Abstract
Cauliflower mosaic virus (CaMV) transactivator/viroplasmin (Tav) is an essential multifunctional viral protein. Dissection of Tav by deletion mutagenesis revealed that the central region is essential for CaMV replication in single cells but that the N- and C-terminal parts are not. Strains with mutations in the central region were defective in the translational transactivator function and could be complemented by coexpressing Gag (capsid protein precursor) and Pol (polyprotein with protease, reverse transcriptase, and RNase H activity) from separate monocistronic plasmids. In contrast, total omission of Tav was only partially complemented by Gag and Pol overexpression from separate plasmids. These results indicate that CaMV basic replication requires both Tav-activated polycistronic translation and some posttranslational function(s) of Tav that is not affected by the deletions in the central region of Tav.
Multifunctionality of viral proteins is one of the most important strategies allowing viruses to survive despite their minimal genome sizes. Furthermore, multifunctional proteins offer attractive model systems to study basic processes of host and nonhost cells; e.g., papovavirus T antigens, which activate viral genome replication and transform cells (3), have helped our understanding of transcription, DNA replication, and cell transformation. The basic strategy in studying a multifunctional protein relies on mutant proteins in which only one (or few) of the multiple functions is affected.
Cauliflower mosaic virus (CaMV), the type member of plant pararetroviruses (Caulimoviridae) (16), encodes a specific multifunctional protein, the transactivator/viroplasmin (Tav). CaMV genomic DNA codes for at least six functional proteins; open reading frames (ORFs) I, II, III, IV, V, and VI encode cell-to-cell movement factor (Mov); aphid transmission factor (Atf); virion-associated protein (Vap); the capsid protein precursor (Gag); a polyprotein with protease, reverse transcriptase, and RNase H domains (Pol); and Tav, respectively (33). Of these six proteins, Mov, Atf, and Vap have roles in virus transport and are dispensable for basic CaMV replication (15, 20, 21, 39). In contrast, Gag and Pol are probably essential for basic replication based on their similarity to retroviral Gag and Pol proteins. We have shown that Tav is also required for CaMV replication in transfected protoplasts (19).
CaMV Tav was first identified as the major component of CaMV inclusion bodies (4), also called viroplasms, which are known to be the site of CaMV protein synthesis (14), reverse transcription (27), and virion assembly (38). Tav has also been shown to transactivate polycistronic translation of the CaMV pregenomic RNA (1). The CaMV genome is transcribed into two major transcripts: the 35S RNA (10), which serves as a template for the reverse transcription step of viral DNA replication (32), and the 19S RNA, which encodes Tav (4). ORFs I, II, III, IV, and V are translated from the polycistronic 35S pregenomic RNA or its spliced derivatives (18); this polycistronic translation has long been known to be promoted by Tav (13). Tav also interacts with a processed form of Gag and has been implicated in virion assembly (11). We have found that it stabilizes Gag and Pol and is essential for virus replication in single cells (19). Tav has also been shown to play a central role in pathogenesis in solanaceous plants (5, 6, 29, 35-37). A recent study has successfully separated the avirulence function and the translational transactivation function by using an agroinfiltration system (30) but did not address virus replication. To elucidate which functions of Tav (including as yet undiscovered functions) are essential for basic virus replication, we established a sensitive detection system to study CaMV replication in transfected cells and performed a deletion mutagenesis of Tav.
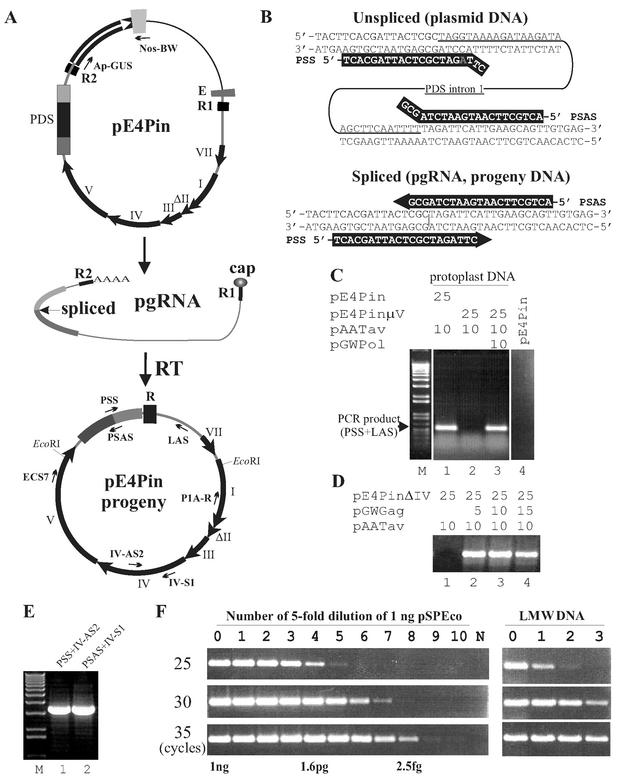
The CaMV vector pE4Pin (Fig. 1A), which has a PCR reporter gene (Pin) in place of the Tav gene, was used in protoplast transfection experiments as a “wild-type” viral construct. Details of the construction of pE4Pin are available on our website (K. Kobayashi, CaMV molecular clones; Kappei's collection, http://www.fmi.ch/members/kappei.kobayashi/KpCT.htm, 2003). In pE4Pin, the Tav gene has been replaced by an Arabidopsis thaliana phytoene desaturase gene segment covering the efficient intron 1 (28) to serve as a reporter for the transcription-reverse transcription replication cycle; while the original replicon contains the intron, the progeny do not (Fig. 1A). CaMV DNA replication was examined by PCR with a primer overlapping the splice junction of the PCR reporter (Fig. 1B and Table 1). Transfection of Brassica rapa cv. Saishin protoplasts was performed as described previously (39). Total low-molecular-weight (LMW) DNA was extracted as described previously (12) after proteinase K treatment. PCR analysis of LMW DNA from transfected protoplasts revealed that pE4Pin did not give rise to replicated progeny unless cotransfected with Tav-expressing plasmid pAATav (Fig. 1C, lane 1, and 2C, lane 14), confirming that Tav is essential for CaMV replication. CaMV DNA replication was not observed when mutant vectors encoding defective Pol or Gag (pE4PinμV or pE4PinΔIV) were cotransfected instead of pE4Pin together with pAATav, indicating that three CaMV gene products, Gag, Pol, and Tav, are required for basic CaMV replication. The last two mutant vectors could be fully complemented by coexpressing Pol and Gag from separate monocistronic plasmids (Fig. 1C and D) (K. Kobayashi, http://www.fmi.ch/members/kappei.kobayashi/KpCT.htm), confirming the fidelity of the detection system. Two different primer pairs amplified two overlapping DNA fragments derived from full-length pE4Pin progeny, indicating that pE4Pin can give rise to complete CaMV DNA replication (Fig. 1E).
FIG. 1.
CaMV vector and reporter-targeted PCR for studying replication. (A) Schematic representation of CaMV genome replication from the vector pE4Pin. Thick black arrows, CaMV ORFs I to VII; ΔII, deleted version of ORF II as in CaMV strain CM4-184; E, synthetic enhancer; R1 and R2, small terminal repeats of the CaMV replicon in provirus-like form that become R in the circular form; white arrow and the accompanying gray box, β-glucuronidase coding sequence and nopaline synthase gene polyadenylation signal; dark gray, black, and light gray boxes, A. thaliana phytoene desaturase (PDS) coding sequence consisting of exon 1, intron 1, and exon 2; thin black line, bacterial vector sequence; gray line, CaMV noncoding sequence. The primary transcript with small terminal repeats is capped, spliced, and polyadenylated to generate pregenomic CaMV RNA (pgRNA), which serves as the template for reverse transcription. Note that the progeny DNA, like the wild-type virus, has only a single copy of the R sequence. Thin arrows, PCR primers used for analysis of viral replication. The positions of EcoRI sites used for cloning of the spliced version of the PDS sequence into pBluescript (the control plasmid, pSPEco) are also indicated. (B) Reporter-targeted PCR. Spliced-PDS-specific primers (PSS and PSAS; white letters) cannot prime DNA synthesis from the unspliced sequence (top) but can do so from the spliced version (bottom). Gray “A,” internal mismatch. (C) Detection of CaMV replication by reporter-targeted PCR. Total LMW DNA from protoplasts transfected with the plasmids at indicated amounts (micrograms) was analyzed by PCR with primers PSS and LAS. The total amounts of plasmids were adjusted to 50 μg with pBluescript. pE4PinμV, an ORF V frameshift mutant vector, served as negative control and was complemented by coexpression of Pol from a separate plasmid (lanes 2 and 3). Total LMW DNA was isolated 3 days posttransfection and analyzed by 35 cycles of PCR. The input plasmid DNA (pE4Pin; 10 ng/reaction) was analyzed in the same way as the protoplast-derived DNA (lane 4). (D) Complementation of ORF IV defects by coexpression of Gag from a separate plasmid. Protoplasts were transfected with the indicated amounts of ORF IV deletion mutant vector pE4PinΔIV and pAATav, without pGWGag (lane 1) or with different amounts of pGWGag (lanes 2 to 4). (E) Whole-genome amplification of pE4Pin progeny. DNA from the DNase-treated virion fraction from protoplasts transfected with pE4Pin and pAATav was subjected to PCR with the primers indicated. (F) Semiquantitative PCR. (Left) Standard experiment. The control plasmid, pSPEco, was serially diluted (fivefold each time) with 1 ng of pE4Pin/μl to mimic total LMW DNA from protoplasts and subjected to reporter-targeted PCR with PSS and LAS primers and 25, 30, or 35 reaction cycles. (Right) Representative reporter-targeted PCR result. Total LMW DNA from protoplasts transfected with pE4Pin and pAATav was serially diluted fivefold and analyzed.
TABLE 1.
PCR primers and reaction conditions
| Targeta | Primer names, sequencesb | Reaction conditions |
|---|---|---|
| Pin and LIGc | PSS, TCACGATTACTCGCTAGATTC; LAS, GCCTTAGCATCTTTTCCC | 94°C for 5 min; then 94°C for 30 s, 62°C for 30 s, and 72°C for 2 min for 25-35 cycles |
| Whole genomed | PSS, TCACGATTACTCGCTAGATTC; IV-AS2, GGCCTTCTCTTGCTCGTC; PSAS, ACTGCTTCAATGAATCTAGCG; IV-S1, GGTGAAGGACCATCTAGATAC | 94°C for 5 min; then 94°C for 30 s, 62°C for 30 s, and 72°C for 8 min for 35 cycles |
| GUS and nos3′e | Ap-GUS, GCGGATCCGGGGCCCATGTTACGTCCTGTAGAAACCC; nos-BW, GGAATTCCGTACGGCATGCAAATGTATATTGCGGGACTC | 94°C for 5 min; then 94°C for 30 s, 62°C for 30 s, and 72°C for 2 min for 25-35 cycles |
| EcoRI fragmentf | ECS7, CTGGGGAGGTATGTTAAAAGC; I-AS2, GGGCTATATCAATGGCCCAATTGC | 94°C for 5 min; then 94°C for 30 s, 62°C for 30 s, and 72°C for 6 min for 35 cyclesg |
Target sequence to be amplified.
Shown from 5′ to 3′.
LIG, large intergenic region.
Whole pE4Pin progeny genome was amplified as two overlapping fragments.
Present in pE4Pin vector but not in its progeny.
Cloned to give PCR standard plasmid pSPEco.
Pfu DNA polymerase (Promega) was used in place of Taq DNA polymerase.
CaMV DNA replication levels were determined by semiquantitative PCR. The input CaMV vector DNA persisting in the LMW DNA preparation from transfected protoplasts was present at between 0.2 and 1 ng/μl and was constant within a single batch of protoplasts (revealed by PCR with Ap-GUS and nos-BW primers; data not shown). Protoplast-derived LMW DNA was analyzed by PCR using 25, 30, and 35 cycles in parallel with standard DNA clone pSPEco (Fig. 1A legend), which had been serially diluted with 1 ng of pE4Pin/μl (Fig. 1F). The PCR analyses gave proportional amplification in the range of 8 pg to 0.5 fg of pSPEco DNA/reaction, corresponding to 2 × 106 to 125 copies/reaction (Fig. 1F, left). Accumulation of pE4Pin progeny was calculated to be around 8 pg/reaction (Fig. 1F, right). Taking into account that 333 to 1,667 productive cells were analyzed in a single PCR (calculated from the number of transfected cells [106], the portion of transfected cells used for DNA extraction [one-third], the proportion of DNA analyzed [5 μl/50 μl], a protoplast survival rate of 20 to 50% as examined by observing the cell shape under the microscope, and an infection rate of 5 to 10% as determined by immunofluorescence [8]), transfection with pE4Pin and pAATav gave rise to between 1,200 and 6,000 copies of viral DNA accumulated in one productive cell. These levels are about 1 order of magnitude lower than the previously estimated CaMV DNA copy number per cell in CaMV-infected plants (26) or CaMV particle-infected protoplasts (25). The lower progeny accumulation level from pE4Pin is at least partially due to the lower splicing rate of the parental strain, CM4-184, which has a 420-bp deletion within ORF II (18), because a similar construct derived from strain CM1841 replicated to a greater extent (K. Kobayashi and T. Hohn, unpublished data). The detection limit with 35 cycles of PCR was 0.08 to 0.38 copies/cell, indicating that the absence of a specific signal at this point can be regarded as the absence of CaMV DNA replication.
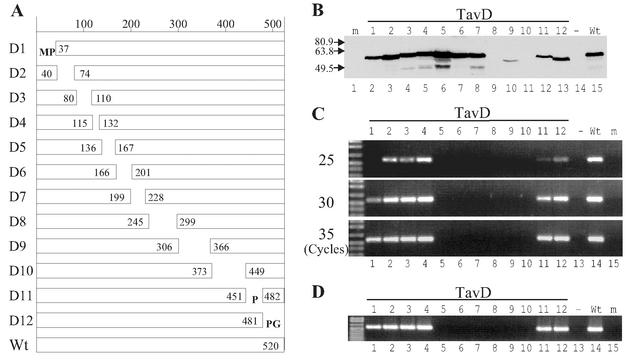
To determine the ability of Tav to support CaMV DNA replication, we constructed a series of deletion mutant proteins (Fig. 2A). The cutoff borders of the deletions were preferentially at proline-glycine dipeptides, because these amino acids are found only rarely within α-helices and therefore such deletions are expected to have a minimal effect on protein conformation. Total omission of Tav resulted in a total block of CaMV replication (Fig. 2C and D, lanes 13), supporting our previous finding that Tav is essential for one or more steps of the CaMV replication cycle (19). TavD1, -D2, -D3, -D4, -D11, and -D12 supported CaMV DNA replication, although to different levels: TavD4, nearly 100% of wild type; TavD2 and -D3, 50 to 70%; TavD12, 20%; TavD11, 5%; TavD1, 1% (Fig. 2C). The regions deleted in TavD1, -D2, -D3, -D11, and -D12 might have some minor role(s) in CaMV basic replication, but it is also possible that the associated mutations slightly alter the conformation of Tav and thus affect functions in which other regions play a central role. The regions deleted in the TavD8, -D9, and -D10 mutant proteins were essential for the stability of the Tav protein (Fig. 2B), and therefore it was not possible to determine whether they are required for replication. Mutant proteins TavD5, -D6, and -D7 were stable but supported CaMV replication only marginally (TavD5; see Fig. 3A, lane 3) or not at all (TavD6 and -D7; Fig. 2C). This suggests that region 5 is important, and that regions 6 and 7 are essential, for virus replication.
FIG. 2.
Ability of Tav to support CaMV replication. (A) Schematic representation of Tav deletion mutant proteins. The amino acid numbers at the cutoff borders are shown. Additional amino acids in mutant TavD1, -D11, and -D12 proteins are shown in single-letter format. Wt, wild type. (B) Western blot analysis for the accumulation of Tav deletion mutant proteins in transfected protoplasts. Lane m, mock-transfected protoplasts; lanes 1 to 12, transfection with pE4Pin (25 μg) and pAATavD1 to -D12 (15 μg), respectively; lane −, transfection with pE4Pin (25 μg) alone; lane Wt, transfection with pE4Pin (25 μg) and pAATav (wild type; 15 μg). In all transfection experiments, the total amounts of plasmids were adjusted to 50 μg with pBluescript. Molecular mass markers (in kilodaltons) are indicated on the left. (C) Semiquantitative reporter-targeted PCR analysis of total LMW DNA from transfected protoplasts. Protoplasts were transfected as in panel B and analyzed by reporter-targeted PCR of 25, 30, and 35 cycles as in Fig. 1F. (D) PCR analysis of encapsidated DNA. DNA from DNase-treated virion fractions from protoplasts transfected as in panel B was analyzed by reporter-targeted PCR (35 cycles).
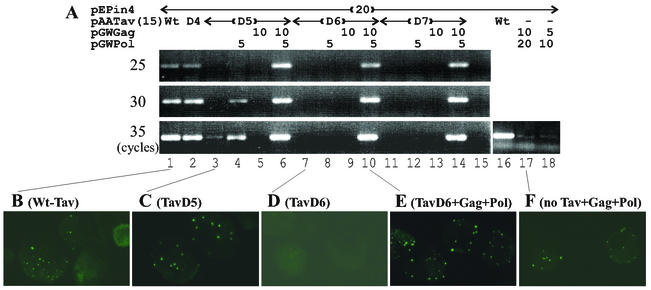
FIG. 3.
The translational transactivation function of Tav is essential for basic CaMV replication. (A) Complementation of translational transactivation-defective Tav mutant proteins by coexpression of Gag and/or Pol. Protoplasts were transfected with pE4Pin, pGWGag, and pGWPol in the amounts indicated together with 15 μg of Tav or Tav mutant plasmids and analyzed as in Fig. 3C. −, no Tav expression plasmid. Wt, wild type. (B to F) Protoplasts transfected as indicated for panel A were stained with an anti-CaMV Gag antibody after 3 days of culture.
To analyze whether the replicated DNA is encapsidated, we treated the virion fraction (9) from transfected protoplasts with DNase I, extracted DNA, and analyzed it by PCR. All mutant Tav proteins that supported CaMV DNA replication also supported encapsidation (Fig. 2D). The results are consistent with the previously proposed model that CaMV DNA replication is coupled with packaging (24, 32).
The regions deleted in TavD6 and -D7 are located within mini-TAV, the minimal region required for transactivation of polycistronic translation (7), suggesting that this function is essential for basic CaMV replication. The translational transactivation activity of mini-TAV mutant proteins was measured in a transient-expression assay using an artificial bicistronic reporter construct (31) for comparison with the pE4Pin-derived CaMV replication levels. Three independent triplicate experiments indicated that TavD4 had wild-type levels (100%) of translational transactivator activity and that TavD5 had quite low but significant levels (5%), while TavD6 and -D7 exhibited only marginal transactivation activity (1.3 and 1.8%, respectively). The results demonstrated good correlation between the translational transactivator function and viral DNA replication levels (Fig. 2C and 3A). However, it remained possible that the translational transactivation function is dispensable for replication and that TavD5, -D6, and -D7 have defects in another essential function of Tav. To test this possibility, we circumvented the need for translation of Gag and Pol from a polycistronic RNA by coexpressing the essential Gag and Pol proteins from the monocistronic constructs pGWGag and pGWPol (K. Kobayashi, http://www.fmi.ch/members/kappei.kobayashi/KpCT.htm) together with wild-type and mutant Tav constructs.
When coexpressed with Gag and Pol, TavD5, -D6, and -D7 supported CaMV DNA replication at wild-type levels or even higher (Fig. 3A, lanes 6, 10, and 14). In the presence of TavD6 or -D7, omission of either Gag or Pol abolished CaMV DNA replication, suggesting that expression of both Gag and Pol from natural CaMV RNA depends on Tav (Fig. 3A, lanes 8, 9, 12, and 13). TavD5 supported a marginal level of CaMV DNA replication (less than 1/1,000 of wild type), and replication was enhanced about 10-fold by coexpression of Pol alone, suggesting that Pol is more dependent on the transactivation function than Gag (Fig. 3A, lanes 3 and 4). Immunofluorescence data confirmed that TavD6 was unable to activate Gag expression (Fig. 3D), while Gag expressed from a separate plasmid accumulated to levels similar to, or even higher than, those in cells transfected with pE4Pin and pAATav (Fig. 3A and E). Gag accumulation was observed in cells transfected with pE4Pin and pAATavD5 (Fig. 3C). Since the transfection efficiency was not high enough to detect Gag by Western blotting, we could not directly compare the amounts of Gag expressed from pE4Pin in the presence of wild-type Tav and TavD5. Coexpression of Gag with TavD5 abolished CaMV DNA replication from pE4Pin (Fig. 3A, lane 5), suggesting that Gag might down-regulate expression of Pol.
The results shown above clearly demonstrate that the translational transactivation function of Tav is essential for basic CaMV replication, confirming that expression of Gag and Pol from CaMV pregenomic RNA and/or its spliced derivatives depends highly on Tav; i.e., Gag and Pol are produced by Tav-activated polycistronic translation.
Since we have shown that Tav can stabilize Gag and Pol in transfected cells (19), we examined if CaMV DNA replication could occur in cells cotransfected with pE4Pin and Gag and Pol expression plasmids but not with the Tav expression plasmid. The level of CaMV DNA replication when Gag and Pol were provided from monocistronic plasmids (in the absence of Tav) was about 10,000 times lower than that when Gag and Pol were expressed from the polycistronic plasmid in the presence of Tav (Fig. 3A, lanes 16 to 18). In contrast, the expression of Gag and Pol from separate plasmids gave rise to high levels of CaMV DNA replication in the presence of a mutant Tav defective in promoting polycistronic translation (Fig. 3A, lanes 6, 10, and 14). In contrast to our previous report (19), Gag accumulation was observed in the absence of Tav when Gag and Pol were provided from separate plasmids, although seemingly to much lower levels than those in the presence of the mutant TavD6 protein (cf. Fig. 3F and E). The results suggest that Tav also has a posttranslational function that is essential for efficient basic replication and that is not affected in the mutant TavD5, -D6, and -D7 proteins.
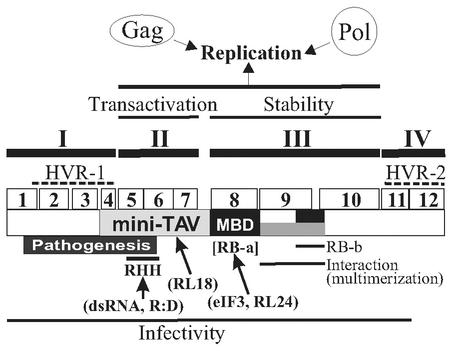
We can distinguish four domains of Tav, each consisting of several regions characterized by individual deletion mutations (Fig. 4): deletions within domains I (regions 1 to 4) and IV (regions 11 and 12), consisting of the first 132 and last 69 amino acids, respectively, did not abolish viral replication in protoplasts; deletions in domain II (regions 5 to 7), consisting of amino acids 137 to 227, caused defects in basic viral replication; deletions in domain III (regions 8 to 10), covering amino acids 246 to 448, gave rise to unstable protein products.
FIG. 4.
Schematic representation of the Tav domain map. I, II, III, and IV, Tav domains distinguished in this study; open boxes 1 to 12, regions deleted in the correspondingly numbered mutant proteins used; black boxes, RNA binding domains (RB-a [MBD] and RB-b); gray box, interaction domain; dotted lines, HVR-1 and -2. Other lines indicate the positions of regions involved in the function or property indicated. RHH, RNase H homology. Interaction partners are shown in parentheses. R:D, RNA-DNA hybrid; RL18 and RL24, ribosomal proteins L18 and L24, respectively; eIF3, eukaryotic translational initiation factor 3; dsRNA, double-stranded RNA.
Intraspecific comparison of Tav amino acid sequences revealed two hypervariable regions (HVRs) (34). They correspond to domains I and IV, determined in this study; domains II and III are more conserved among CaMV isolates. This finding suggested that the conserved domains play a role in basic replication, while the variable domains would be responsible for host selection and/or pathogenesis. This function of the variable domains has been repeatedly shown by recombination experiments with CaMV isolates that have different host ranges or pathogenic phenotypes (see, e.g., references 6, 17, 37, and 40). In contrast, the function(s) of the conserved domains in the CaMV replication cycle remains poorly understood, while their roles in molecular interaction and translational transactivation have been well documented (2, 7, 22, 31).
Region 1, the most N-terminal part of domain I, is conserved among CaMV isolates but is not essential for basic replication. Recently, the N-terminal region of Tav was shown to have a role in viroplasm formation (M. Haas, A. Geldreich, M. Keller, and P. Yot, Abstr. 12th Int. Congr. Virol., p. 197, 2002). The reduced CaMV replication supported by TavD1 might be due to the inability of TavD1 to form normal viroplasm.
Domain II partially overlaps with Mini-TAV, the fragment of Tav which is still able to activate polycistronic translation when available in a large amount (7) and which interacts with ribosomal protein L18 (22, 31) and double-stranded RNA (2). Although region 4 forms part of one of the HVRs, it is included in Mini-TAV (34). Our detailed mutational analysis narrowed the domain essential for transactivation activity to regions 5, 6, and 7 and excluded region 4. Domain II mutant proteins were not active in the transactivation assay with artificial bicistronic mRNA, while TavD4 was still as active as wild-type Tav. In the replication assay, domain II mutations could be suppressed by providing Gag and Pol from separate monocistronic plasmids, showing that the defects are caused by an inability to express Gag and Pol from the polycistronic CaMV RNA.
Domain III has long been known to be multifunctional, although we could not analyze its role in basic CaMV replication due to instability of the mutant proteins. It includes a multiple binding domain (MBD; amino acids 242 to 310) with affinity for both protein (31) and RNA (7), a second RNA binding domain, and an interaction domain, which was mapped by competition experiments for transactivation activity (7). Recently Li and Leisner suggested that a region included in our domain III has a pivotal role in the self-assembly of Tav, although this has been shown only with yeast two-hybrid experiments (23). Because domains I and IV are dispensable for basic replication and because domain II defects can be complemented by coexpressing Gag and Pol from separate plasmids, it is likely that at least part of domain III supplies the posttranslational function required for efficient basic replication. It will be of great interest to discover the relationship between the domain responsible for the posttranslation function and the MBD that interacts with a variety of molecules and that has a role in efficient transactivation. Further mutational analysis to uncover the molecular basis of Tav's posttranslational function and to investigate the correlation between the translational and posttranslational functions of Tav is in progress.
Acknowledgments
We thank Helen Rothnie for critical reading of the manuscript, Matthias Mueller for technical assistance, and all members of our lab for fruitful discussions.
This work was supported by the Novartis Research Foundation.
REFERENCES
- 1.Bonneville, J. M., H. Sanfaçon, J. Fütterer, and T. Hohn. 1989. Posttranscriptional trans-activation in cauliflower mosaic virus. Cell 59:1135-1143. [DOI] [PubMed] [Google Scholar]
- 2.Cerritelli, S. M., O. Y. Fedoroff, B. R. Reid, and R. J. Crouch. 1998. A common 40 amino acid motif in eukaryotic RNases H1 and caulimovirus ORF VI proteins binds to duplex RNAs. Nucleic Acids Res. 26:1834-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole, C. N. 1996. Polyomavirinae: the viruses and their replication, p. 917-945. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 4.Covey, S. N., and R. Hull. 1981. Transcription of cauliflower mosaic virus DNA. Detection of transcripts, properties, and location of the gene encoding the virus inclusion body protein. Virology 111:463-474. [DOI] [PubMed] [Google Scholar]
- 5.Daubert, S. D., and G. Routh. 1990. Point mutations in cauliflower mosaic virus gene VI confer host-specific symptom changes. Mol. Plant-Microbe Interact. 3:341-345. [DOI] [PubMed] [Google Scholar]
- 6.Daubert, S. D., J. Schoelz, L. Debao, and R. J. Shepherd. 1984. Expression of disease symptoms in cauliflower mosaic virus genomic hybrids. J. Mol. Appl. Genet. 2:537-547. [PubMed] [Google Scholar]
- 7.DeTapia, M., A. Himmelbach, and T. Hohn. 1993. Molecular dissection of the cauliflower mosaic virus translational transactivator. EMBO J. 12:3305-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furusawa, I., N. Yamaoka, T. Okuno, M. Yamamoto, M. Kohno, and H. Kunoh. 1980. Infection of turnip protoplasts with cauliflower mosaic virus. J. Gen. Virol. 48:431-435. [DOI] [PubMed] [Google Scholar]
- 9.Gardner, R. C., and R. J. Shepherd. 1980. A procedure for rapid isolation and analysis of cauliflower mosaic virus DNA. Virology 106:159-161. [DOI] [PubMed] [Google Scholar]
- 10.Guilley, H., R. K. Dudley, G. Jonard, E. Balázs, and K. E. Richards. 1982. Transcription of cauliflower mosaic virus DNA: detection of promoter sequences, and characterization of transcripts. Cell 30:763-773. [DOI] [PubMed] [Google Scholar]
- 11.Himmelbach, A., Y. Chapdelaine, and T. Hohn. 1996. Interaction between cauliflower mosaic virus inclusion body protein and capsid protein: implications for viral assembly. Virology 217:147-157. [DOI] [PubMed] [Google Scholar]
- 12.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 13.Hohn, T., J. M. Bonneville, J. Fütterer, K. Gordon, J. Jiricny, S. Karlsson, H. Sanfaçon, M. Schultze, and M. DeTapia. 1990. The use of 35S RNA as either messenger or replicative intermediate might control the CaMV replication cycle, p. 67-78. In T. P. Pirone and J. G. Shaw (ed.), Viral genes and plant pathogens. Springer, New York, N.Y.
- 14.Hohn, T., and J. Fütterer. 1991. Pararetroviruses and retroviruses: a comparison of expression strategies. Semin. Virol. 2:55-69. [Google Scholar]
- 15.Howarth, A. J., R. C. Gardner, J. Messing, and R. J. Shepherd. 1981. Nucleotide sequence of naturally occurring deletion mutants of cauliflower mosaic virus. Virology 112:678-685. [DOI] [PubMed] [Google Scholar]
- 16.Hull, R. 2002. Matthews' plant virology. Academic Press, London, United Kingdom.
- 17.Király, L., A. B. Cole, J. E. Bourque, and J. E. Schoelz. 1999. Systemic cell death is elicited by the interaction of a single gene in Nicotiana clevelandii and gene VI of cauliflower mosaic virus. Mol. Plant-Microbe Interact. 12:919-925. [Google Scholar]
- 18.Kiss-László, Z., S. Blanc, and T. Hohn. 1995. Splicing of cauliflower mosaic virus 35S RNA is essential for viral infectivity. EMBO J. 14:3552-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi, K., S. Tsuge, H. Nakayashiki, K. Mise, and I. Furusawa. 1998. Requirement of cauliflower mosaic virus open reading frame VI product for viral gene expression and multiplication in turnip protoplasts. Microbiol. Immunol. 42:377-386. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi, K., S. Tsuge, L. Stavolone, and T. Hohn. 2002. The cauliflower mosaic virus virion-associated protein is dispensable for viral replication in single cells. J. Virol. 76:9457-9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leh, V., E. Jacquot, A. Geldreich, T. Hermann, D. Leclerc, M. Cerutti, P. Yot, M. Keller, and S. Blanc. 1999. Aphid transmission of cauliflower mosaic virus requires the viral PIII protein. EMBO J. 18:7077-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leh, V., P. Yot, and M. Keller. 2000. The cauliflower mosaic virus translational transactivator interacts with the 60S ribosomal subunit protein L18 of Arabidopsis thaliana. Virology 266:1-7. [DOI] [PubMed] [Google Scholar]
- 23.Li, Y., and S. M. Leisner. 2002. Multiple domains within the cauliflower mosaic virus gene VI product interact with the full-length protein. Mol. Plant-Microbe Interact. 15:1050-1057. [DOI] [PubMed] [Google Scholar]
- 24.Marsh, L., and T. J. Guilfoyle. 1987. CaMV replication intermediates are encapsidated into virion-like particles. Virology 161:129-137. [DOI] [PubMed] [Google Scholar]
- 25.Maule, A. J. 1983. Infection of protoplasts from several Brassica species with cauliflower mosaic virus following inoculation using polyethylene glycol. J. Gen. Virol. 64:2655-2660. [Google Scholar]
- 26.Maule, A. J., R. Hull, and J. Donson. 1983. The application of spot hybridization to the detection of DNA and RNA viruses in plant tissue. J. Virol. Methods 6:215-224. [DOI] [PubMed] [Google Scholar]
- 27.Modjtahedi, N., M. Volovitch, L. Sossountzov, Y. Habricot, J. M. Bonneville, and P. Yot. 1984. CaMV induced viroplasms support viral DNA synthesis in a cell free system. Virology 133:289-300. [DOI] [PubMed] [Google Scholar]
- 28.Noad, R. J., D. S. Turner, and S. N. Covey. 1997. Expression of functional elements inserted into the 35S promoter region of infectious cauliflower mosaic virus replicons. Nucleic Acids Res. 25:1123-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palanichelvam, K., A. B. Cole, M. Shababi, and J. E. Schoelz. 2000. Agroinfiltration of cauliflower mosaic virus gene VI elicits hypersensitive response in Nicotiana species. Mol. Plant-Microbe Interact. 13:1275-1279. [DOI] [PubMed] [Google Scholar]
- 30.Palanichelvam, K., and J. E. Schoelz. 2002. A comparative analysis of the avirulence and translational transactivator functions of gene VI of cauliflower mosaic virus. Virology 293:225-233. [DOI] [PubMed] [Google Scholar]
- 31.Park, H. S., A. Himmelbach, K. S. Browning, T. Hohn, and L. A. Ryabova. 2001. A plant viral “reinitiation” factor interacts with the host translational machinery. Cell 106:723-733. [DOI] [PubMed] [Google Scholar]
- 32.Pfeiffer, P., and T. Hohn. 1983. Involvement of reverse transcription in the replication of cauliflower mosaic virus: a detailed model and test of some aspects. Cell 33:781-789. [DOI] [PubMed] [Google Scholar]
- 33.Rothnie, H. M., Y. Chapdelaine, and T. Hohn. 1994. Pararetroviruses and retroviruses: a comparative review of viral structure and gene expression strategies. Adv. Virus Res. 44:1-67. [DOI] [PubMed] [Google Scholar]
- 34.Sanger, M., S. D. Daubert, and R. M. Goodman. 1991. The regions of sequence variation in caulimovirus gene VI. Virology 182:830-834. [DOI] [PubMed] [Google Scholar]
- 35.Schoelz, J. E., K. B. Goldberg, and J. M. Kiernan. 1991. Expression of cauliflower mosaic virus (CaMV) gene VI in transgenic Nicotiana bigelovii complements a strain of CaMV defective in movement in nontransformed N. bigelovii. Mol. Plant-Microbe Interact. 4:350-355. [Google Scholar]
- 36.Schoelz, J. E., R. J. Shephard, and R. D. Richins. 1986. Properties of an unusual strain of cauliflower mosaic virus. Phytopathology 76:451-454. [Google Scholar]
- 37.Schoelz, J. E., and R. J. Shepherd. 1988. Host range control of cauliflower mosaic virus. Virology 162:30-37. [DOI] [PubMed] [Google Scholar]
- 38.Shepherd, R. J. 1976. DNA viruses of higher plants. Adv. Virus Res. 20:305-339. [DOI] [PubMed] [Google Scholar]
- 39.Tsuge, S., K. Kobayashi, H. Nakayashiki, T. Okuno, and I. Furusawa. 1994. Replication of cauliflower mosaic virus ORF 1 mutants in turnip protoplasts. Ann. Phytopathol. Soc. Japan 60:27-35. [Google Scholar]
- 40.Wintermantel, W. M., E. J. Anderson, and J. E. Schoelz. 1993. Identification of domains within gene VI of cauliflower mosaic virus that influence systemic infection of N. bigelovii in a light-dependent manner. Virology 196:789-798. [DOI] [PubMed] [Google Scholar]