) are colored in magenta for V3 and orange for V2. The V2 and V3 from one receptor molecule bind to sites on two adjacent VP1s, presumably preventing relative movement of the VP1s. (G) and (H) Models of the two binding modes of VLDL-R minireceptors on HRV2. In both modes, V3 is strongly bound. V2 is bound when there is no receptor attached to the neighboring site (panel G). V2 is not bound when there is a receptor bound to the neighboring site (panel H). Panels I and J show schematic diagrams of the footprint of VLDL-R on HRV2 illustrating the residues involved. Panels I and J correspond to the binding patterns shown in panels G and H, respectively. Only the dome on the fivefold axes is represented, and the BC, HI, and DE loops of VP1 are indicated.
RESULTS
Cryoelectron microscopy of HRV2 complexed with minireceptors of VLDL-R.
Cryoelectron microscope images of HRV2-V123 complexes are practically indistinguishable visually from native HRV2 particles. Both appear as smooth spheres with only slight surface texture discernible at high defocus (15). MBP fused to the N terminus of the receptor greatly assisted bacterial expression and subsequent folding of the VLDL-R fragments (30); the MBP was not cleaved off the minireceptor and the entire fusion protein was used in all but one experiment with V123, as reported previously (15). Thus, the MBP prefix is omitted in the following for simplicity; only when MBP was cleaved off is this explicitly noted in the text.
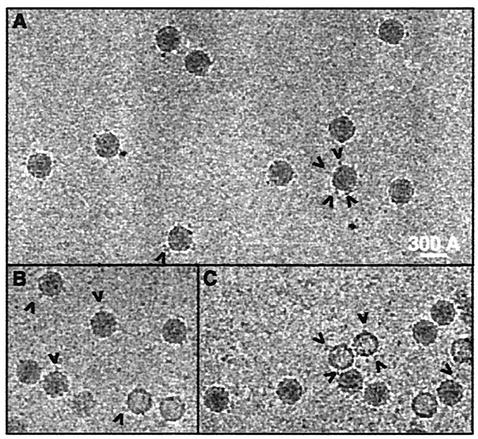
With MBP present, the complexes were seen to be decorated at high defocus (Fig. 3). We attribute this visible decoration to the relatively large MBP (42.7 kDa). There are also a few empty capsids, which are all decorated as for the native capsid. As empty capsids remaining after uncoating do not bind the receptor (21, 23), these must be the “natural top component”—structures that have failed to incorporate RNA and migrate behind the native virus upon sucrose density gradient centrifugation (25). There were, however, too few empty capsids to make a reconstruction.
FIG. 3.
Highly underfocused 3- to 4-μm cryoelectron photomicrographs of HRV2 in complex with a receptor fragment fused to MBP. Note that a few empty capsids (most probably the “natural top component”) are also seen to be decorated. (A) Images of the HRV2-V333 complex; (B and C), the HRV2-V123 complex.
We previously established that, at the resolution attained, the reconstructions of the complex of HRV2 with V123—with or without MBP—are essentially identical (15). While the MBP is sometimes visible in the raw images, it is only faintly visible, if at all, in the reconstructions. Apparently, the MBP does not cause any steric hindrance in this case and must be free to move about.
Overview of binding patterns of the different minireceptors on HRV2.
The smallest receptor fragment investigated in our previous study was V123, in which just two of the three repeats were in contact with the viral surface (Fig. 4A and B). Studying the complex formed between HRV2 and V12 and V23 was thus the obvious first choice to identify the repeats involved. However, bacterial expression of V12 was low, resulting in an insufficient amount of material to study the complex. V23 was expressed at good yields, but it formed a complex with only one repeat attached per asymmetric unit (Fig. 4C, I, M, N, and P). (The footprint of a single repeat is of interest because it defines a minimal binding site.) However, these results do not give a clear answer as to which repeats bind, so we chose to verify binding of each repeat separately.
The concatemer V333 formed a complex with HRV2 with two repeats bound per asymmetric unit (Fig. 4K and L; Table 3. Thus, we conclude that V3 can bind. This result also shows that identical repeats of VLDL-R can bind to different viral epitopes. The reconstruction of the complex with V333 is more diffuse and covers the fivefold axis. This is probably because any one of the three identical repeats can bind, giving several different binding patterns, which are averaged in the reconstruction. In the complex of HRV2 with V122, two repeats are also bound. This may be interpreted as binding of V12 or V22. In either case, the second repeat is bound. Finally, we found that V111 fails to bind to HRV2. In view of the importance of this result, we checked whether V111 is correctly folded. Ca2+ binding was used as a criterion for assessing whether a repeat was in its native conformation (7). We believe that V111 is correctly folded because its 45Ca2+ binding is comparable to that of V123. The failure of V111 to bind to HRV2 at all strongly favors the contention that it is the second and third repeats that bind.
TABLE 3.
List of the constructs of VLDL-R studied and the number of repeats bound to HRV2 per asymmetric unita
| Construct | No. of repeats bound per asymmetric unit | Occupancy (%) |
|---|---|---|
| V12 | No result | |
| V23 | 1 | 30 |
| V33 | 1 | 20 |
| V123 | 2 | 70 |
| V123 (no MBP) | 2 | 70 |
| V333 | 2 | 50 |
| V3333333 | 2 | 50 |
| V12345678 | 2 | 40 |
| V122 | 2 | 50 |
| V111 | 0 |
Except where indicated, MBP is present at the N terminus and a hexa-His tag at the C terminus. An estimate of the occupancy of each receptor fragment was made by measuring the ratio of the maximum density in the receptor domain to the mean density in the capsid. This can only be a rough estimate since, at this resolution, the density of a large domain will be higher than that of a small domain with the same occupancy (35). This is why we used the mean density of the larger capsid proteins and the maximum density in the receptor domain.
We also studied the complex with V12345678 since it is the most closely related to the native VLDL-R we have produced. Again, two repeats are bound, and for this larger construct there is some extra density that points toward the fivefold axis (Fig. 4D and G). The low density in the bound repeats is consistent with a reduction of the number of molecules bound due to steric hindrance (Table 3). The relatively high occupancy of V123 indicates that the MBP does not cause any steric hindrance. The steric hindrance in the V12345678 minireceptor must therefore result from the extra VLDL-R repeats. Table 3 shows the number of repeats that bind for each minireceptor and a rough estimate of the occupancy of each minireceptor.
Comparison of the footprints of different minireceptors bound to HRV2.
In a previous report, we demonstrated that the footprint of V123 lies on an annulus on the dome of the fivefold axis and that the density attributed to V123 corresponds to two repeats only (15). We have now shown that for each case in which two repeats are bound to HRV2 (i.e., for V123 and for V123 without MBP, V122, V12345678, V333, and V3333333), the footprint is essentially the same. By the term “footprint,” we refer to the area on the viral surface covered by the bound receptor. This area includes the epitope—that is, the viral residues that interact with the receptor. In Fig. 4H the footprints of V123 without MBP and V12345678 are compared, and in Fig. 4L the footprints of V122 and V333 are compared. When the minireceptor contains more than three repeats, some additional density is always present and must therefore correspond to the additional repeats. For V12345678, the density points toward the fivefold axis, so it appears that steric hindrance between receptors may limit their occupancy. Some of the visible density can probably be attributed to the fourth and fifth repeats. However, the footprints of the minireceptors on the viral surface always lie on an annulus around the fivefold axis, where there is only space for two repeats to bind per asymmetric unit.
Since in the HRV2 complex with V33 only one repeat binds (Table 3) while two repeats of V333 bind, the question arises as to whether the MBP (and/or the C-terminal hexa-His tag) can prevent attachment. Any such steric hindrance will depend on the position of the MBP and the hexa-His tag with respect to the binding site (Fig. 2C). This observation helps to explain why, contrary to the expectation, only one repeat of V23 binds. We suppose that V3 is bound and that V2 is prevented from binding because of steric hindrance of the MBP.
Resolution of the individual repeats of V123 in the HRV2-V123 complex.
By using a cryoelectron microscope equipped with an FEG and being more selective as to the quality of the particle images retained for reconstruction, we produced a map of the HRV2 in complex with V123 for which the individual binding repeats are resolved (Fig. 5A and B). We used the PFT model-based approach to find the origin and orientation of each particle image. The particle images are selected for inclusion in the reconstruction based on the value of their correlation coefficient with the projected image of the model. By using the selected particle images, a new model is computed and the cycle was repeated to convergence. In general, we retained roughly half the particles in the final reconstruction (Table 2). Roughly one quarter of the particles analyzed were retained in the final reconstruction to give a resolution of 16 Å (Fig. 5A, B, and E). Part of the difficulty in improving the resolution comes from the fact that, as will be seen in the following, the receptor can be bound in two different modes, i.e., with only one repeat (V3) or with two repeats (V2 and V3) bound. V1 is not bound and is free to move in both cases, while V2 is also free to move in the first case. The final reconstruction is the average over these two different modes and so represents the sum of both modes in proportion to their occupancy. When half of the particle images analyzed are retained, the reconstruction has the same resolution, 16 Å, but the receptor density is more diffuse, as is seen in Fig. 5D.
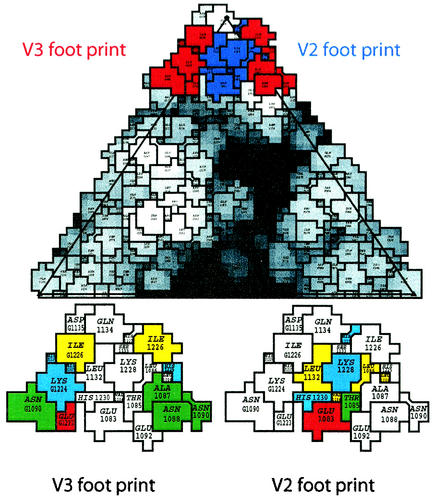
The repeats are arranged on an approximately straight line (Fig. 5D and E). This means that they must be numbered sequentially, and since we have established that the second and third repeats bind but the first does not, the attribution of the domains is unambiguous. The situation must be as shown in Fig. 5D. The V3 repeat is strongly bound since it has a high density and the volume of a single binding repeat. In Fig. 5D, the middle repeat,V2, is of much lower density and is spread out over a larger area. In Fig. 5E, V2 is seen as separated into two much smaller parts—one labeled V2 and which is in contact with HRV2 and one labeled V2* and which is not in contact with HRV2—thus indicating that it is sometimes bound and sometimes not. The repeat V1 is not in contact with the capsid and is only weakly represented, per Fig. 5D. Thus, V1 is not bound to HRV2 and is free to move. V3 binds most strongly with a footprint that includes residues TEKHI of the HI loop and ANYN of the BC loop of one VP1 molecule, i.e., Thr-1222 to Ile-1226 and Ala-1087 to Asn-1090. The footprint of V2 includes residues HKVH of the HI loop and EVTL of the BC loop of the adjacent VP1, i.e., His-1227 to His-1230 and Glu-1083 to Leu-1086. Leu-1132 of the DE loop is also probably included (Fig. 5F, I, and J). The roadmap representation of the V2 and V3 footprints shows that there is a prominent lysine in both cases (Fig. 6).
FIG. 6.
Footprint of V123 on HRV2. The individual footprints V2 (blue) and V3 (red) are indicated on a roadmap (5) of HRV2 at the top. The distance of the amino acid residues from the center of the virion is displayed in different shades of gray to visualize the canyon. The footprints of V3 and V2 (shown at bottom) with the amino acid residues are color coded red for acidic, blue for basic, yellow for hydrophobic, green for hydrophilic neutral, and light blue for histidine. Note that V3 and V2 of one receptor molecule bind to sites on two different symmetry-related VP1 molecules. The first digit signifies VP1; the other digits represent the amino acid number as in the PDB database entry 1FPN.
We suppose that the second repeat is only bound when the neighboring VP1 is not occupied by another receptor molecule. When a V3 is bound to the neighboring VP1, there is steric hindrance between it and V2 (Fig. 5G and H). The similarity model of the structure of V3 is the correct size to fit into the region of density attributed to it in the cryoelectron microscopy map, though we cannot determine its orientation. As expected, the two regions of weak density attributed to V2 are too small to accommodate the model of the structure of V2, since each is only partially occupied. In this experiment we added an excess of the receptor to give a maximum occupancy so that in the majority of cases the second repeat will not be bound. In vivo, there may be fewer receptor molecules available and it is thus highly probable that, in general, both the second and third repeats will be bound.
DISCUSSION
Our technique of using selected soluble fragments of a receptor to determine which repeats bind proved to be more difficult to interpret than we first imagined. Steric hindrance due to the MBP and the hexa-His tag is only one of the steric problems encountered. The relative lack of specificity, as demonstrated by the attachment of two V3 repeats per asymmetric unit is another source of difficulty. Nevertheless, by combining these results with a map of the HRV2-receptor complex, in which the repeats are resolved, it was possible to determine the binding pattern of the receptor and propose an unambiguous model. We have determined the amino acid residues at the viral surface that are covered by the binding repeats—the footprints—however, determination of the subset of these residues that interact with the receptor must await resolution of the atomic structure of the complex.
We have shown explicitly that the second and third repeats of the VLDL-R can bind to HRV2 and that there is no direct evidence that the first repeat binds to HRV2. In fact, V111 does not bind. All our results can be explained by supposing that it is the second and third repeats that bind in the native VLDL-R and that in certain cases V2 or V3 is prevented from binding by steric hindrance of the MBP. In an earlier experiment, it was shown that V123 is among the most effective fragments in regard to the inhibition of HRV2 infection (Moser et al., unpublished). Also, V456 was inactive in a cell protection assay. We have thus ruled out the participation of repeats other than V2 and V3.
The apparent lack of specificity, as manifested by the attachment of two copies of the third repeat, correlates with the supposed electrostatic nature of the interaction. It is also in line with the ability of HRV2 to bind to several members of the LDL receptor family that are composed of various numbers of repeats with similar but not identical amino acid sequences. It is also reminiscent of the large number of structurally and functionally unrelated ligands (e.g., lactoferrin, Pseudomonas exotoxin, and tissue plasminogen activator-inhibitor complexes, among many others) that bind VLDL-R and LRP (34).
An alignment of the eight ligand-binding repeats of human VLDL-R reveals that only the six cysteines and the residues involved in Ca2+ coordination are strictly conserved. Enzyme-linked immunosorbent type assays of phage displaying single repeats indicate low but detectable virus binding of V2 and V3. In this assay, V3 bound more strongly than V2 (Nizet et al., unpublished results); however, we do not know whether both repeats use the same binding site in this test. V2 and V3 have 40% amino acid sequence identity, but it is difficult to explain these differences in binding affinity while the residues contacting the viral surface remain unidentified.
By combining the results from HRV2 in complex with eight different soluble fragments and the higher-resolution structure of the HRV2-V123 complex, we determined the binding pattern of the VLDL-R on HRV2. We have shown that the footprint of the third repeat extends down the north side of the canyon and that this repeat is strongly bound to the charged and polar residues of the HI and BC loops of VP1. The second repeat is more weakly bound to the HI and BC loops of the neighboring VP1 and can be prevented from binding when another receptor molecule is bound on the neighboring VP1. The first repeat is not bound and must be quite mobile, as it is only weakly visible. The fourth and fifth repeats apparently point toward the fivefold axis, which results in steric hindrance and limits the number of receptor molecules that can be bound. In vivo, it is most likely that no more than one or two receptor molecules are bound on any one fivefold axis.
Since each receptor molecule can be bound to two adjacent copies of VP1, and the receptor does not bind to empty capsids remaining after the RNA has been released, we suggest that the bound receptor may “staple” the VP1s together around the fivefold axis, thus inhibiting relative movement of the VP1s. Since uncoating of HRV2 involves a cooperative movement of the capsid proteins around the fivefold axis to open a channel for release of the RNA (13), the receptor must be released before uncoating can occur. It was suggested previously (13) that the virus must dissociate from the receptor in the endosome since the receptor is about 150 Å in length and so would tend to hold the virion at a distance from the membrane. The role of maintaining the virion close to the membrane during transfer of the RNA was then supposed to be fulfilled by the hydrophobic N terminus of VP1 that anchors itself in the membrane. These results reinforce the idea that the only function of the LDL receptors lies in attaching the virion to the host cell for internalization. In contrast to the major-group HRV receptor, ICAM-1, they do not act as catalysts in uncoating.
Acknowledgments
We thank F. Metoz for assistance in operating the computers, J. F. Conway for the CTFmix programs, T. S. Baker for supplying the PFT programs we employed, and I. Goesler for preparing HRV2.
This work was supported in part by the Austrian Science Foundation (grant number P14503-MOB) to D.B.
REFERENCES
- 1.Andersen, O. M., L. L. Christensen, P. A. Christensen, E. S. Sorensen, C. Jacobsen, S. K. Moestrup, M. Etzerodt, and H. C. Thogersen. 2000. Identification of the minimal functional unit in the low density lipoprotein receptor-related protein for binding the receptor-associated protein (RAP). J. Biol. Chem. 275:21017-21024. [DOI] [PubMed] [Google Scholar]
- 2.Andries, K., B. Dewindt, J. Snoeks, L. Wouters, H. Moereels, P. J. Lewi, and P. A. J. Janssen. 1990. Two groups of rhinoviruses revealed by a panel of antiviral compounds present sequence divergence and differential pathogenicity. J. Virol. 64:1117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120-130. [DOI] [PubMed] [Google Scholar]
- 4.Brown, M. S., and J. L. Goldstein. 1986. A receptor-mediated pathway for cholesterol homeostasis. Science 232:34-47. [DOI] [PubMed] [Google Scholar]
- 5.Chapman, M. S. 1993. Mapping the surface properties of macromolecules. Protein Sci. 2:459-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conway, J. F., and A. C. Steven. 1999. Methods for reconstructing density maps of “single” particles from cryoelectron micrographs to subnanometer resolution. J. Struct. Biol. 128:106-118. [DOI] [PubMed] [Google Scholar]
- 7.Dirlam-Schatz, K. A., and A. D. Attie. 1998. Calcium induces a conformational change in the ligand binding domain of the low density lipoprotein receptor. J. Lipid. Res. 39:402-411. [PubMed] [Google Scholar]
- 8.Esnouf, R. M. 1997. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph Model. 15:132-134 and 112-113. [DOI] [PubMed] [Google Scholar]
- 9.Esser, V., L. E. Limbird, M. S. Brown, J. L. Goldstein, and D. W. Russell. 1988. Mutational analysis of the ligand binding domain of the low density lipoprotein receptor. J. Biol. Chem. 263:13282-13290. [PubMed] [Google Scholar]
- 10.Fass, D., S. Blacklow, P. S. Kim, and J. M. Berger. 1997. Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature 388:691-693. [DOI] [PubMed] [Google Scholar]
- 11.Greve, J. M., C. P. Forte, C. W. Marlor, A. M. Meyer, H. Hooverlitty, D. Wunderlich, and A. McClelland. 1991. Mechanisms of receptor-mediated rhinovirus neutralization defined by two soluble forms of ICAM-1. J. Virol. 65:6015-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hadfield, A. T., W. M. Lee, R. Zhao, M. A. Oliveira, I. Minor, R. R. Rueckert, and M. G. Rossmann. 1997. The refined structure of human rhinovirus 16 at 2.15 Å resolution: implications for the viral life cycle. Structure 5:427-441. [DOI] [PubMed] [Google Scholar]
- 13.Hewat, E., E. Neumann, and D. Blaas. 2002. The concerted conformational changes during human rhinovirus 2 uncoating. Mol. Cell 10:317-326. [DOI] [PubMed] [Google Scholar]
- 14.Hewat, E. A., and D. Blaas. 1996. Structure of a neutralizing antibody bound bivalently to human rhinovirus 2. EMBO J. 15:1515-1523. [PMC free article] [PubMed] [Google Scholar]
- 15.Hewat, E. A., E. Neumann, J. F. Conway, R. Moser, B. Ronacher, T. C. Marlovits, and D. Blaas. 2000. The cellular receptor to human rhinovirus 2 binds around the 5-fold axis and not in the canyon: a structural view. EMBO J. 19:6317-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofer, F., M. Gruenberger, H. Kowalski, H. Machat, M. Huettinger, E. Kuechler, and D. Blaas. 1994. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc. Natl. Acad. Sci. USA 91:1839-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hooverlitty, H., and J. M. Greve. 1993. Formation of rhinovirus-soluble ICAM-1 complexes and conformational changes in the virion. J. Virol. 67:390-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeon, H., and G. G. Shipley. 2000. Localization of the N-terminal domain of the low density lipoprotein receptor. J. Biol. Chem. 275:30465-30470. [DOI] [PubMed] [Google Scholar]
- 19.Kim, K. H., P. Willingmann, Z. X. Gong, M. J. Kremer, M. S. Chapman, I. Minor, M. A. Oliveira, M. G. Rossmann, K. Andries, G. D. Diana, F. J. Dutko, M. A. McKinley, and D. C. Pevear. 1993. A comparison of the anti-rhinoviral drug binding pocket in HRV14 and HRV1A. J. Mol. Biol. 230:206-227. [DOI] [PubMed] [Google Scholar]
- 20.Kolatkar, P. R., J. Bella, N. H. Olson, C. M. Bator, T. S. Baker, and M. G. Rossmann. 1999. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 18:6249-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korant, B. D., K. Lonberg Holm, J. Noble, and J. T. Stasny. 1972. Naturally occurring and artificially produced components of three rhinoviruses. Virology 48:71-86. [DOI] [PubMed] [Google Scholar]
- 22.Kurniawan, N. D., A. R. Atkins, S. Bieri, C. J. Brown, I. M. Brereton, P. A. Kroon, and R. Smith. 2000. NMR structure of a concatemer of the first and second ligand-binding modules of the human low-density lipoprotein receptor. Protein Sci. 9:1282-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lonberg-Holm, K., and B. D. Korant. 1972. Early interaction of rhinoviruses with host cells. J. Virol. 9:29-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merritt, E. A., and D. J. Bacon. 1997. Raster3D photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 25.Noble, J. N., and K. Lonberg-Holm. 1973. Interactions of components of human rhinovirus type 2 with HeLa cells. Virology 51:270-278. [DOI] [PubMed] [Google Scholar]
- 26.North, C. L., and S. C. Blacklow. 1999. Structural independence of ligand-binding modules five and six of the LDL receptor. Biochemistry 38:3926-3935. [DOI] [PubMed] [Google Scholar]
- 27.Okun, V. M., B. Ronacher, D. Blaas, and E. Kenndler. 1999. Analysis of common cold virus (human rhinovirus serotype 2) by capillary zone electrophoresis: the problem of peak identification. Anal. Chem. 71:2028-2032. [DOI] [PubMed] [Google Scholar]
- 28.Olson, N. H., P. R. Kolatkar, M. A. Oliveira, R. H. Cheng, J. M. Greve, A. McClelland, T. S. Baker, and M. G. Rossmann. 1993. Structure of a human rhinovirus complexed with its receptor molecule. Proc. Natl. Acad. Sci. USA 90:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prchla, E., E. Kuechler, D. Blaas, and R. Fuchs. 1994. Uncoating of human rhinovirus serotype 2 from late endosomes. J. Virol. 68:3713-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ronacher, B., T. C. Marlovits, R. Moser, and D. Blaas. 2000. Expression and folding of human very-low-density lipoprotein receptor fragments: neutralization capacity toward human rhinovirus HRV2. Virology 278:541-550. [DOI] [PubMed] [Google Scholar]
- 31.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H. J. Hecht, J. E. Johnson, G. Kamer, M. Luo, A. G. Mosser, R. R. Rueckert, B. Sherry, and G. Vriend. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145-153. [DOI] [PubMed] [Google Scholar]
- 32.Skern, T., W. Sommergruber, D. Blaas, C. Pieler, and E. Kuechler. 1984. Relationship of human rhinovirus strain 2 and poliovirus as indicated by comparison of the polymerase gene regions. Virology 136:125-132. [DOI] [PubMed] [Google Scholar]
- 33.Smith, T. J., M. J. Kremer, M. Luo, G. Vriend, E. Arnold, G. Kamer, M. G. Rossman, M. A. McKinlay, G. D. Diana, and M. J. Otto. 1986. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science 233:1286-1293. [DOI] [PubMed] [Google Scholar]
- 34.Strickland, D. K., M. Z. Kounnas, and W. S. Argraves. 1995. LDL receptor-related protein: a multiligand receptor for lipoprotein and proteinase catabolism. FASEB J. 9:890-898. [DOI] [PubMed] [Google Scholar]
- 35.Thouvenin, E., G. Schoehn, F. Rey, I. Petitpas, M. Mathieu, M. C. Vaney, J. Cohen, E. Kohli, P. Pothier, and E. Hewat. 2001. Antibody inhibition of the transcriptase activity of the rotavirus DLP: a structural view. J. Mol. Biol. 307:161-172. [DOI] [PubMed] [Google Scholar]
- 36.Tomassini, J. E., D. Graham, C. M. DeWitt, D. W. Lineberger, J. A. Rodkey, and R. J. Colonno. 1989. cDNA cloning reveals that the major group rhinovirus receptor on HeLa cells is intercellular adhesion molecule 1. Proc. Natl. Acad. Sci. USA 86:4907-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]