Abstract
Viruses encode proteins that disrupt chemokine responses. The murine gammaherpesvirus 68 gene M3 encodes a chemokine binding protein (vCKBP-3) which has no sequence similarity to chemokine receptors but inhibits chemokine receptor binding and activity. We have used a panel of CXCL8 analogs to identify the structural requirements for CXCL8 to bind to vCKBP-3 in a scintillation proximity assay. Our data suggest that vCKBP-3 acts by mimicking the binding of chemokine receptors to CXCL8.
Chemokines orchestrate leukocyte migration from blood to sites of injury or infection. Their central importance is underscored by the fact that many viruses (e.g., herpesvirus, poxvirus, and retrovirus) subvert the chemokine system during infection by encoding chemokine binding proteins (vCKBPs) or chemokine and chemokine receptor homologues (1, 17). During the course of their evolution, viruses have finely tuned their immunomodulatory proteins and offer us potential therapeutics for disease and greater insight into chemokine biology.
Three types of vCKBPs have been identified: vCKBP-1 (encoded by myxoma virus) binds with low specificity to chemokines from all subfamilies (CC, CXC, C, and CX3C), vCKBP-2 (encoded by several poxviruses) binds with high affinity to only CC chemokines, and vCKBP-3 (unique to murine gammaherpesvirus 68) binds with high affinity to chemokines from all subfamilies (2, 11, 14, 18, 21, 22). Identification of the chemokine epitopes involved in vCKBP-1 and vCKBP-2 binding explain their different specificities. While vCKBP-1 binds weakly to the glycosaminoglycan (GAG) binding domain present in the majority of chemokines (14), vCKBP-2 specifically interacts with residues within the N-loop of certain CC chemokines (5, 20). Only the structural requirements for vCKBP-3 to bind to the CC chemokine, CCL2 (monocyte chemoattractant protein 1), have been determined (3). Like vCKBP-2, vCKBP-3 uses the N-loop of CCL2 for binding, with Tyr13 playing a key role. Both vCKBP-2 and vCKBP-3 bind to most CC chemokines. However, in contrast to vCKBP-2, vCKBP-3 is able to bind to some CXC chemokines with significant affinity (18, 22). It has been shown to bind CXCL1, CXCL8, CXCL10, and CXCL13 in a species-dependent manner (18, 22). It shows selectivity in the CXC chemokines in that it binds to many CXC chemokines with no significant affinity for vCKBP-3. As yet, the CXC epitopes used for vCKBP-3 binding remain unknown. vCKBP-3 can bind and inhibit CXCL8 (interleukin-8) (18, 22). Inactivation of the M3 gene (which encodes vCKBP-3 from murine gammaherpesvirus 68) has shown that vCKBP-3 is critical for establishing latency in splenic B cells and for induction of lethal meningitis (6, 23).
We have used chemically synthesized analogs of CXCL8 to map the vCKBP-3 binding site (Fig. 1). Chemokine analogs are powerful tools that have been extensively used to define key residues and domains required for chemokine activity (8-10, 12, 13, 15, 24). Analogs were synthesized with tertiary-butyloxycarbonyl chemistry and automated solid-phase methods (9). Purified recombinant vCKBP-3 protein containing a COOH-terminal six-histidine tag was produced in the baculovirus system and provided by Campbell Bunce and Mark Wilson (Xenova, Cambridge, United Kingdom) (18).
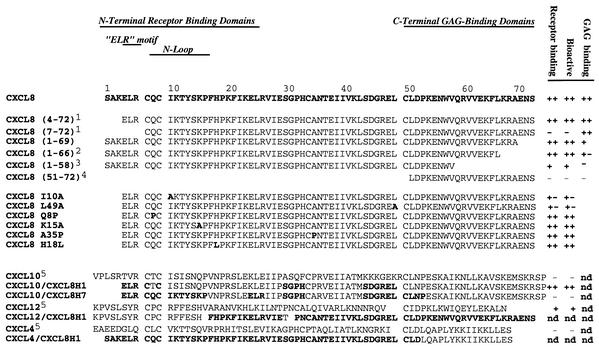
FIG. 1.
Amino acid sequences of the following hybrid analogs used in this study: CXCL8 (8, 9, 10, 15), CXCL8 residues 4 to 72 (8, 9, 10, 15, 19), CXCL8 residues 7 to 72, (8, 15, 24), CXCL8 residues 1 to 69 (8, 24), CXCL8 residues 1 to 66 (14, 18, 24), CXCL8 residues 1 to 58 (8, 24), CXCL8 residues 51 to 72 (8, 24), CXCL8 I10A (10, 19), CXCL8 L49A (10), CXCL8 Q8P (9), CXCL8 K15A (9), CXCL8 A35P (9), CXCL8 H18L (9), CXCL10 (9), CXCL10-CXCL8H1 (9), CXCL10-CXCL8H7 (9), CXCL12 (4), CXCL12-CXCL8H1, CXCL4 (4), and CXCL4-CXCL8H1. The first group comprises the analogs with either N- or C-terminal truncation. The analogs in the second group are analogs of CXCL8 with substitutions (substituted residues are shown in boldface type). The third group contains hybrid analogs with CXCL8 residues shown in boldface type. 1, analysis of 1H nuclear magnetic resonance parameters has shown that the core structure is the same as that for WT CSCL8, thus differences in binding and function cannot be attributed to incorrect folding of the protein (19); 2, unable to bind vCKBP-1 (14); 3, able to bind neutrophils and induce chemotaxis but with 30 times less potency (8); 4, able to restore bioactivity to CXCL8 residues 1 to 51 (18); 5, CXCL12 and CXCL4 are unable to bind vCKBP-3 and CXCL10 can bind vCKBP-3 but with reduced affinity (Table 1); ND, not determined; +, medium; ++, high; −, undetectable; +−, low.
To investigate CXCL8 binding to vCKBP-3, we determined equilibrium competition binding in a scintillation proximity assay with Flashplates (Perkin Elmer Life Sciences) (7). His-tagged vCKBP-3 (40 ng/ml) was incubated with 200 pM 125I-CXCL8 (wild type [WT]) and a dose response of cold CXCL8 analog in a total volume of 100 μl in 0.1% bovine serum albumin-phosphate-buffered saline. Background 125I-CXCL8 binding was measured without vCKBP-3 and was similar to binding in the presence of excess unlabeled CXCL8. Data were analyzed by subtracting the background binding for each assay. Affinities were calculated with the LIGAND program (16). Scatchard analysis of a saturation curve was used to first calculate the affinity of CXCL8 for vCKBP-3 (7.4 pM) (data not shown). The differences between the CXCL8 affinities for vCKBP-3 obtained with saturation and competition assays (23.5 pM) (Table 1) are due to different chemical sources of CXCL8 and assays used.
TABLE 1.
Affinities of different CXCL8 analogs for vCKBP-3a
| Analog | Kd (pM) |
|---|---|
| WT CXCL8 (residues 1-72) | 23.54 ± 8.61 |
| Truncations of CXCL8 (residues contained) | |
| 4-72 | 33.38 ± 9.39 |
| 7-72 | 37.85 ± 1.81 |
| 1-69 | 25.42 ± 4.41 |
| 1-66 | 34.5 ± 8.90 |
| 1-58 | 520.33 ± 8.09 |
| Substitutions within CXCL8 | |
| I10A | 288.25 ± 67.86 |
| L49A | 118.06 ± 18.22 |
| Q8P | 71.85 ± 14.41 |
| K15A | 45.53 ± 9.25 |
| A35P | 33.68 ± 9.30 |
| H18L | 34.69 ± 1.14 |
| Hybrid analogs | |
| WT CXCL10 | 232.48 ± 37.76 |
| CXCL10-CXCL8H1 | 241.03 ± 24.2 |
| CXCL10-CXCL8H7 | 33.49 ± 5.36 |
| WT CXCL4 | >b |
| CXCL4-CXCL8H1 | 69.05 ± 19.73 |
| WT CXCL12 | > |
| CXCL12-CXCL8H1 | > |
Analogs that have substitutions within CXCL8 are based on the truncated CXCL8 containing residues 4 to 72.
>, undetectable binding.
We first used CXCL8 analogs with progressive deletions in the N terminus to compete with 125I-CXCL8 for binding to vCKBP-3 in scintillation proximity assays. Removal of the first six residues of CXCL8 decreased its ability to compete for vCKBP-3 binding (Fig. 2A and B; Table 1). This region contains the ELR motif which binds to the CXCL8 receptors (8). The N-loop of CXCL8 (residues 8 to 16) is also important for receptor interactions. Within the N-loop, Ile10 is crucial for receptor binding and activation (9). The only analog within the N-loop to show a significant drop in vCKBP-3 affinity was I10A (Fig. 2C and Table 1). Leu49 packs into the bulge in the N-loop and provides optimal conformation and binding for CXCR1 (10). We also found that this analog had a reduced affinity for vCKBP-3 (Fig. 2C and Table 1).
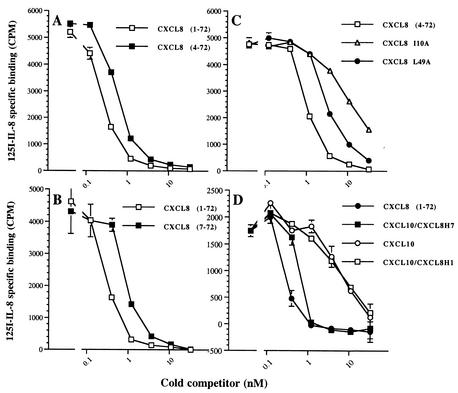
FIG. 2.
Effect of N-terminal truncations in CXCL8 on vCKBP-3 binding. A competition assay was performed to assess the role of the N terminus of CXCL8 for vCKBP-3 binding. This figure shows the effect of removal of the first 3 N-terminal residues of CXCL8 compared to WT CXCL8 (A), removal of the first 6 residues of CXCL8 compared to WT CXCL8 (B), and substitution of Ile for Ala at position 10 and substitution of Leu for Ile at position 49 compared to CXCL8 (C). Panel D shows competition assays with WT CXCL8 and WT CXCL10 compared to CXCL10-CXCL8H1 and CXCL10-CXCL8H7.
The similar tertiary structure of chemokines has enabled the construction of hybrid analogs to identify the minimal CXCL8 structure required for activity (8). We used hybrids of CXCL8 with CXCL10 (interferon-inducible protein 10) (Fig. 1) to confirm the role of the N-loop for vCKBP-3 binding. CXCL10 has no neutrophil binding or activating properties and has a significantly lower affinity for vCKBP-3 than CXCL8 (Table 1; Fig. 2D) (8). CXCL10-CXCL8H1 differs from CXCL10-CXCL8H7 by lacking the N-loop of CXCL8 (residues 4 to 15 and 24 to 26) (Fig. 1). We found that CXCL10-CXCL8H7 competed with 125I-CXCL8 for vCKBP-3 as effectively as CXCL8, whereas CXCL10-CXCL8H1 behaved like CXCL10 (Fig. 2D).
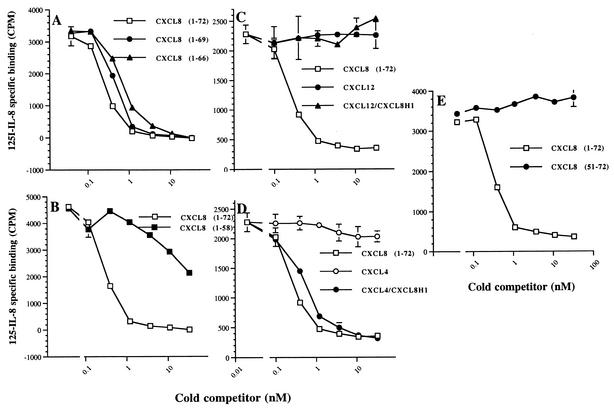
We next assessed the importance of the C-terminal alpha helix of CXCL8 for vCKBP-3 binding. This region does not directly participate in receptor binding but is critical for GAG binding and provides a platform for the receptor binding domains (Fig. 1) (24). We used analogs in which the C terminus was progressively truncated. Removal of the last three or six residues of CXCL8 resulted in a slight decrease in affinity (Fig. 3A; Table 1). This is in contrast to vCKBP-1, which is unable to bind the analog of CXCL8 containing residues 1 to 66, providing a structural explanation for its ability to prevent the chemokine-GAG interaction (14). There was a significant loss in affinity for vCKBP-3 when the last 14 residues of CXCL8 were removed (analog containing residues 1 to 58) (Fig. 3B; Table 1). To determine whether the C terminus of CXCL8 interacts directly with vCKBP-3 or is simply required for optimal conformation of CXCL8, we used hybrid analogs of CXCL8 with either CXCL12 (stroma-derived factor 1α) or CXCL4 (platelet factor 4) (Fig. 1). Neither CXCL12 nor CXCL4 could bind vCKBP-3 (Table 1; Fig. 3C and D) (18). As shown in Fig. 3C, CXCL12-CXCL8H1 was unable to bind vCKBP-3, showing that the C-terminal region of CXCL8 (residues 18 to 72) is insufficient for vCKBP-3 binding. The hybrid CXCL4-CXCL8H1 bound vCKBP-3 with high affinity (Table 1 and Fig. 3D), showing that while the C terminus of CXCL8 is not important for vCKBP-3 binding, the N terminus is critical. Unlike CXCL10-CXCL8H1, CXCL4-CXCL8H1 contains the N-loop of CXCL8 and binds to vCKBP-3 with high affinity. This experiment not only shows that the C terminus of CXCL8 is dispensable for vCKBP-3 binding but also restates the importance of the N-loop of CXCL8 for vCKBP-3 binding. In agreement with this, the hybrid CXCL10-CXCL8H7 (which contains the N-loop of CXCL8 and the C terminus of CXCL10) behaved like CXCL8 (Fig. 2D). We also examined the role of the C-terminal alpha helix of CXCL8 alone in vCKBP-3 binding. This analog was unable to bind vCKBP-3 (Fig. 3E).
FIG. 3.
Role of the C terminus of CXCL8 for vCKBP-3 binding. Panel A shows the effect of removal of the last 3 residues and removal of the last 6 residues compared to WT CXCL8. Panel B shows the effect of removal of the last 14 residues of CXCL8 compared to WT CXCL8. Panel C shows competition assays with WT CXCL8, WT CXCL12-1α, and CXCL12-1α-CXCL8H1. Panel D shows competition assays with WT CXCL8 and WT CXCL4 compared to CXCL4-CXCL8H1. Panel E shows a competition assay comparing WT CXCL8 with an analog containing only the C terminus (residues 51 to 72). The means ± standard deviations for triplicate samples are shown.
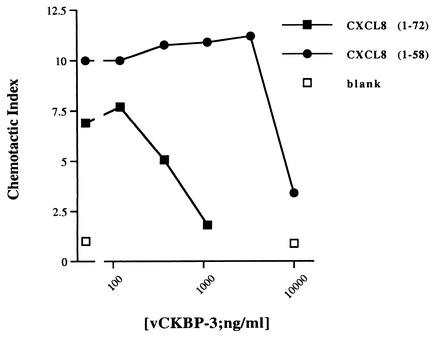
Finally, we examined the effect of vCKBP-3 on chemotaxis induced by either WT or C-terminally truncated CXCL8 (residues 1 to 58). Using the values obtained in our competition assays for the affinity of vCKBP-3 for WT CXCL8 and CXCL8 containing residues 1 to 58, we were able to predict how much vCKBP-3 would be required to inhibit chemotaxis. We estimated that 69 μM vCKBP-3 would be required to inhibit 50% of the chemotaxis induced by 30 ng of CXCL8/ml and 46.8 mM vCKBP-3 would be required to inhibit 50% of the chemotaxis induced by 900 ng of CXCL8/ml, assuming a 1:1 binding of vCKBP-3 to CXCL8. WT CXCL8 (30 ng/ml) or CXCL8 containing residues 1 to 58 (900 ng/ml) was preincubated with various doses of vCKBP-3 in a neutrophil chemotaxis assay. The higher dose of CXCL8 (residues 1 to 58) was used to obtain a similar induction of chemotaxis as WT CXCL8 since it is 30 times less potent than WT CXCL8 (9). vCKBP-3 inhibited chemotaxis induced by both WT CXCL8 and CXCL8 containing residues 1 to 58, showing that binding to the N terminus is sufficient to block CXCL8 activity (Fig. 4). The higher concentration of vCKBP-3 required for inhibition of CXCL8 (residues 1 to 58) reflects the larger amount of chemokine present. The functional 50% inhibitory concentration of vCKBP-3 for blocking WT CXCL8 and residues 1 to 58 of CXCL8 were 66.7 μM and 68.7 mM, respectively, calculated with the Prism Program (GraphPad Prim). This showed that the observed inhibition of chemotaxis correlated well with our in vitro competition assays.
FIG. 4.
Chemotaxis to both WT and C-terminally truncated CXCL8 is inhibited by vCKBP-3. Neutrophil chemotaxis to WT (residues 1 to 72) or C-terminally truncated (residues 1 to 58) CXCL8 was measured in the presence of various doses of vCKBP-3. Chemotaxis to WT CXCL8 or to C-terminally truncated CXCL8 (residues 1 to 58) compared to a blank is shown. The means ± standard deviations for duplicate samples are shown. No effect of a control His-tagged protein at this concentration of CXCL8-induced chemotaxis was observed (data not shown).
Our data show that it is the N terminus of CXCL8 that binds to vCKBP-3, with the greatest contribution coming from the N-loop. This is in contrast to vCKBP-2, which uses only the N-loop of CC chemokines for binding, since removal of the N terminus of CCL2 actually increases vCKBP-2 affinity. This may explain the broader specificity of vCKBP-3. Unlike vCKBP-2, vCKBP-3 binds to some (but not all) CXC chemokines (18, 22). The crystal structure of vCKBP-3 bound to CCL2 has been determined and shows that, like CXCL8, the N-loop of CCL2 contributes most to vCKBP-3 binding (3). The N-loop is the secondary receptor binding site and is thought to confer specificity. Within the N-loop, Ile10 in CXCL8 is important for both receptor and vCKBP-3 binding. This residue aligns in the same position as Tyr13 within CCL2, which is important for the interaction of CCL2 with vCKBP-3 and vCKBP-2 (3, 20). The observation that similar binding epitopes are used for recognition of vCKBP-3 and chemokine receptors provides a structural basis for the ability of vCKBP-3 to occlude the binding of chemokines to their host receptors.
In conclusion, we have defined the key residues of a CXC chemokine, CXCL8, that interact with vCKBP-3. This provides valuable information on the recognition of chemokines by viral proteins and will allow the rational design of chemokine inhibitors.
Acknowledgments
We acknowledge the support of the Wellcome Trust (grant 051087/Z/97/Z). A. A. is a Wellcome Trust Senior Research Fellow.
We thank Campbell Bunce and Mark Wilson for providing recombinant vCKBP-3.
Footnotes
This paper is dedicated to the memory of Ian Clark-Lewis (1955 to 2002).
REFERENCES
- 1.Alcami, A. 2003. Viral mimicry of cytokines, chemokines and their receptors. Nat. Rev. Immunol. 3:36-50. [DOI] [PubMed] [Google Scholar]
- 2.Alcami, A., J. A. Symons, P. D. Collins, T. J. Williams, and G. L. Smith. 1998. Blockade of chemokine activity by a soluble chemokine binding protein from vaccinia virus. J. Immunol. 160:624-633. [PubMed] [Google Scholar]
- 3.Alexander, J. M., C. A. Nelson, V. van Berkel, E. K. Lau, J. M. Studts, T. J. Brett, S. H. Speck, T. M. Handel, H. W. Virgin, and D. H. Fremont. 2002. Structural basis of chemokine sequestration by a herpesvirus decoy receptor. Cell 111:343-356. [DOI] [PubMed] [Google Scholar]
- 4.Baggiolini, M. 1998. Chemokines and leukocyte traffic. Nature 392:565-568. [DOI] [PubMed] [Google Scholar]
- 5.Beck, C. G., C. Studer, J. F. Zuber, B. J. Demange, U. Manning, and R. Urfer. 2001. The viral CC chemokine-binding protein vCCI inhibits monocyte chemoattractant protein-1 activity by masking its CCR2B-binding site. J. Biol. Chem. 276:43270-43276. [DOI] [PubMed] [Google Scholar]
- 6.Bridgeman, A., P. G. Stevenson, J. P. Simas, and S. Efstathiou. 2001. A secreted chemokine binding protein encoded by murine gammaherpesvirus-68 is necessary for the establishment of a normal latent load. J. Exp. Med. 194:301-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryant, N. A., N. Davis-Poynter, A. Vanderplasschen, and A. Alcami. 2003. Glycoprotein G isoforms from some alphaherpesviruses function as broad-spectrum chemokine binding proteins. EMBO J. 22:833-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark-Lewis, I., B. Dewald, M. Loetscher, B. Moser, and M. Baggiolini. 1994. Structural requirements for interleukin-8 function identified by design of analogs and CXC chemokine hybrids. J. Biol. Chem. 269:16075-16081. [PubMed] [Google Scholar]
- 9.Clark-Lewis, I., C. Schumacher, M. Baggiolini, and B. Moser. 1991. Structure-activity relationships of interleukin-8 determined using chemically synthesised analogs. Critical role of NH2-terminal residues and evidence for uncoupling of neutrophil chemotaxis, exocytosis, and receptor binding activities. J. Biol. Chem. 266:23128-23134. [PubMed] [Google Scholar]
- 10.Fairbrother, W. J., and H. B. Lowman. 1997. Molecular approaches to structure-function analysis of interleukin-8. Methods Enzymol. 287:45-58. [DOI] [PubMed] [Google Scholar]
- 11.Graham, K. A., A. S. Lalani, J. L. Macen, T. L. Ness, M. Barry, L. Y. Liu, A. Lucas, I. Clark-Lewis, R. W. Moyer, and G. McFadden. 1997. The T1/35kDa family of poxvirus-secreted proteins bind chemokines and modulate leukocyte influx into virus-infected tissues. Virology 229:12-24. [DOI] [PubMed] [Google Scholar]
- 12.Jones, S. A., B. Dewald, I. Clark-Lewis, and M. Baggiolini. 1997. Chemokine antagonists that discriminate between interleukin-8 receptors. Selective blockers of CXCR2. J. Biol. Chem. 272:16166-16169. [DOI] [PubMed] [Google Scholar]
- 13.Kuschert, G. S., A. J. Hoogewerf, A. E. Proudfoot, C. W. Chung, R. M. Cooke, R. E. Hubbard, T. N. Wells, and P. N. Sanderson. 1998. Identification of a glycosaminoglycan binding surface on human interleukin-8. Biochemistry 37:11193-11201. [DOI] [PubMed] [Google Scholar]
- 14.Lalani, A. S., K. Graham, K. Mossman, K. Rajarathnam, I. Clark-Lewis, D. Kelvin, and G. McFadden. 1997. The purified myxoma virus gamma interferon receptor homolog M-T7 interacts with the heparin-binding domains of chemokines. J. Virol. 71:4356-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moser, B., B. Dewald, L. Barella, C. Schumacher, M. Baggiolini, and I. Clark-Lewis. 1993. Interleukin-8 antagonists generated by N-terminal modification. J. Biol. Chem. 268:7125-7128. [PubMed] [Google Scholar]
- 16.Munson, P. J., and D. Rodbard. 1980. Ligand: a versatile computerized approach for characterization of ligand-binding systems. Anal. Biochem. 107:220-239. [DOI] [PubMed] [Google Scholar]
- 17.Murphy, P. M. 2001. Viral exploitation and subversion of the immune system through chemokine mimicry. Nat. Immunol. 2:116-122. [DOI] [PubMed] [Google Scholar]
- 18.Parry, C. M., J. P. Simas, V. P. Smith, C. A. Stewart, A. C. Minson, S. Efstathiou, and A. Alcami. 2000. A broad spectrum secreted chemokine binding protein encoded by a herpesvirus. J. Exp. Med. 191:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajarathnam, K., I. Clark-Lewis, and B. D. Sykes. 1994. 1H NMR studies of interleukine 8 analogs: characterisation of the domains essential for function. Biochemistry 33:6623-6630. [DOI] [PubMed] [Google Scholar]
- 20.Seet, B. T., R. Singh, C. Paavola, E. K. Lau, T. M. Handel, and G. McFadden. 2001. Molecular determinants for CC-chemokine recognition by a poxvirus CC-chemokine inhibitor. Proc. Natl. Acad. Sci. USA 98:9008-9013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith, C. A., T. D. Smith, P. J. Smolak, D. Friend, H. Hagen, M. Gerhart, L. Park, D. J. Pickup, D. Torrance, K. Mohler, K. Schooley, and R. G. Goodwin. 1997. Poxvirus genomes encode a secreted, soluble protein that preferentially inhibits beta chemokine activity yet lacks sequence homology to known chemokine receptors. Virology 236:316-327. [DOI] [PubMed] [Google Scholar]
- 22.van Berkel, V., J. Barrett, H. L. Tiffany, D. H. Fremont, P. M. Murphy, G. McFadden, S. H. Speck, and H. I. Virgin. 2000. Identification of a gammaherpesvirus selective chemokine binding protein that inhibits chemokine action. J. Virol. 74:6741-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Berkel, V., B. Levine, S. B. Kapadia, J. E. Goldman, S. H. Speck, and H. W. Virgin. 2002. Critical role for a high-affinity chemokine-binding protein in gamma-herpesvirus-induced lethal meningitis. J. Clin. Investig. 109:905-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webb, L. M., M. U. Ehrengruber, I. Clark-Lewis, M. Baggiolini, and A. Rot. 1993. Binding to heparan sulfate or heparin enhances neutrophil responses to interleukin 8. Proc. Natl. Acad. Sci. USA 90:7158-7162. [DOI] [PMC free article] [PubMed] [Google Scholar]