Abstract
E1B-55K-mutant or E4orf6-mutant adenoviruses replicate more effectively after infecting cells in S phase than after infecting cells in G1. Enhanced S-phase replication of the E4orf6-mutant viruses requires the E4orf3 gene. This report demonstrates that the E4orf3 gene is also required for enhanced S-phase replication of the E1B-55K-mutant virus.
Adenoviruses (Ads) defective in the coding regions of the early region 1B 55-kDa (E1B-55K) protein or early region 4 open reading frame 6 (E4orf6) are restricted for growth by the cell cycle. These mutant viruses were more likely to produce progeny virus in cells infected during S phase than in cells infected during G1. Furthermore, the yields of mutant viruses from S-phase-infected cells were significantly greater than the yields from G1-infected cells. By contrast, every cell infected with the wild-type virus produced progeny to an equivalent level irrespective of the stage of the cell cycle at the time of infection (5, 6). For this reason, viruses defective in either the E1B-55K or E4orf6 genes can be considered G1 restricted for replication. The similarity in the cell cycle restrictions of these two mutant viruses is consistent with other genetic and biochemical data that suggest that some of the functions of these two proteins are mediated by a complex of E1B-55K and E4orf6 proteins.
The ability of the E4orf6-mutant virus to replicate better upon infecting a cell in S phase depended on another early gene, the E4orf3 gene. Mutant viruses that failed to express both E4orf6 and E4orf3 were severely defective for replication. Moreover, the replication of these viruses was not enhanced in S-phase-infected cells (6). These findings suggested that E4orf3 is necessary for the enhanced S-phase replication of the E4orf6-mutant virus. This result was unusual because deletion of E4orf3 has no effect on virus yield or viral gene expression of viruses with wild-type E4orf6 (2, 7).
The purpose of this work was to determine if E4orf3 is required for the replication of the cell cycle-restricted E1B-55K-mutant virus, similar to the E4orf6-mutant virus. Alternatively, E4orf3 could be required in the absence of E4orf6, but not in the absence of E1B-55K. An E1B-55K/E4orf3 double-mutant recombinant virus was created by ligating a DNA fragment containing map units 0 to 37 of the E1B-55K-null virus dl1520 (1) to a DNA fragment containing map units 37 to 100 of the E4orf3-mutant virus dl341 (13). The recombinant viral DNA was introduced into the E1-complementing 293 cell line to recover infectious virus, and a recombinant virus was easily recovered by successive rounds of plaque isolation.
Asynchronously growing HeLa cells were infected with the wild-type virus dl309 (8), the E1B-55K-mutant virus dl1520, the E4orf3-mutant virus dl341, or the E1B-55K/E4orf3 double-mutant virus. At various times after infection, the total virus present in the culture was measured by plaque assay on 293 cells (Fig. 1). By 48 h after infection, the E1B-55K/E4orf3 double-mutant-virus-infected cells exhibited a 1-log-unit decrease in virus yield compared to E1B-55K single-mutant-virus-infected cells and a 3-log-unit difference in growth compared to the wild-type virus-infected cells. This difference remained consistent over time. As noted previously (2, 7), replication of the E4orf3-mutant virus was nearly indistinguishable from that of wild-type virus. Thus, although the E4orf3 protein appears to contribute little to the replication of an otherwise wild-type virus, these results show that the E4orf3 protein contributes to the replication of an E1B-55K-mutant virus.
FIG. 1.
The E1B-55K/E4orf3 double-mutant virus exhibits similar growth kinetics but is more defective for replication than the E1B-55K single-mutant virus. Asynchronously growing HeLa cells were infected with the viruses indicated in the legend at a multiplicity of 10 PFU per cell. The cells and culture medium were collected at 1, 6, 12, 18, 24, 48, and 96 h postinfection, the cells were lysed, and the virus yield was measured by plaque assay with 293 cells. The value plotted at 0 h postinfection is the amount of virus used to infect the cells. Data shown are from a representative experiment performed three times with similar results.
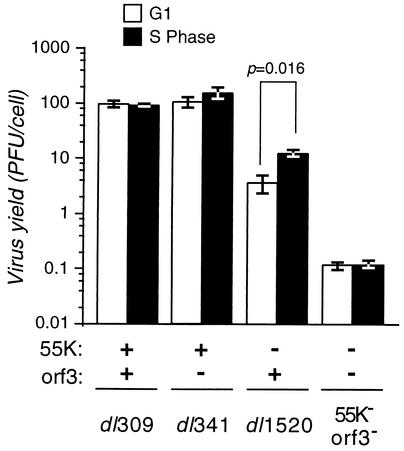
To determine if E4orf3 contributes to the replication of the E1B-55K-mutant virus in a cell cycle-dependent manner, HeLa cells were synchronized to G1 or S phase as previously described (6) and infected with each virus of interest, and the virus yield at 48 h was measured by plaque assay (Fig. 2). The wild-type virus and E4orf3-mutant virus produced nearly equivalent amounts of virus in G1- and S-phase-infected cells. The E1B-55K-mutant virus grew to a higher yield in cells infected during S phase and was more restricted for growth in G1-infected cells. However, the E1B-55K/E4orf3 double-mutant virus showed an overall decrease in virus yield in cells infected in either G1 or S phase, with no significant difference between G1- and S-phase-infected cells. These data show that the contribution of E4orf3 to replication of the E1B-55K-mutant virus was indeed cell cycle dependent.
FIG. 2.
The E1B-55K/E4orf3 double-mutant virus shows comparable growth in S-phase-infected cells and G1-infected cells. HeLa cells were synchronized to G1 or S phase and infected with the indicated viruses at a multiplicity of 10 PFU per cell. The status of the relevant viral genes (E1B-55K or E4orf3) is indicated below the histogram. Virus yield at 48 h postinfection was measured by plaque assay with 293 cells. The results shown are averages for three to five independent infections in three independent experiments. Error bars indicate the standard errors of the means. The difference in virus yield between G1- and S-phase-infected cells was significant only for dl1520-infected cells (P = 0.016, two-tailed Student's t test).
To determine the fraction of cells containing viral progeny, synchronously growing HeLa cells were infected with the viruses of interest, and approximately 100 cells were examined by transmission electron microscopy (Fig. 3). Cells that contained one or more viral particles were scored as positive. Nearly every G1- and S-phase cell infected with wild-type virus or E4orf3-mutant virus contained viral progeny, confirming that these viruses are not restricted for growth by the cell cycle. By contrast, 55% of cells infected during S phase with the E1B-55K-mutant virus contained viral progeny while only 24% of G1-infected cells contained viral progeny, similar to previous findings. However, only 9 to 12% of cells infected with the E1B-55K/E4orf3 double-mutant virus contained viral particles irrespective of the stage of the cell cycle at the time of infection. These data indicate that E4orf3 increases the number of cells capable of producing progeny virus particles in the absence of E1B-55K, and this effect is particularly prominent in cells infected in S phase.
FIG. 3.
A comparable fraction of cells infected in G1 or S phase with the E1B-55K/E4orf3 double-mutant virus contain progeny. HeLa cells were synchronized to either G1 or S phase and infected with the indicated viruses at a multiplicity of 10 PFU per cell. The status of the relevant viral genes (E1B-55K or E4orf3) is indicated below the histogram. Cells that were clearly infected were scored for the presence or absence of progeny virus particles. At least 100 cells per experiment were evaluated. Each value represents the average percent ± standard deviation from three or four independent experiments.
A limiting dilution assay for infectious centers was used to determine the total number of E1B-55K/E4orf3 double-mutant-virus-infected cells producing virus in order to confirm the electron microscopy data. Unlike electron microscopy, which enumerates cells containing viral particles, the infectious centers assay reveals the number of cells containing infectious virus. For these studies, HeLa cells synchronized to either G1 or S phase were infected with each of the viruses indicated in Table 1, and the fraction of infected cells producing virus was determined as previously described (5). The results of three independent experiments are summarized in Table 1. Each HeLa cell infected with the wild-type virus or E4orf3-mutant virus produced infectious progeny virus, in agreement with observations made with the electron microscope (Fig. 3). As anticipated, a greater fraction of S-phase cells infected with the E1B-55K-mutant virus contained infectious virus than G1-infected cells (45 and 16%, respectively). Finally, the infectious centers assay shows that only 4% of G1-infected cells and 2% of S-phase-infected cells permitted replication of the double-mutant virus. Collectively, the plaque assay for virus yield, electron microscopy, and the infectious centers assay support the conclusion that E4orf3 is necessary for the enhanced S phase growth shown by the E1B-55K single-mutant virus.
TABLE 1.
Frequency of infectious centers from cells infected in G1 or S phase
| Virus | Cell cyclea | Frequency of infectious centers
|
||
|---|---|---|---|---|
| Replicate valuesb | Avg | Fraction wild type (%)c | ||
| dl309 | G1 | 1.2 | 1.1 ± 0.2 | 100 |
| 0.7 | ||||
| 1.5 | ||||
| S | 1.5 | 1.1 ± 0.2 | 105 | |
| 1.0 | ||||
| 0.7 | ||||
| dl1520 | G1 | 3.6 | 6.9 ± 1.6 | 16 |
| 7.2 | ||||
| 10.0 | ||||
| S | 2.9 | 2.5 ± 0.2 | 45 | |
| 2.4 | ||||
| 2.1 | ||||
| dl341 | G1 | 1.5 | 1.0 ± 0.2 | 110 |
| 0.8 | ||||
| 0.7 | ||||
| S | 0.5 | 0.7 ± 0.1 | 156 | |
| 0.8 | ||||
| 0.8 | ||||
| 55K−/orf3− | G1 | 28.6 | 25.8 ± 4.1 | 4 |
| 17.7 | ||||
| 31.1 | ||||
| S | 44.2 | 46.6 ± 5.6 | 2 | |
| 38.4 | ||||
| 57.2 | ||||
HeLa cells were infected at the stage of the cell cycle indicated.
The number of cells required to obtain an infectious center ± standard error was determined from three independent replicates.
The fraction of infectious centers (1/number of cells required to obtain an infectious center) is expressed as a percentage of the fraction of infectious centers for G1 cells infected with the wild-type virus.
Although the absence of the E4orf3 protein has little apparent impact on a productive wild-type Ad infection in vitro, the E4orf3 gene is required for enhanced S phase growth when cells are infected with viruses defective in either the E1B-55K gene, as shown here, or the E4orf6 gene, as shown previously (6). Additionally, the E4orf3 protein promotes efficient viral replication of the E1B-55K-mutant virus in both G1- and S-phase-infected cells. Together, these results suggest a shared role for the E4orf3 protein with the complex of E1B-55K and E4orf6 proteins. The E1B-55K-E4orf6 protein complex can apparently compensate for the loss of E4orf3. However, if either E1B-55K or E4orf6 is absent, E4orf3 is required for viral replication, and the E4orf3 function is more effective in S-phase-infected cells than in G1-infected cells. Potential functions for E4orf3 in this context may include the regulation of mRNA processing (11), the promotion of mRNA transport (2), the inhibition of host translation, the suppression of host antiviral responses, or the prevention of viral DNA concatemer formation (14). The findings reported here may also be significant for the development of oncolytic Ads for the treatment of cancer. It is possible that the selective replication in cancer cells shown by viruses that lack the E1B-55K gene (9) requires the E4orf3 gene.
The ability of the E4orf3 protein to enhance replication of the E1B-55K-deficient virus may be significant in the course of a natural Ad infection in humans. Recently, Garnett and associates recovered group C Ad from mucosal T lymphocytes of children (4). Studying the regulation of the E1A and E3 promoter in T-cell lines, Mahr et al. have suggested that upregulation of the E3 promoter may occur in activated T cells in an E1A-independent manner (10). Because the E1B promoter depends strongly on E1A for activation (3), a virus that reactivates in the absence of E1A expression may express little if any E1B-55K. This virus could potentially establish a productive infection independent of the E1 region through products of the E4 region (12). Under this circumstance, we would predict that the E4orf3 protein is required to enhance replication in the absence of expression of E1B.
Acknowledgments
This work was supported in part by Public Health Service grant CA 77342 and a supplement from the National Cancer Institute. Tissue culture reagents and services were provided by the Tissue Culture Core Laboratory, and electron microscopy was performed through the Micromed facility, both services of the Comprehensive Cancer Center of Wake Forest University, which is supported in part by the National Cancer Institute, grant CA 12197.
We gratefully acknowledge Pat Hearing (SUNY Stony Brook) for the E4orf3-mutant virus, dl341, and Arnie Berk (UCLA) for the E1B-55K-mutant virus, dl1520D. We also acknowledge the help of Ken Grant of the Micromed facility for assistance with electron microscopy. We thank Thomas Dobner (University of Regensburg, Regensburg, Germany) for kindly providing E4orf3-specific antibodies used during the creation of the E1B-55K/E4orf3 double-mutant virus. We thank Doug Lyles, Griff Parks, and Linda Gooding for providing valuable advice on the work in progress and on the manuscript.
REFERENCES
- 1.Barker, D. D., and A. J. Berk. 1987. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology 156:107-121. [DOI] [PubMed] [Google Scholar]
- 2.Bridge, E., and G. Ketner. 1989. Redundant control of adenovirus late gene expression by early region 4. J. Virol. 63:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dery, C. V., C. H. Herrmann, and M. B. Mathews. 1987. Response of individual adenovirus promoters to the products of the E1A gene. Oncogene 2:15-23. [PubMed] [Google Scholar]
- 4.Garnett, C. T., D. Erdman, W. Xu, and L. R. Gooding. 2002. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J. Virol. 76:10608-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodrum, F. D., and D. A. Ornelles. 1997. The early region 1B 55-kilodalton oncoprotein of adenovirus relieves growth restrictions imposed on viral replication by the cell cycle. J. Virol. 71:548-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodrum, F. D., and D. A. Ornelles. 1999. Roles for the E4 orf6, orf3, and E1B 55-kilodalton proteins in cell cycle-independent adenovirus replication. J. Virol. 73:7474-7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halbert, D. N., J. R. Cutt, and T. Shenk. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56:250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683-689. [DOI] [PubMed] [Google Scholar]
- 9.Kirn, D. 2001. Oncolytic virotherapy for cancer with the adenovirus dl1520 (Onyx-015): results of phase I and II trials. Expert Opin. Biol. Ther. 1:525-538. [DOI] [PubMed] [Google Scholar]
- 10.Mahr, J. A., J. M. Boss, and L. R. Gooding. 2003. The adenovirus E3 promoter is sensitive to activation signals in human T cells. J. Virol. 77:1112-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nordqvist, K., K. Ohman, and G. Akusjarvi. 1994. Human adenovirus encodes two proteins which have opposite effects on accumulation of alternatively spliced mRNAs. Mol. Cell. Biol. 14:437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Connor, R. J., and P. Hearing. 2000. The E4-6/7 protein functionally compensates for the loss of E1A expression in adenovirus infection. J. Virol. 74:5819-5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarnow, P., P. Hearing, C. W. Anderson, N. Reich, and A. J. Levine. 1982. Identification and characterization of an immunologically conserved adenovirus early region 11,000 Mr protein and its association with the nuclear matrix. J. Mol. Biol. 162:565-583. [DOI] [PubMed] [Google Scholar]
- 14.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature (London) 418:348-352. [DOI] [PubMed] [Google Scholar]