Abstract
The late stages of assembly of herpes simplex virus (HSV) and other herpesviruses are not well understood. Acquisition of the final virion envelope apparently involves interactions between viral nucleocapsids coated with tegument proteins and the cytoplasmic domains of membrane glycoproteins. This promotes budding of virus particles into cytoplasmic vesicles derived from the trans-Golgi network or endosomes. The identities of viral membrane glycoproteins and tegument proteins involved in these processes are not well known. Here, we report that HSV mutants lacking two viral glycoproteins, gD and gE, accumulated large numbers of unenveloped nucleocapsids in the cytoplasm. These aggregated capsids were immersed in an electron-dense layer that appeared to be tegument. Few or no enveloped virions were observed. More subtle defects were observed with an HSV unable to express gD and gI. A triple mutant lacking gD, gE, and gI exhibited more severe defects in envelopment. We concluded that HSV gD and the gE/gI heterodimeric complex act in a redundant fashion to anchor the virion envelope onto tegument-coated capsids. In the absence of either one of these HSV glycoproteins, envelopment proceeds; however, without both gD and gE, or gE/gI, there is profound inhibition of cytoplasmic envelopment.
Herpes simplex virus (HSV) is in many respects the best studied of the herpesviruses and the paradigm for α-herpesvirus replication and assembly. As with other herpesviruses, viral DNA is packaged into nucleocapsids in the nucleus (43). To escape the nucleus, capsids become enveloped by regions of the inner nuclear envelope and then move into the cytoplasm by fusion with the outer lamellae of the nuclear envelope (45, 47). In the cytoplasm, nucleocapsids bind several tegument proteins (reviewed in reference 35). Tegument-coated capsids bind onto cytoplasmic membranes, including the trans-Golgi network (TGN) and endosomes, enriched in viral glycoproteins (reviewed in reference 24). Nascent virions bud into the lumen of these cytoplasmic vesicles as the virion envelope wraps around tegument-coated capsids. Enveloped virions are transported to the cell surface where fusion of cytosolic vesicles with the plasma membrane delivers virus particles into the extracellular environment or onto the cell surface.
The molecular details of how α-herpesviruses become enveloped at either nuclear or cytoplasmic membranes are poorly characterized. In the case of cytoplasmic envelopment, it is clear that viral membrane glycoproteins accumulate extensively in the TGN or endosomes and this likely promotes incorporation into the virion envelope (reviewed in references 24 and 35). The cytosolic domains of these membrane glycoproteins probably provide a surface on which the last stage of assembly, i.e., acquisition of the virion envelope, occurs. However, it is not clear which of the viral membrane proteins are important for tethering of tegument-coated capsids onto the envelope. HSV mutants lacking individual glycoproteins known to be essential for virus replication, gD, gB, gH, and gL, display no obvious defects in envelopment (8, 19, 31, 44). In each case, normal numbers of enveloped particles are produced, although these particles are not infectious because they lack the machinery to enter cells. When either gD or gH is modified with endoplasmic reticulum localization signals so that they do not reach the TGN, the glycoproteins are not incorporated into nascent virions and normal numbers of enveloped particles are produced but these particles lack gD or gH (45, 52). Therefore, any one of the so-called essential glycoproteins is not necessary for cytoplasmic envelopment. Moreover, no major defects in envelopment have been noted with HSV mutants lacking other so-called nonessential glycoproteins: gE, gG, gI, gJ, and gM (2, 3, 13, 32, 49). Recently, clues as to how cytoplasmic envelopment occurs came from studies of the pig α-herpesvirus, pseudorabies virus (PRV). Large numbers of nucleocapsids accumulated in the cytoplasm of cells infected with PRV double mutants lacking gE, or just the cytoplasmic (CT) domain of gE, and a second glycoprotein gM (6, 7). These cytoplasmic capsids were immersed in an electron-dense array that appeared to be tegument and which stained with anti-UL49 antibodies (7). Therefore, the CT domains of PRV gE and gM act in a redundant fashion to interact with tegument proteins coating the surface of nucleocapsids; when one or the other is present, there is still significant envelopment.
The tegument proteins that bridge the virion envelope onto nucleocapsids are also ill defined, although some candidates have recently come to light. Many HSV or PRV tegument proteins can be deleted without affecting virus assembly (35). The PRV UL49, which encodes a homologue of HSV VP22, interacts with the CT domains of both gE and gM (20). Nevertheless, PRV UL49 can be deleted with only minor effects on virus envelopment or growth and without affecting the subcellular localization of gE/gI or gM (20). An HSV tegument protein, VP16, the product of the UL48 gene, interacts with gH in vitro (S. Gross, C. A. Harley, and D. W. Wilson, Proc. 27th Int. Herpesvirus Workshop 2002, abstr. 7.05, 2002). Moreover, an HSV VP16 mutant exhibited significant defects in cytoplasmic envelopment, accumulating unenveloped nucleocapsids (37). HSV VP16 was also found to associate with gD in mild detergent extracts of purified preparations of HSV virions (28). Therefore, it appears that several tegument proteins may interact with more than a single α-herpesvirus membrane protein to promote cytoplasmic envelopment.
HSV glycoprotein gD is essential for virus entry into all cells studied to date, and virus mutants lacking gD must be propagated on cells that express gD (25, 31). By expression cloning, several gD receptors have been defined and one of these, nectin-1, is expressed extensively on biologically relevant epithelial and neuronal cells (reviewed in reference 46). gD-null mutants produce relatively normal numbers of enveloped virus particles in noncomplementing cells (31). An HSV type 1 (HSV-1) mutant that lacks the gD CT domain was able to access virus receptors and enter cells, but it produced smaller plaques and there was evidence for reduced yields of infectious virus (31). This was consistent with subtle effects of the gD CT domain in virus replication or cell-to-cell spread.
HSV and PRV glycoproteins gE and gI form a heterodimeric complex that promotes the cell-to-cell spread of viruses, especially in epithelial and neuronal tissues, but gE/gI does not play a role in virus entry when extracellular virions are applied to cells (1, 3, 10, 13, 14, 15, 17, 22, 24, 50, 51, 54). It was recently proposed that HSV gE/gI can act in two phases of the process of virus cell-to-cell spread in epithelial cells (24, 27). In one aspect of this, gE/gI apparently interacts with cellular components of epithelial junctions in a manner that promotes movement of virions across the cell junctions (11, 34). By a distinct mechanism, gE/gI promotes virus spread by directing nascent virions to epithelial cell junctions (27). We hypothesized that gE/gI drives envelopment so that nascent virions are delivered into cytoplasmic vesicles that specifically traffic to lateral surfaces of epithelial cells, which would promote HSV cell-to-cell spread (24, 27). During studies of HSV gE− and gI− mutants in epithelial cells, we observed subtle defects in envelopment, so that more cytosolic nucleocapsids accumulated (27).
Recently, an HSV-1 gD mutant, designated F-US6kan, in which a kanamycin gene cassette was placed between the gD promoter and coding sequences, was characterized (25). Surprisingly, F-US6kan could spread between keratinocytes and form plaques. However, it became clear that F-US6kan expressed 1/500 of the level of gD compared with wild-type HSV (21). Moreover, HaCaT keratinocytes express high levels of the gD receptor nectin-1, which, in part, may explain the ability of F-US6kan to spread between these cells but not other cells. There still remained the possibility that gE/gI also contributed substantially to the ability of F-US6kan to spread on keratinocytes. In order to test this we replaced the gE gene in F-US6kan with a green fluorescent protein (GFP) gene. F-US6kan/gE-GFP did not spread beyond a single infected cell, an observation that was consistent with the notion that gE/gI promoted cell-to-cell spread in the absence of gD. However, F-US6kan/gE-GFP exhibited major defects in the late stages of virus assembly. Large numbers of unenveloped nucleocapsids accumulated in the cytoplasm of cells infected with this mutant. Two other HSV-1 mutants in which gD and gE or gD, gE, and gI were replaced with β-galactosidase or GFP sequences also failed to produce enveloped virus particles. We concluded that gD and gE/gI act in a redundant fashion to promote cytoplasmic envelopment of HSV.
MATERIALS AND METHODS
Cells and viruses.
HEC-1A, human endometrial epithelial cells (4), were grown in RPMI medium (BioWhittaker, Inc., Walkersville, Md.) supplemented with 8% fetal bovine serum (FBS). HaCaT, human keratinocyte cells derived from a culture of primary keratinocyes (5), and Vero cells were grown in Dulbecco's modified Eagle's medium (BioWhittaker, Inc.) supplemented with 8% FBS. VD60 cells were maintained in Dulbecco's modified Eagle's medium lacking histidine and supplemented with 0.4 to 1.0 mM histidinol and 8% FBS (Sigma Chemical Co., St. Louis, Mo.). Wild-type HSV-1 strain F (originally from Pat Spear, Northwestern Medical School) and the HSV-1 gE− recombinants derived from F: F-gEβ (14) and F-gE-GFP (produced here) were propagated and their titers were determined on Vero cells. gD− recombinants derived from F, F-US6kan (21, 25), F-gDβ, which lacks gD and gI sequences (31), and vRR1097, which has much of the gD coding sequences replaced by GFP sequences (41), as well as the gD−/gE− recombinants described below, were propagated and their titers were determined on gD-expressing VD60 cells (31).
Construction of gD−/gE− recombinants.
In order to insert GFP sequences in place of gE sequences, a plasmid, designated pgE-GFP, was constructed from pUCUS7/8, which contains a 3.1-kb fragment of the US region of HSV-1 F DNA encompassing the US7 (gI) and US8 (gE) genes (53). Plasmid pUCUS7/8 was digested to completion with restriction endonuclease SmaI, removing the first 700 bp of coding sequence of US8, creating the plasmid pUCUS7/8ΔSmaI. GFP coding sequences were subcloned from the plasmid pEGFP-C1 (GenBank accession no. U55763; Clontech), in which GFP sequences were coupled to the human cytomegalovirus immediate early promoter by PCR. Two primers were employed, ACGCCCCGGGTGCATTAGTTATTAATAGTAATCAAT and TCCCCCCGGGTTACTTGTACAGCTCGTCCATGC, adding SmaI sites at either end of the GFP sequences. The PCR product was sequenced, digested with SmaI, and inserted into the SmaI site of pUCUS7/8ΔSmaI, creating pgE-GFP. Three gE− derivatives were produced by using pgE-GFP. VD60 cells were cotransfected with wild-type HSV-1 F, F-US6kan, or F-gDβ DNA as well as plasmid pgE-GFP and viruses that expressed isolated GFP, producing three recombinants, F-gE-GFP, F-US6kan/gE-GFP, and F-gDβ/gE-GFP. vRR1097/gEβ was constructed from vRR1097 (41) by cotransfecting VD60 cells with vRR1097 DNA and plasmid pgEβ, in which β-galactosidase sequences replace gE coding sequences (13), and viruses that expressed characterized β-galactosidase. A rescued version of this virus was created by cotransfection of vRR1097/gEββ DNA and plasmid pSS17 (31) Vero cells.
Staining of HSV-infected cells.
HaCaT cells with HSV plaques were washed, fixed, and stained with polyclonal anti-HSV-1 antibodies, peroxidase-conjugated secondary antibodies, and peroxidase substrate as previously described (53). Recombinant viruses expressing β-galactosidase were identified after overlaying cells with agarose containing 5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside as previously described (31). Recombinant viruses expressing GFP were identified by exposing infected cell monolayers to 521-nm fluorescent light visualized with a Nikon Optiphot fluorescence microscope.
Radiolabeling of infected cells and immunoprecipitation.
HaCaT cells were left uninfected or were infected with wild-type or mutant HSV-1 with 5 PFU/cell. After 2 h, the cells were labeled with [35S]methionine-cysteine (Amersham) (50 μCi/ml) for 5 h, cell extracts were made using NP-40-deoxycholate lysis buffer (100 mM NaCl, 50 mM Tris-HCl [pH 7.5], 1.0% NP-40, 0.5% deoxycholate) supplemented with 2 mg of bovine serum albumin/ml and 1 mM phenylmethylsulfonyl fluoride, and the extracts were frozen at −70°C. HSV-1 gD was immunoprecipitated by using monoclonal antibody (MAb) DL6 (16), gE was precipitated by using MAb 3114 (13), and gI was precipitated by using MAb 3104 (23). The immunoprecipitated cell extracts were eluted and subjected to electrophoresis as described previously (48). The dried gels were placed in contact with a phosphorimager, and viral proteins were quantified.
Electron microscopy.
HEC-1A or HaCaT cells were infected with wild-type or mutant HSV-1 for 16 h and then washed with 0.1 M sodium cacodylate buffer (pH 7.2) and fixed in Ito and Karnovsky's fixative (1.6% paraformaldehyde, 2.5% glutaraldehyde, and 0.5% picric acid in 0.1 M sodium cacodylate). Samples were postfixed in 1.5% osmium tetroxide, rinsed, and then postfixed in 4% paraformaldehyde. Samples were dehydrated in a graded acetone series and embedded in epoxy resin, and ultrathin sections were double stained in uranyl acetate and lead citrate and viewed with a Philips EM 300 electron microscope.
RESULTS
Characterization of a virus derived from F-US6kan and lacking gE.
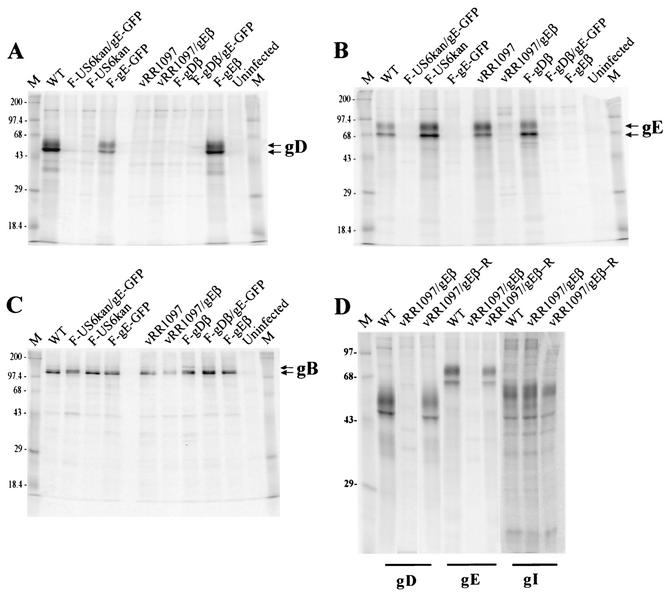
F-US6kan expresses extremely low levels of gD, yet it can spread between human keratinocytes, forming small plaques (21). Anti-gE antibodies reduced the number of plaques formed on these cells, suggesting the possibility that gE/gI might function in entry or in cell-to-cell spread in the context of these low levels of gD (21). To characterize further the effects of gE/gI, we replaced gE coding sequences with GFP sequences in F-US6kan. The virus used to construct this double mutant was derived from a nonsyncytial version of F-US6kan (21). A GFP gene cassette was inserted in place of most of the gE coding sequences in a plasmid designated pgE-GFP (Fig. 1 and Table 1). Plasmid pgE-GFP was cotransfected along with DNA derived from F-US6kan into VD60 cells which express gD. Viruses derived from this transfection were screened for GFP expression and further plaque purified. HSV recombinants were characterized for expression of gB, gD, and gE by infecting HaCaT cells and labeling the cells with [35S]methionine-cysteine. HSV glycoproteins were immunoprecipitated with anti-gB, -gD, or -gE MAb. As expected, a virus derived from this transfection and designated F-US6kan/gE-GFP expressed gB but not gE and only a trace of gD that was not readily apparent in these exposures (Fig. 2), as was described previously for F-US6kan (22).
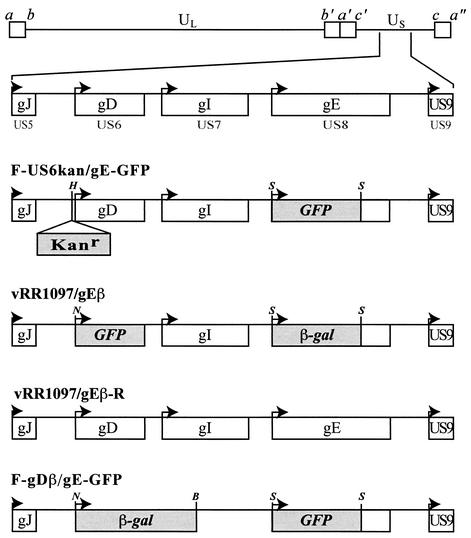
FIG. 1.
Construction of recombinant viruses. The genome of HSV-1, including the US5 (gJ), US6 (gD), US7 (gI), US8 (gE), and US9 genes, is depicted in the upper part of the figure (33). F-US6kan/gE-GFP was derived from F-US6kan (which contains a kanamycin gene cassette inserted between the gD promoter and coding sequences) by replacing gE coding sequences with GFP sequences between two SmaI restriction sites. vRR1097/gEβ was derived from vRR1097 (in which GFP sequences replace gD coding sequences) by replacing gE coding sequences with β-galactosidase sequences. vRR1097/gEβ-R was derived from vRR1097/gEβ by restoring both gD and gE sequences. F-gDβ/gE-GFP was derived from F-gDβ (a mutant in which β-galactosidase sequences replace gD and gI coding sequences) by additionally replacing gE coding sequences with GFP sequences. H represents a HindIII restriction site, S represents SmaI sites, B represents BalI, and N represents an NcoI restriction site.
TABLE 1.
Expression of viral glycoproteins by HSV recombinants
| Virus | Expression level of:
|
||
|---|---|---|---|
| gD | gE | gI | |
| FUS6kan | Lowa | WTb | WT |
| FUS6kan/gE-GFP | Lowa | Null | WT |
| vRR1097 | Null | WT | WT |
| vRR1097/gEβ | Null | Null | WT |
| F-gDβ | Null | WT | Null |
| F-gDβ/gE-GFP | Null | Null | Null |
| vRR1097/gEβR | WT | WT | WT |
As shown by Huber et al. gD is expressed at approximately 1/500 of the level observed in wild-type HSV (21).
WT, wild type.
FIG. 2.
Expression of gD, gE, gB, and gI by recombinant HSV-1. Human keratinocyte HaCaT cells were infected with wild-type HSV-1, F-US6kan/gE-GFP, F-US6kan, F-gE-GFP, vRR1097, vRR1097/gEβ, F-gDβ, F-gDβ/gE-GFP, or F-gEβ or were left uninfected. The cells were labeled with [35S]methionine-cysteine, and gD, gE, gB, or gI was immunoprecipitated from cell extracts by using MAb DL6, 3114, 15βB2, or 3104, respectively. Antigen-antibody complexes were collected by using protein A-Sepharose, and eluted proteins were subjected to electrophoresis on 12% polyacrylamide gels. Panels A, B, and C include all the mutants except vRR1097/gEβR; gD was precipitated in panel A, gE was precipitated in panel B, and gB was precipitated in panel C. In panel D, vRR1097/gEβR was characterized for gD, gE, and gI. The positions of mature and immature forms of gE, gD, and gB as well as marker proteins of 200, 97.4, 68, 43, 29, and 18.4 kDa are indicated.
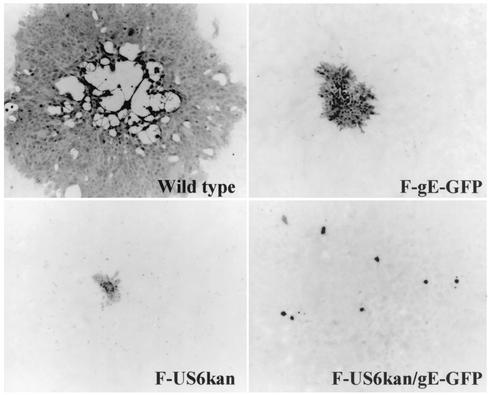
To determine whether F-US6kan/gE-GFP could spread on human keratinocytes, we infected HaCaT cells and determined the sizes of plaques. As previously reported (21), F-US6kan produced on complementing VD60 cells could produce small plaques on HaCaT cells (Fig. 3). The numbers of plaques were similar in each case (data not shown), suggesting that entry of complemented virus into the HaCaT cells was not compromised. Similarly, F-gE-GFP, which lacks gE, formed plaques, although these were of markedly reduced size. By contrast, F-US6kan/gE-GFP was consistently unable to spread beyond a single infected cell. The numbers of these single infected cells that stained with anti-HSV antibodies were similar to the numbers of plaques formed by F-US6kan. This suggested that F-US6kan/gE-GFP produced on gD-complementing cells produced relatively normal numbers of infectious viruses and that these entered HaCaT cells and were able to initiate an infection efficiently. However, without gE and with low levels of gD, F-US6kan/gEβ could not replicate or spread beyond a single infected cell.
FIG. 3.
Plaques formed by F-US6kan and F-US6kan/gEβ. HaCaT cells were infected with wild-type HSV-1, F-gE-GFP, F-US6kan, or F-US6kan/gE-GFP. In the case of F-US6kan and F-US6kan/gE-GFP, virus stocks were prepared by using gD-complementing VD60 cells. After 2 days, the cells were stained with anti-HSV antibodies and peroxidase-conjugated secondary antibodies.
F-US6kan/gEGFP is severely restricted in the production of enveloped virus particles.
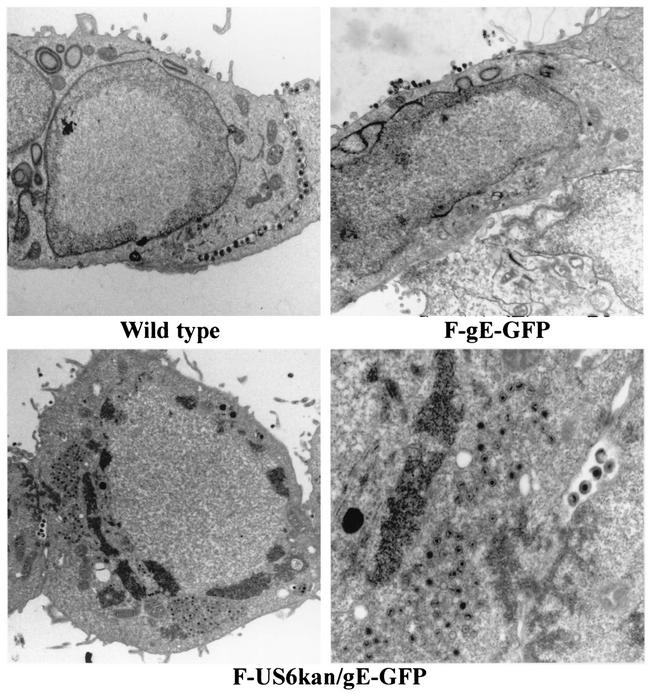
To determine whether F-US6kan/gE-GFP could replicate normally in cells, electron microscopy was used. HEC-1A human endometrial epithelial cells were employed in these and previous electron microscopic studies (27) because the morphology of these epithelial cells allows detailed characterization of virus subcellular localization. As before, HEC-1A cells infected with wild-type HSV-1 displayed relatively large numbers of enveloped virus particles on the cell surfaces, with the majority at cell junctions (Fig. 4, upper left panel). Relatively few unenveloped or enveloped particles were observed in the cytoplasm of wild-type-infected cells, suggesting, as in previous studies (26, 27), that late stages of assembly and acquisition of the final envelope are rapid, as is movement to the cell surface. Cells infected with F-gE-GFP, a recombinant lacking gE but expressing gD, displayed a similar pattern of enveloped virions at the cell surface, although these were frequently on apical rather than lateral surfaces (Fig. 4, upper right panel). In stark contrast, F-US6kan/gE-GFP-infected HEC-1A cells contained large aggregates of virus particles in the cytoplasm and very few enveloped particles (Fig. 4, lower panels). These aggregated particles were not enveloped (Fig. 4, lower right panel), and the particles were immersed in an electron-dense structure which appeared to be tegument. There were very few enveloped particles observed either in the cytoplasm or on the cell surface. In some cases, a few enveloped particles were observed, as in Fig. 4, but often no enveloped particles were observed in sections that contained 50 or more unenveloped particles. Therefore, the loss of most gD and all gE markedly inhibited HSV cytoplasmic envelopment.
FIG. 4.
Electron micrographs of F-US6kan/gE-GFP-infected HEC-1A cells. HEC-1A cells were infected with wild-type HSV-1 (upper left panel), F-gE-GFP (upper right panel), or F-US6kan/gE-GFP (lower panels) for 16 h. The cells were fixed and processed for electron microscopy.
Construction of HSV mutants unable to express gD and gE.
F-US6kan expresses low levels of gD, which confuses the interpretation of the results. To determine whether similar defects in late virus assembly occurred when both gD and gE were not expressed, we constructed two different gD−/gE− recombinants in which the gD and gE genes were entirely or largely replaced with marker genes (refer to Fig. 1 and Table 1). Previously, Rauch et al. (41) constructed a recombinant HSV-1, designated vRR1097, in which GFP sequences replaced most of the gD gene. To remove gE sequences in vRR1097, we cotransfected VD60 cells (gD-complementing cells) with viral DNA from vRR1097 and a plasmid pgEβ in which gE coding sequences were replaced by β-galactosidase sequences (13). An isolate expressing β-galactosidase and GFP was selected and designated vRR1097/gEβ. A rescued version of vRR1097/gEβ able to express gD and gE was also constructed.
A second recombinant unable to express gD and gE, and also missing gI coding sequences, was derived from F-gDβ, in which all of the gD gene and the 5′ end of the gI gene were replaced with β-galactosidase sequences (31). F-gDβ DNA was cotransfected with plasmid pgE-GFP into VD60 cells, and virus isolates which expressed both β-galactosidase and GFP were selected. To characterize all of these recombinant viruses further, HEC-1A cells were infected with each virus, the cells were radiolabeled with [35S]methionine-cysteine, and gD, gE, or gB was immunoprecipitated. As expected, cells infected with vRR1097 did not express gD but expressed gE and gB (Fig. 2A to C). vRR1097/gEβ derived from this virus expressed gB but not gD and gE. F-gDβ expressed gB and gE but not gD, and F-gDβ/gE-GFP expressed gB but not gD and gE (Fig. 2A to C). The rescued virus, vRR1097/gEβ-R, expressed gD, gE, and gI (Fig. 2D). These data are summarized in Table 1.
Assembly of HSV gD−/gE− recombinants in HEC-1A endometrial cells.
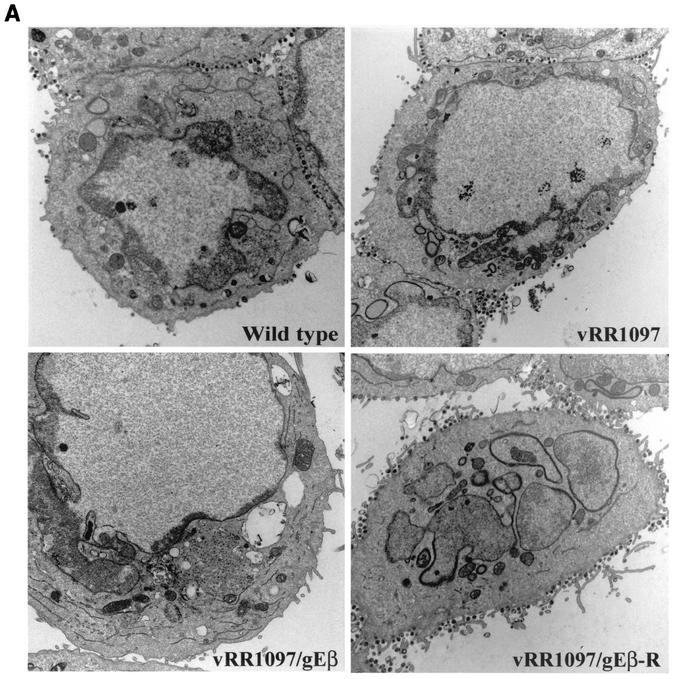
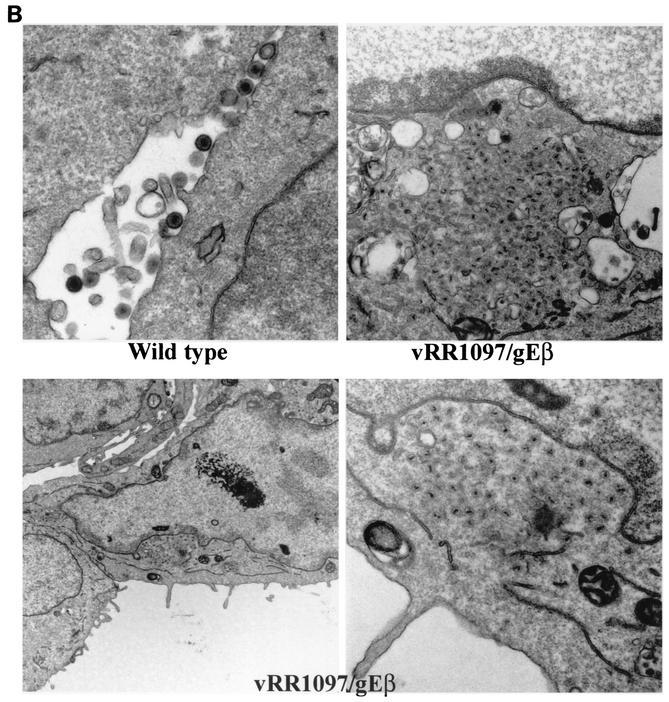
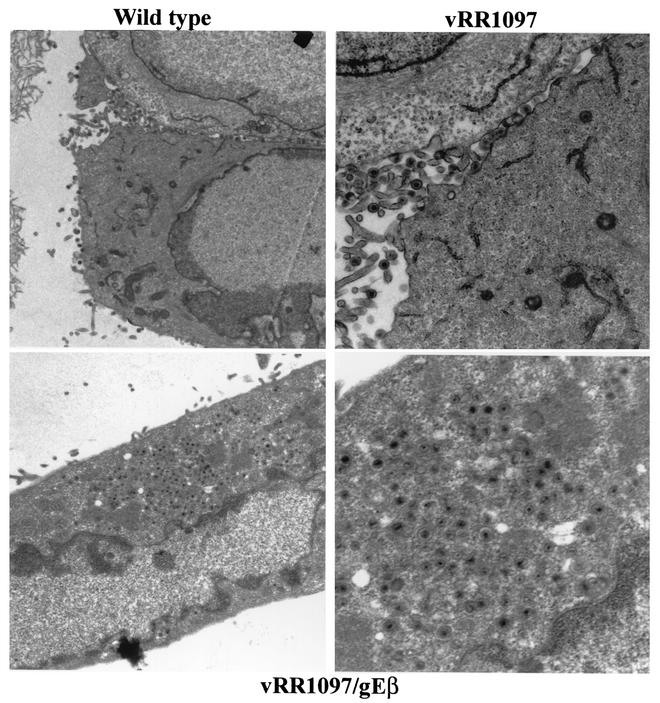
The double gD−/gE− mutant vRR1097/gEβ was characterized by electron microscopy. As before, HEC-1A cells infected with either wild-type HSV-1 displayed numerous enveloped particles largely on the cell surface at cell junctions (Fig. 5A, upper left panel). Cells infected with the gD− mutant vRR1097 also exhibited numerous cell surface virions, although in some cells there were slightly more unenveloped particles in the cytoplasm (Fig. 5A, upper right panel). By contrast, vRR1097/gEβ-infected cells accumulated large numbers of virus particles in cytoplasmic aggregates (Fig. 5A, lower left panel). At higher magnifications, the aggregated vRR1097/gEβ particles clearly lacked an envelope and were found in aggregates immersed in an electron-dense material, probably composed of tegument proteins (Fig. 5B, right panels). There were enveloped virions in vRR1097/gEβ-infected cells, but these were markedly reduced in numbers and some sections showed only unenveloped particles. This was quantified by counting approximately 750 to 1,000 virus particles, unenveloped nucleocapsids, and enveloped virions in the cytoplasm and on the cell surface in numerous cell sections (Table 2). Cytoplasmic unenveloped nucleocapsids were increased by 23-fold in vRR1097/gEβ-infected cells compared with wild-type HSV-1-infected cells, and the numbers of cell surface enveloped virions were reduced by 6-fold in vRR1097/gEβ-infected cells. We believe that some of the enveloped particles that were present on surfaces of vRR1097/gEβ-infected cells were derived from input infecting viruses. This likely relates to the observation that vRR1097/gEβ stocks derived from VD60 cells were significantly lower in titer than other viruses so that higher input doses were required. Cells infected with vRR1097/gEβ-R, derived from vRR1097/gEβ by restoring the gD and gE genes, displayed numerous enveloped viruses on the cell surface, similar to the pattern observed with wild-type HSV-1 (Fig. 5A, lower right panel, and Table 2).
FIG. 5.
Electron micrographs of HEC-1A cells infected with F-gDβ/gE-GFP. (A) HEC-1A cells were infected with wild-type HSV-1, vRR1097 (gD−), vRR1097/gEβ (gD−/gE−), or vRR1097/gEβ-R (a rescued version of vRR1097/gEβ). (B) HEC-1A cells were infected with wild-type HSV-1 or vRR1097/gEβ. After 16 to 18 h, the cells were fixed and processed for electron microscopy.
TABLE 2.
Distribution of virus particles produced by wild-type and mutant HSV-1 in HEC-1A cellsa
| Virus | No. (%) ofb:
|
|||
|---|---|---|---|---|
| Nucleocapsids in:
|
Enveloped virions:
|
|||
| Nucleus | Cytoplasm | In cytoplasm | On cell surface | |
| Wild type | 393 (43.9) | 30 (3.3) | 36 (4.0) | 435 (48.6) |
| F-gEβ | 335 (44.5) | 75 (9.9) | 97 (12.8) | 246 (32.6) |
| vRR1097 | 414 (52.5) | 36 (4.6) | 45 (5.7) | 293 (37.1) |
| vRR1097/gEβ | 268 (25.2) | 695 (65.6) | 12 (1.2) | 85 (8) |
| F-gDβ/gE-GFP | 374 (22.4) | 1,227 (73.4) | 32 (1.9) | 38 (2.3) |
| VRR1097/gEβ-R | 389 (56.3) | 44 (6.3) | 38 (5.4) | 220 (31.8) |
Human HEC-1A cells were infected with wild-type or mutant HSV-1 for 16 h. The cells were fixed, sectioned, and then examined by electron microscopy.
The numbers of unenveloped nucleocapsids in the nucleus and in the cytoplasm and enveloped virions in the cytoplasm and on the cell surface were counted in approximately 10 to 15 randomly chosen cells. The numbers in parentheses are percentages of the total numbers of particles.
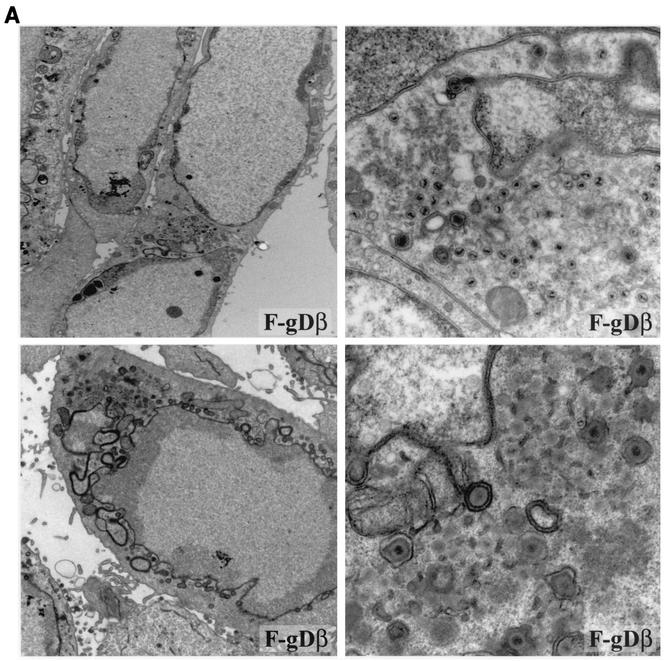
Previously, minor defects were reported in the assembly of F-gEβ, an HSV-1 recombinant in which β-galactosidase sequences replace those of gE in HEC-1A cells (27). In those studies and the present studies, there were more-subtle two- to threefold increases in numbers of unenveloped nucleocapsids observed in the cytoplasm (Table 2). Here, we characterized a second gE− mutant, F-gE-GFP, a recombinant in which GFP sequences replace gE sequences, and there were subtle increases in unenveloped particles, although most cells primarily displayed enveloped particles on the cell surface (Fig. 4). Somewhat more-striking defects in late assembly were observed with the gD−/gI− mutant F-gDβ. HEC-1A cells infected with F-gDβ displayed aggregates of unenveloped particles in many, but not all, sections, and enveloped particles were also observed in both the cytoplasm and on the cell surfaces (Fig. 6A). This was not previously noted in Vero cells (31), and we did not observe obvious defects in the assembly of F-gDβ in HaCaT cells (data not shown). Therefore, it appears that deletion of either gE alone or gD and gI causes more-subtle but still apparent defects in virus envelopment so that increased numbers of unenveloped particles accumulate, but this is dependent on the host cells studied.
FIG. 6.
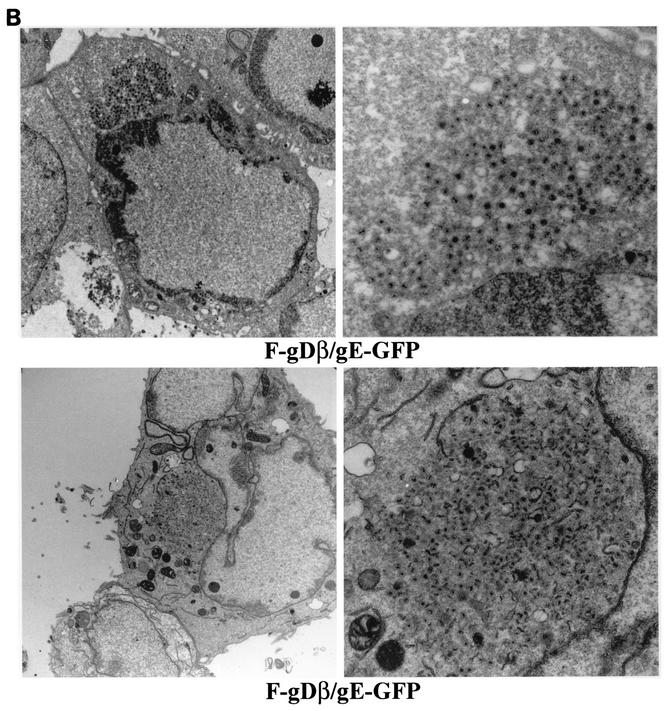
Electron micrographs of F-gDβ- and F-gDβ/gE-GFP-infected HEC-1A cells. HEC-1A cells infected with the gD−/gI− mutant F-gDβ (A) or the gD−/gI−/gE− mutant F-gDβ/gE-GFP (B) were fixed and processed for electron microscopy.
HEC-1A cells infected with the triple mutant F-gDβ/gE-GFP, lacking gD, gE, and gI, displayed large aggregates of cytoplasmic nucleocapsids (Fig. 6B, lower panels). These aggregates were similar to those observed with vRR1097/gEβ, except, in most cases, the F-gDβ/gE-GFP aggregates were greater in size. Moreover, there were fewer examples of enveloped particles on the surfaces of F-gDβ/gE-GFP-infected cells (Table 2). Therefore, it appeared that the additional loss of gI in a virus also missing gD and gE further decreased the production of enveloped virions and increased the accumulation of unenveloped capsids in the cytoplasm.
Assembly of a gD−/gE− HSV in HaCaT keratinocytes.
To extend these observations to other cells, we infected the human keratinocyte cell line HaCaT. These cells are derived from primary keratinocytes and are able to form differentiated epidermal tissue when transplanted into nude mice (5), and we believe that these cells are particularly relevant for studies of HSV replication. HSV gE− mutants produce plaques that are reduced by five- to eightfold in the numbers of infected HaCaT cells compared with wild-type HSV (53). HaCaT cells infected with wild-type HSV-1 showed extensive numbers of enveloped virions at cell junctions (Fig. 7, upper left panel). Similarly, vRR1097 (lacking gD) also produced relatively normal numbers of virus particles and these were again largely on the cell surface (Fig. 7, upper right panel). The double gD−/gE− mutant vRR1097/gEβ produced large numbers of unenveloped virions that accumulated in an electron-dense matrix in the cytoplasm of HaCaT cells (Fig. 7, lower panels). Few or no enveloped particles were produced. Therefore, as with HEC-1A cells, a gD−/gE− virus failed to produce unenveloped, cytoplasmic nucleocapsids in HaCaT cells.
FIG. 7.
Electron micrographs of vRR1097/gEβ-infected HaCaT cells. HaCaT cells were infected with wild-type HSV-1 (upper left panel), vRR1097 (upper right panel), or vRR1097/gEβ (lower panels) for 16 to 18 h. The cells were fixed and processed for electron microscopy.
DISCUSSION
HSV acquires two different envelopes. Nucleocapsids interact with the inner nuclear membrane, budding into the space between the inner and outer nuclear envelopes. Fusion of the virion envelope with the outer nuclear membrane delivers nucleocapsids into the cytoplasm. Tegument-coated proteins interact with the CT domains of HSV membrane glycoproteins that accumulate in the TGN or endosomes, and there is budding into cytoplasmic vesicles. This secondary envelopment apparently occurs relatively rapidly and efficiently with HSV-1, as few nucleocapsids are observed in the cytoplasm of most cultured cells (9, 26, 27, 45) Apparently, no single HSV-1 glycoprotein is essential for either nuclear or cytoplasmic envelopment. There are no major defects in the assembly of enveloped particles with HSV-1 mutants lacking any of the membrane glycoproteins considered essential for replication, gB, gD, and gH/gL (8, 19, 31, 44). Similarly, mutants lacking glycoproteins not required for replication in cultured cells show, at most, subtle defects in envelopment (reviewed in reference 24). The important work of Brack et al. (6, 7) showing that PRV depends upon two glycoproteins, gM and gE, suggested a hypothesis that two or more α-herpesvirus membrane proteins interact with more than a single tegument protein to anchor the virion envelope onto nucleocapsids.
Here, we demonstrated that HSV-1 depends upon gD and gE for secondary envelopment. HSV-1 mutants expressing low levels of or no gD and no gE produced very few enveloped virions. Another HSV-1 recombinant lacking gD, gE, and gI exhibited a similar, but apparently more profound, phenotype. We note that this reduction in enveloped particles between gD−/gE− and gD−/gE−/gI− mutants and wild-type HSV was most pronounced in enumerating virions on the cell surface, where most enveloped virions accumulate, rather than counting enveloped virions in the cytoplasm, which were few in number. Moreover, in gD−/gE− mutants, large numbers of unenveloped nucleocapsids accumulated in the cytoplasm in an electron-dense milieu which appeared to be tegument. There were no obvious effects on nuclear envelopment, and no obvious accumulation of capsids in the nucleus or perinuclear space was observed. Therefore, gD and gE serve essential but redundant functions in the cytoplasmic envelopment.
Previously, minor defects were noted in envelopment when characterizing HSV-1 mutants lacking gE or just the CT domain of gE (27). There were two- to threefold-increased numbers of cytoplasmic nucleocapsids compared with wild-type HSV-1, and here we observed similar defects with a second gE− mutant. Moreover, deletion of the CT domain of gD also subtly reduced production of infectious virus and plaque size (18). However, the effects observed with gD and gE single mutants were relatively subtle compared to the more severe phenotype of a mutant lacking both gD and gE.
There also appeared to be a contribution of the gI glycoprotein. Careful analysis of a mutant, F-gDβ, which lacks gD and gI showed that increased numbers of unenveloped nucleocapsids accumulated in HEC-1A cells compared with wild-type HSV, although significant numbers of enveloped virions were also observed. Nevertheless, this reduced envelopment was not observed in F-gDβ-infected HaCaT (this study) or in Vero cells (31). Defects in virus assembly associated with HSV-1 gD−/gI− double mutants were never as obvious as with gD−/gE− viruses. The loss of gD and gE produced a massive accumulation of unenveloped capsids in virtually every cell, whereas many cells infected with F-gDβ exhibited few unenveloped capsids. However, the loss of gD, gE, and gI in the triple mutant F-gDβ/gE-GFP led to a more severe phenotype than was observed with the gD−/gE− mutant, and even fewer numbers of enveloped capsids were observed. Therefore, we concluded that gI also contributes, perhaps in a more subtle manner than gE, to cytoplasmic envelopment.
In contrast to these results with HSV-1, a PRV quadruple, gD−, gE−, gI−, and gG−, mutant produced relatively normal numbers, or at least not drastically reduced numbers, of enveloped virions that could be purified and characterized by immunoelectron microscopy (36). Evidently, HSV and PRV differ in their requirements for envelopment. There have been other differences described for how gD functions for HSV and PRV. PRV gD is not required for the spread of viruses between cells once gD-complemented virus particles are able to enter cells (38, 39, 42). Moreover, gD− PRV can spread in animal tissues. By contrast, HSV gD− mutants do not spread beyond a single infected cell either in cultured cells or in animal models (14, 21, 31). Therefore, these differences may be as illuminating as the many similarities between PRV and HSV but caution against drawing parallels in every case.
To begin to understand how the virion envelope interacts with tegument, Fuchs et al. (20) have characterized interactions between the PRV UL49 gene, a homologue of HSV VP22, and gE and gM. Both gE and gM interacted with UL49 in yeast two-hybrid screens, and a PRV gE−/gM− deletion virus failed to incorporate UL49 into virions. This might have suggested that UL49 was essential for envelopment, as PRV gE and gM appear to be. However, PRV VP22 can be deleted without affecting envelopment or even neurovirulence (12, 20). Thus, interactions between gE and gM and other PRV tegument proteins are also apparently important for late assembly. In other α-herpesviruses, bovine herpesvirus type 1 VP22 is important for virus replication and neurovirulence (29, 30). With HSV, the C terminus of VP22 appears to be critical for incorporation into virions and for cell-to-cell spread (40). However, there is presently no HSV VP22-null mutant to test the notion that VP22 is essential for envelopment. It appears that HSV gE, like the PRV counterpart, interacts with VP22, as we detected a 37-kDa viral protein, corresponding to the size of HSV-1 VP22, bound onto a glutathione S-transferase fusion protein containing the gE CT domain (T. Wisner and D. C. Johnson, unpublished data). Recently, the cytoplasmic tail of HSV gD was found to interact with VP22 in vitro (Gross et al., Proc. 27th Int. Herpesvirus Workshop 2002), which provides the possibility that HSV gD and gE both interact with VP22. Unfortunately, the contribution of gM in envelopment has not been tested in HSV by the construction of either gE−/gM− or gD−/gM− double mutants.
A second HSV tegument protein VP16 appears to play an important role in cytoplasmic envelopment. An HSV-1 recombinant unable to express VP16 (and the virion host shutoff protein, vhs) produced few enveloped virions, and unenveloped particles accumulated in the cytoplasm, suggesting that VP16 is essential or important for cytoplasmic envelopment but not for nuclear envelopment (37). There is evidence that the cytoplasmic domain of HSV gH interacts with VP16 in vitro (Gross et al., Proc. 27th Int. Herpesvirus Workshop 2002), supporting the notion that gH may also play a role in envelopment. Moreover, VP16 was found to be physically associated with gD extracted from virions with mild nonionic detergents and purified by antibody affinity chromatography (28). Therefore, it is plausible that HSV cytoplasmic envelopment involves interactions between gE and gD with VP22 and between gH and gD with VP16, although loss of any individual interaction may not be fatal. Clearly, loss of both gE and gD dramatically diminishes envelopment. However, it is also reasonable that HSV glycoproteins other than gD and gE/gI are involved in cytoplasmic envelopment, e.g., gB and gH, and there may be yet other interactions for nuclear envelopment. It will require other double and triple glycoprotein mutants to define this further.
Acknowledgments
We thank Michael Webb and Keith Mayo for assistance with the electron microscopy experiments. Todd Wisner provided a great deal of technical support during this work.
The research was funded by a grant from the National Institutes of Health (CA73996).
REFERENCES
- 1.Babic, N., B. Klupp, A. Brack, T. C. Mettenleiter, G. Ugolini, and A. Flamand. 1996. Deletion of glycoprotein gE reduces the propagation of pseudorabies virus in the nervous system of mice after intranasal inoculation. Virology 219:279-284. [DOI] [PubMed] [Google Scholar]
- 2.Baines, J. D., and B. Roizman. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J. Virol. 65:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balan, P., N. Davis-Poynter, S. Bell, H. Atkinson, H. Browne, and T. Minson. 1994. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J. Gen. Virol. 75(Pt 6):1245-1258. [DOI] [PubMed] [Google Scholar]
- 4.Ball, J. M., Z. Moldoveaunu, L. R. Melsen, P. A. Kozlowski, S. Jackson, M. J. Mulligan, J. F. Mestecky, and R. W. Compans. 1995. A polarized human endometrial cell line that binds and transports polymeric IgA in vitro. Cell. Dev. Biol. 31:196-206. [DOI] [PubMed] [Google Scholar]
- 5.Boukamp, P., R. T. Petrussevska, D. Breitkreutz, J. Hornung, A. Markham, and N. E. Fusenig. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106:761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, W. H., B. Gu, and S. Person. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 62:2596-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campadelli-Fiume, G., F. Farabegoli, S. Di Gaeta, and B. Roizman. 1991. Origin of unenveloped capsids in the cytoplasm of cells infected with herpes simplex virus 1. J. Virol. 65:1589-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Card, J. P., M. E. Whealy, A. K. Robbins, and L. W. Enquist. 1992. Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat visual system. J. Virol. 66:3032-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, W. J., and D. C. Johnson. 2003. Herpes simplex virus gE/gI expressed in epithelial cells interferes with cell-to-cell spread. J. Virol. 77:2686-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.del Rio, T., H. C. Werner, and L. W. Enquist. 2002. The pseudorabies virus VP22 homologue (UL49) is dispensable for virus growth in vitro and has no effect on virulence and neuronal spread in rodents. J. Virol. 76:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dingwell, K. S., C. R. Brunetti, R. L. Hendricks, Q. Tang, M. Tang, A. J. Rainbow, and D. C. Johnson. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J. Virol. 68:834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dingwell, K. S., L. C. Doering, and D. C. Johnson. 1995. Glycoproteins E and I facilitate neuron-to-neuron spread of herpes simplex virus. J. Virol. 69:7087-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dingwell, K. S., and D. C. Johnson. 1998. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J. Virol. 72:8933-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenberg, R. J., D. Long, M. Ponce de Leon, J. T. Matthews, P. G. Spear, M. G. Gibson, L. A. Lasky, P. Berman, E. Golub, and G. H. Cohen. 1985. Localization of epitopes of herpes simplex virus type 1 glycoprotein D. J. Virol. 53:634-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enquist, L. W., P. J. Husak, B. W. Banfield, and G. A. Smith. 1998. Infection and spread of α-herpesviruses in the nervous system. Adv. Virus Res. 51:237-347. [DOI] [PubMed] [Google Scholar]
- 18.Feenstra, V., M. Hodaie, and D. C. Johnson. 1990. Deletions in herpes simplex virus glycoprotein D define nonessential and essential domains. J. Virol. 64:2096-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis-Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66:341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. R. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huber, M. T., T. W. Wisner, N. R. Hegde, K. A. Goldsmith, D. A. Rauch, R. J. Roller, C. Krummenacher, R. J. Eisenberg, G. H. Cohen, and D. C. Johnson. 2001. Herpes simplex virus with highly reduced gD levels can efficiently enter and spread between human keratinocytes. J. Virol. 75:10309-10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, D. C., and V. Feenstra. 1987. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J. Virol. 61:2208-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, D. C., M. C. Frame, M. W. Ligas, A. M. Cross, and N. D. Stow. 1988. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J. Virol. 62:1347-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, D. C., and M. T. Huber. 2002. Directed egress of animal viruses promotes cell-to-cell spread. J. Virol. 76:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, D. C., and M. W. Ligas. 1988. Herpes simplex viruses lacking glycoprotein D are unable to inhibit virus penetration: quantitative evidence for virus-specific cell surface receptors. J. Virol. 62:4605-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, D. C., and P. G. Spear. 1982. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J. Virol. 43:1102-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, D. C., M. Webb, T. W. Wisner, and C. Brunetti. 2001. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J. Virol. 75:821-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, D. C., M. Wittels, and P. G. Spear. 1984. Binding to cells of virosomes containing herpes simplex virus type 1 glycoproteins and evidence for fusion. J. Virol. 52:238-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang, X., B. Chow, and L. A. Babiuk. 1997. Study of immunogenicity and virulence of bovine herpesvirus 1 mutants deficient in the UL49 homolog, UL49.5 homolog and dUTPase genes in cattle. Vaccine 15:1057-1064. [DOI] [PubMed] [Google Scholar]
- 30.Liang, X., C. Pyne, Y. Li, L. A. Babiuk, and J. Kowalski. 1995. Delineation of the essential function of bovine herpesvirus 1 gD: an indication for the modulatory role of gD in virus entry. Virology 207:429-441. [DOI] [PubMed] [Google Scholar]
- 31.Ligas, M. W., and D. C. Johnson. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 62:1486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longnecker, R., and B. Roizman. 1987. Clustering of genes dispensable for growth in culture in the S component of the HSV-1 genome. Science 236:573-576. [DOI] [PubMed] [Google Scholar]
- 33.McGeoch, D. J., A. Dolan, S. Donald, and F. J. Rixon. 1985. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J. Mol. Biol. 181:1-13. [DOI] [PubMed] [Google Scholar]
- 34.McMillan, T. N., and D. C. Johnson. 2000. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J. Virol. 75:1928-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mettenleiter, T. C., B. G. Klupp, F. Weiland, and N. Visser. 1994. Characterization of a quadruple glycoprotein-deleted pseudorabies virus mutant for use as a biologically safe live virus vaccine. J. Gen. Virol. 75:1723-1733. [DOI] [PubMed] [Google Scholar]
- 37.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peeters, B., N. de Wind, M. Hooisma, F. Wagenaar, A. Gielkens, and T. Kimman. 1992. Pseudorabies virus envelope glycoproteins gp50 and gII are essential for virus penetration, but only gII is involved in membrane fusion. J. Virol. 66:894-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peeters, B., J. Pol, A. Gielkens, and R. Moormann. 1993. Envelope glycoprotein gp50 of pseudorabies virus is essential for virus entry but is not required for viral spread in mice. J. Virol. 67:170-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pomeranz, L. E., and J. A. Blaho. 2000. Assembly of infectious herpes simplex virus type 1 virions in the absence of full-length VP22. J. Virol. 74:10041-10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rauch, D. A., N. Rodriguez, and R. J. Roller. 2000. Mutations in herpes simplex virus glycoprotein D distinguish entry of free virus from cell-cell spread. J. Virol. 74:11437-11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rauh, I., and T. C. Mettenleiter. 1991. Pseudorabies virus glycoproteins gII and gp50 are essential for virus penetration. J. Virol. 65:5348-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roizman, B., and A. E. Sears. 1996. Herpes simplex viruses and their replication, p. 1043-1107. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fundamental virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 44.Roop, C., L. Hutchinson, and D. C. Johnson. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J. Virol. 67:2285-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment → deenvelopment → reenvelopment pathway. J. Virol. 75:5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 47.Stackpole, C. W. 1969. Herpes-type virus of the frog renal adenocarcinoma. I. Virus development in tumor transplants maintained at low temperature. J. Virol. 4:75-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomazin, R., A. B. Hill, P. Jugovic, I. York, P. van Endert, H. L. Ploegh, D. W. Andrews, and D. C. Johnson. 1996. Stable binding of the herpes simplex virus ICP47 protein to the peptide binding site of TAP. EMBO J. 15:3256-3266. [PMC free article] [PubMed] [Google Scholar]
- 49.Weber, P. C., M. Levine, and J. C. Glorioso. 1987. Rapid identification of nonessential genes of herpes simplex virus type 1 by Tn5 mutagenesis. Science 236:576-579. [DOI] [PubMed] [Google Scholar]
- 50.Weeks, B. S., P. Sundaresan, T. Nagashunmugam, E. H. Kang, and H. M. Friedman. 1997. The herpes simplex virus-1 glycoprotein E (gE) mediates IgG binding and cell-to-cell spread through distinct gE domains. Biochem. Biophys. Res. Commun. 235:31-35. [DOI] [PubMed] [Google Scholar]
- 51.Whealy, M. E., J. P. Card, A. K. Robbins, J. R. Dubin, H. J. Rziha, and L. W. Enquist. 1993. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J. Virol. 67:3786-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whiteley, A., B. Bruun, T. Minson, and H. Browne. 1999. Effects of targeting herpes simplex virus type 1 gD to the endoplasmic reticulum and trans-Golgi network. J. Virol. 73:9515-9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wisner, T. W., C. Brunetti, K. Dingwell, and D. C. Johnson. 2000. The extracellular domain of herpes simplex virus gE is sufficient for accumulation at cell junctions but not for cell-to-cell spread. J. Virol. 74:2278-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuckermann, F., T. C. Mettenleiter, C. Schreurs, N. Sugg, and T. Ben-Porat. 1988. Complex between glycoproteins gI and gp63 of pseudorabies virus: its effect on virus replication. J. Virol. 62:4622-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]